Abstract
Symptomatic and non-symptomatic leaf samples of cucumber, zucchini, melon and watermelon plants were collected from three locations (Al-Ahsa in the East, Jizan in the South and Tabuk in the North) in the Kingdom of Saudi Arabia during 2013–2014. The detection of begomovirus infection was commenced with serological assay, rolling circle amplification and PCR amplification with universal begomovirus primers. The zucchini and watermelon samples tested positive for the presence of begomovirus infection in 53 and 39.6%, and 54.5 and 41.5% samples using serological and PCR-based detection, respectively. Based upon the core coat protein sequence, the full-length genomes of DNA-A and DNA-B were amplified and subsequently sequenced. The sequence analysis revealed that the two clones of DNA-A shared 96.9% nucleotide (nt) sequence identity with each other while they shared 95.9–98.4% nt sequence identity with watermelon chlorotic stunt virus (WmCSV) isolates reported so far. Similarly, the two DNA-B clones showed 91.8% mutual nt sequence identity, while sharing 95.2–95.5% nt sequence identity with all other reported DNA-B sequences. The phylogenetic dendrogram grouped both DNA-A clones into separate sub-groups, suggesting that both isolates were introduced separately into Saudi Arabia. The identification of WmCSV from zucchini in Saudi Arabia and other hosts in the neighbouring countries suggests that this virus is becoming an emerging threat to cucurbits in this region.
Résumé
En 2013-2014, des échantillons de feuilles symptomatiques et asymptomatiques de concombre, de courgette, de melon et de pastèques ont été collectés en trois endroits (Al-Ahsa dans l’est, Jizan dans le sud et Tabuk dans le nord) du royaume d’Arabie Saoudite. La détection d’une infection causée par un bégomovirus a été amorcée avec un test sérologique, une amplification par cercle roulant et une amplification par PCR avec amorces universelles pour la détection de bégomovirus. Le test effectué sur les échantillons de courgette et de pastèque s’est révélé positif quant à l’infection causée par le bégomovirus chez 53 et 39.6% ainsi que 54.5 et 41.5% des échantillons soumis au test sérologique et à l’analyse par PCR, respectivement. En se basant sur la séquence du cœur de la protéine de l’enveloppe, les génomes entiers de l’ADN-A et de l’ADN-B ont été amplifiés et, par la suite, séquencés. L’analyse des séquences a révélé que les deux clones de l’ADN-A partageaient une identité de séquence nucléotidique de 96.9%, tandis qu’ils en partageaient une de 95.9 à 98.4% avec les isolats du virus du nanisme chlorotique de la pastèque (VNCP) rapportés jusqu’à présent. De même, les deux clones de l’ADN-B ont affiché une identité réciproque de séquence nucléotidique de 91.8%, tandis qu’ils partageaient une identité de 95.2 à 95.5% avec toutes les autres séquences rapportées d’ADN-B. Le dendogramme phylogénétique a regroupé les deux clones d’ADN-A dans des sous-groupes distincts, ce qui suggère que les deux isolats ont été introduits séparément en Arabie Saoudite. L’identification du VNCP à partir de courgettes en Arabie Saoudite et d’autres hôtes dans les pays voisins suggère que ce virus est une nouvelle menace pour les cucurbitacées dans cette région du monde.
Keywords:
Mots clés:
Introduction
Geminiviruses (family Geminiviridae) are the most important plant pathogens of food and fibre crops with global distribution. The family Geminiviridae has been further characterized into nine genera: Becurtovirus, Begomo-virus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocovirus and Turncurotovirus (Zerbini et al., Citation2017). The whitefly-borne begomoviruses are responsible for huge economic losses to different dicotyledonous crops and weeds in the tropical to temperate regions (Brown et al., Citation2015). Collectively, begomoviruses constitute the largest genus of the family Geminiviridae and are further categorized into bipartite and monopartite begomoviruses. Their genome is encapsidated in twinned icosahedrons either as two (bipartite) similar sized circular, single-stranded (css) DNA (DNA-A & DNA-B; ~2.8 kb in length) or a single (monopartite) cssDNA equivalent to DNA-A (Hanley-Bowdoin et al., Citation2013). The DNA-A of bipartite begomoviruses in the Old World (OW) usually encodes two genes in virion-sense (AV1 and AV2) while four genes are encoded in the complementary-sense orientation (AC1, AC2, AC3 and AC4) (Sattar, Citation2012). However, the DNA-A component of bipartite begomovirus genome in the New World (NW) lacks AV2 gene. A successful begomovirus infection, encapsidation, in planta movement, countering host immunity and whitefly-mediated transmission necessitate the protein products from the viral encoded genes and certain host factors (Iqbal et al., Citation2012; Hanley-Bowdoin et al., Citation2013). On the other hand, the DNA-B component of a bipartite begomovirus genome encompasses two genes (BV1 and BC1) in the opposite orientation to each other. The DNA-B is essentially important to regulate inter- and intra-cellular viral trafficking in planta (Hanley-Bowdoin et al., Citation2013). The only shared region between DNA-A and DNA-B is a small (~200–400 bp) common region (CR), which is comprised of a stem-loop with nona-nucleotides (TAATATTAC) and a bi-directional promotor (Nawaz-ul-Rehman & Fauquet, Citation2009). Due to rapid recombination and evolution, the host adaptability of begomoviruses has been increasing at a rapid pace.
Most of the begomoviruses prevailing in the OW have a monopartite genome whereas some bipartite begomoviruses are also reported. Watermelon chlorotic stunt virus (WmCSV) is a bipartite begomovirus first reported from Yemen in 1988 (Walkey et al., Citation1990). In subsequent years, WmCSV rapidly spread across the borders in other Middle-Eastern countries and North Africa (Kheyr-Pour et al., Citation2000). Later, it spread to Lebanon (Samsatly et al., Citation2012), Oman (Khan et al., Citation2012), Palestine (Ali-Shtayeh et al., Citation2014), Israel (Abudy et al., Citation2010) and Jordan (Al-Musa et al., Citation2011). In Saudi Arabia the first identification of WmCSV was reported recently from watermelon plants in the Leith region (Al-Saleh et al., Citation2014). In another study, Alhudaib et al. (Citation2018) partially characterized WmCSV DNA-A infecting zucchini plants in Al-Ahsa region of Saudi Arabia.
In the present study, we isolated and characterized full-length WmCSV isolates from watermelon (Citrullus lanatus) and zucchini (Cucurbita pepo) leaves showing typical begomovirus infection symptoms in the eastern and southern regions of the Kingdom of Saudi Arabia.
Materials and methods
Collection of samples
A field survey was carried out in cucurbit fields at three locations (Al-Ahsa in the East, Jizan in the South and Tabuk in the North) in the Kingdom of Saudi Arabia during the growing season of spring 2013–2014. During the survey, 730 leaf samples of non-symptomatic plants or the plants showing begomovirus-like symptoms of chlorosis, dwarfing, yellowing and mosaic were collected from 23 fields of cucumber, zucchini, melon and watermelon (; ). The collected leaf samples were immediately transferred to plastic bags and stored at −20°C until further analysis.
Table 1. Serological and molecular screening of cucumber, melon, watermelon and zucchini plants for identification of WmCSV in Saudi Arabia.
Serological analysis
The crude sap was extracted from leaf samples using phosphate-buffered saline (0.02 M phosphate, 0.15 M NaCl, pH 7.4) containing 0.05% (v/v) Tween 20 (PBS-T) as described by Mowat et al. (Citation1987). The sap was subjected to serological analysis for initial viral detection (). The polyclonal antibodies for cucurbit infecting begomoviruses and immunoglobulin G (IgG) alkaline phosphates were obtained from BIOREBA, Switzerland. Twenty-fold dilution fractions of the sap samples in the extraction buffer were subjected to double antibody sandwich-ELISA (DAS-ELISA) using polyclonal antibodies as described earlier (Alhudaib et al., Citation2018). The colour reactions were observed photometrically at 405 nm using a dual filter reader (Awareness Technology, Inc. USA).
Genomic DNA isolation, PCR amplification, cloning and sequencing
Total genomic DNA was extracted from watermelon and zucchini samples using DNeasy plant mini extraction kit (Qiagen, Germany) according to the manufacturer’s instructions. The extracted total genomic DNA was subjected to rolling circle amplification (RCA) using Phi29 DNA polymerase in Illustra TempliPhi Kit (GE Healthcare, Buckinghamshire, UK) to amplify circular DNA-molecules, as described earlier (Inoue-Nagata et al., Citation2004). The resultant RCA products were employed to carry out PCR amplification using degenerate primers (AVcore/ACcore) to amplify ~550 bp partial core CP gene of begomovirus genome (Brown et al., Citation2001). The PCR amplicons were electrophoresed in a 0.7% agarose gel in TBE buffer and visualized using UV transilluminator after staining with ethidium bromide. The obtained amplicons were purified from excessive salts and impurities using QIAquick PCR Purification Kit (Qiagen, Germany). The purified DNA was cloned into pGEM-T Easy Vector (Promega, USA) according to manufacturer’s instructions and afterwards, transformed into DH5α high library-efficiency Escherichia coli competent cells (Invitrogen, USA). The selected clones were sequenced using Sanger sequencing platform (Perkin-Elmer, Applied Biosystems, USA). An abutting primer pair Wm891F (5ʹ-TCTGGGATGAAGGAGCAGGCGC-3ʹ)/Wm893R (AGAGGGGCCACCGACCACGG) was designed and synthesized to amplify the complete DNA-A component of the begomovirus genome sequence. The full-length DNA-B component of the suspected begomovirus genome was amplified using the primer pair WmB672F/WmB695R (Ali-Shtayeh et al., Citation2014). The PCR specifications were: initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation (94°C for 1 min), annealing (55°C for 2 min) and extension (72°C for 2 min), followed by a final extension step at 72°C for 10 min. One clone each for DNA-A and DNA-B from watermelon (Wm1) and zucchini plant (Zu1) were determined in their entirety, as described above.
Comparative sequence and phylogenetic analysis
All the complete sequences of DNA-A and DNA-B were initially compared with the most closely related begomovirus sequences using BLASTn available in NCBI GenBank database (https://www.ncbi.nlm.nih.gov/). The highly similar begomovirus DNA-A and DNA-B nucleotide (nt) sequences were retrieved and employed in the Sequence Demarcation Tool (SDT) (Muhire et al., Citation2014) to determine the pairwise nt sequence identities of all obtained sequences, respectively. The evolutionary relationships were inferred in Mega7 software (Kumar et al., Citation2016) with Neighbour-joining algorithm and Kimura two-parameter model with 1000-bootstrap iterations. The integrity of the open reading frames (ORFs) in virion and complementary sense orientation were also determined using ORF finder tool in the NCBI GenBank database.
Results and discussion
Sample collection and serological assays by DAS-ELISA
The 730 plant leaf samples from four major cucurbit crops (cucumber, zucchini, melon and watermelon) were tested for the presence of begomoviruses by DAS-ELISA from three geographic locations in Saudi Arabia. The serological test confirmed that 39.6% of the watermelon and 53% of the zucchini plants collected from Jizan and Al-Ahsa tested positive for begomovirus infection, respectively (). However, the rest of the samples were negative in DAS-ELISA and hence, could not be used to amplify a full-length genome of WmCSV.
PCR amplification and sequencing analysis
All samples were further subjected to RCA and PCR amplifications, respectively. The expected fragment of ~550 bp CP gene was amplified from 41.5% of watermelon and 54.5% of zucchini leaf samples using degenerate primer pairs, respectively. The greater number of samples that were positive through PCR reflected the higher sensitivity of PCR-based detection methods over serological assays. The resultant amplicons from 8 watermelon and 19 zucchini plant samples were randomly selected for sequencing. The sequence analysis of the core CP confirmed the presence of WmCSV infection (data not shown). The two clones representing the full-length begomovirus DNA-A component from watermelon (pGWW8) and zucchini (pGWZ5) were randomly selected for sequencing and their complete sequences were deposited in the GenBank database (accession numbers KJ958911 and KJ958912), respectively. The full-length DNA-A clones were 2752 and 2753 nt in length, respectively, and have genome organization similar to the OW bipartite begomoviruses. Both clones shared 96.9% sequence identity with each other and the clone pGWW8 had its highest nt sequence identity at 97.7% with previously reported WmCSV (KJ939448) from Saudi Arabia. However, with all other WmCSV isolates, it shared 95.9–97.6% nt sequence identities. On the other hand, the clone pGWZ5 shared highest nt sequence identity at 98.4% with WmCSV isolates identified from Iran (KT272765) and Sudan (AJ245652), respectively (Kheyr-Pour et al., Citation2000). Whereas, with all other WmCSV isolates, it showed 96.3–98.1% nt sequence identity, respectively. The phylogenetic dendrogram grouped both DNA-A isolates into a well-supported (100% bootstrap value) clade with other WmCSV isolates (). Furthermore, when we resolved WmCSV clade with only WmCSV isolates, we found that the clone pGWW8 grouped into a separate branch with previously reported isolates from Saudi Arabia. However, clone pGWZ5 grouped together with WmCSV isolates from the Middle East and Iran, showing the high diversity of WmCSV in Saudi Arabia (Fig. 2a1). Thus, we predict that the WmCSV isolates in Eastern province and Jizan have different evolutionary origins. The isolate pGWZ5 may have been introduced into the cucurbit fields in Saudi Arabia from Iran, whereas the isolate pGWW8 may have spread from Yemen by whiteflies or exchange of vegetative material. In Saudi Arabia, the dominating whitefly species in the Eastern province (including Al-Ahsa region) has been reported to be the Middle East Asia Minor1 (or B-biotype) (Alhudaib et al., Citation2014). Whereas, in the Fayfa region near Yemen border, the Mediterranean species (or Q-biotype) dominate the whitefly population (Ragab, Citation2013). Thus, most probably the distinct isolates of WmCSV have been spread into Saudi Arabia through the exchange of whiteflies in the neighbouring areas with the Yemen border.
Fig. 2 The phylogenetic dendrograms of the full-length DNA-A (a) and DNA-B (b) components of watermelon chlorotic stunt virus (WmCSV) isolates with the selected begomoviruses based upon the multiple nucleotide (nt) sequence alignments. The WmCSV clade was further resolved to reveal the diversity of begomovirus isolates in this study (a1). All the phylogenetic trees were constructed in Mega7 using Neighbour-joining algorithm. The isolates identified from Saudi Arabia in this study have been highlighted in black with white text. The horizontal lines are showing nt substitutions per site with numeric branch nodes representing per cent (%) bootstrap values more than 60% and 1000 replicates. All the isolates used in this study were acronyms and represented by their respective accession numbers following Brown et al. (Citation2015).
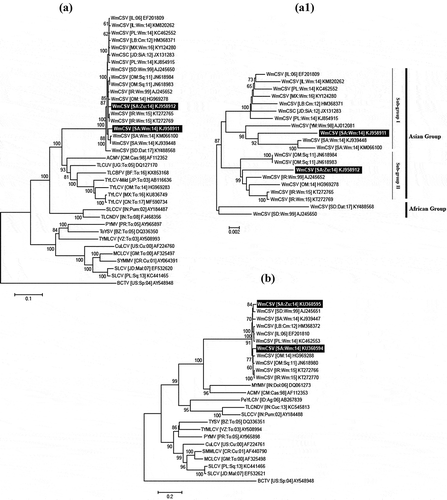
The complete nt sequences of two clones pGWWB8 and pGWZB5 for DNA-B from watermelon and zucchini were 2728 and 2760 nt in length (accession numbers KU360594 and KU360595) and shared 91.8% mutual nt sequence identities, respectively. The clone pGWWB8 shared highest individual nt sequence identity at 95.2% with WmCSV DNA-B from Sudan (AJ245651), whereas the clone pGWZB5 showed highest nt sequence identity at 95.5% with WmCSV DNA-B from Oman (HG969288), respectively. Both DNA-B clones were grouped into a well-supported clade (100% bootstrap) with the previously reported WmCSV DNA-B isolates in the phylogenetic dendrogram ().
A more detailed sequence analysis revealed a ~ 200 nt common region (CR) containing the predicted stem loop sequence (TAATATTAC) and a predicted iteron sequence (TGGAGAC) repeatedly present to the left of the TATA box at the nt positions 2640–2646, 2675–2681 and 2682–2686 in pGWZ5 and pGWW8, respectively, as described earlier by Ali-Shtayeh et al. (Citation2014). Whereas, in the clones pGWWB8 and pGWZB5, the tentative iteron sequence was predicted on the left of the TATA box at nt coordinates 2615–2621, 2650–2656 and 2657–2663, respectively.
Begomoviruses are capable of rapid diversification over time (Ge et al., Citation2007) and thus, regular population diversity surveys are needed. The cucurbit production in the Middle East region has experienced the invasion of two cucurbit-infecting begomoviruses during the last decade, i.e. squash leaf curl virus (SLCV) from the NW and WmCSV from the OW (Lapidot et al., Citation2014). Both of these viruses have been now established in the cucurbit fields parallel to each other in Middle Eastern countries. The high incidence of WmCSV in the watermelon and zucchini plants suggests the virus inoculum is very high and the whitefly vector is highly efficient in these regions. The identification of WmCSV from watermelon (Al-Saleh et al., Citation2014) and zucchini (Alhudaib et al., Citation2018; this study) indicates that WmCSV has been spreading in the cucurbit fields in Saudi Arabia. Moreover, a recent report of WmCSV from Mexico (Domínguez-Duran et al., Citation2018) has further aggravated this scenario. The global trade of virus infected fruits and other vegetative plant parts and/or whiteflies alongside can be the major cause of the geographic distribution of viral diseases (Kamberoglu et al. Citation2016). Such spread of begomovirus-related infections are common between two countries (Sattar et al., Citation2013) or even across two different continents (Tahir et al., Citation2011; Sattar et al., Citation2017). Apart from the appearance of WmCSV as a probable epidemic to the cucurbit production in the Middle East, no detailed dataset is available to predict the factors promoting this spread. We may postulate from the introduction of WmCSV in Africa (Kheyr-Pour et al., Citation2000) and Mexico (Domínguez-Duran et al., Citation2018), that it may become pandemic to the cucurbit production in near future. A detailed study is needed to determine the population structure, sequence variations and the factors promoting the spread of WmCSV in Middle East.
Acknowledgements
The authors would like to thank Mr Wael Alarby and Mr Mostufa Almgasla at Pests & Plant Diseases Unit (PPDU) for their assistance during the sampling for the research.
Additional information
Funding
References
- Abudy A, Sufrin-Ringwald T, Dayan-Glick C, Guenoune-Gelbart D, Livneh O, Zaccai M, Lapidot M. 2010. Watermelon chlorotic stunt and Squash leaf curl begomoviruses-new threats to cucurbit crops in the Middle East. Israel J Plant Sci. 58:33–42.
- Alhudaib KA, Rezk AA, Abdel-Bana BMA, Soliman AM. 2014. Molecular identification of the biotype of whitefly (Bemisia tabaci) inhabiting the Eastern region of Saudi Arabia. J Biol Sci. 14:494–500.
- Alhudaib KA, Rezk AA, Soliman AM. 2018. Current status of watermelon chlorotic stunt virus (WmCSV) on some cucurbit plants (Cucurbitaceae) in Alahsa region of Saudi Arabia. Sci J KFU. 19:31–38.
- Ali-Shtayeh MS, Jamous RM, Mallah OB, Abu-Zeitoun SY. 2014. Molecular characterization of Watermelon chlorotic stunt virus (WmCSV) from Palestine. Viruses. 6:2444–2462.
- Al-Musa A, Anfoka G, Al-Abdulat A, Misbeh S, Haj Ahmed F, Otri I. 2011. Watermelon chlorotic stunt virus (WmCSV): a serious disease threatening watermelon production in Jordan. Virus Genes. 43:79–89.
- Al-Saleh MA, Ahmad MH, Al-Shahwan IM, Brown JK, Idris AM. 2014. First report of Watermelon chlorotic stunt virus infecting watermelon in Saudi Arabia. Plant Dis. 98:1451.
- Brown JK, Idris AM, Torres-Jerez I, Banks GK, Wyatt SD. 2001. The core region of the coat protein gene is highly useful for establishing the provisional identification and classification of begomoviruses. Arch Virol. 146:1581–1598.
- Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Ramos-Sobrinho R, Silva JCF, Fiallo-Olive E, Briddon RW, Hernandez-Zepeda C, et al. 2015. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol. 160:1593–1619.
- Domínguez-Duran G, Rodríguez-Negrete EA, Morales-Aguilar JJ, Camacho-Beltran E, Romero-Romero JL, Rivera-Acosta MA, Leyva-Lopez NE, Arroyo-Becerra A, Mendez-Lozano J. 2018. Molecular and biological characterization of Watermelon chlorotic stunt virus (WmCSV): an Eastern Hemisphere begomovirus introduced in the Western Hemisphere. Crop Prot. 103:51–55.
- Ge LM, Zhang JT, Zhou XP, Li HY. 2007. Genetic structure and population variability of Tomato yellow leaf curl China virus. J Virol. 81:5902–5907.
- Hanley-Bowdoin L, Bejarano E, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 11:777–788.
- Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T. 2004. A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J Virol Methods. 116:209–211.
- Iqbal Z, Sattar MN, Kvarnheden A, Mansoor S, Briddon RW. 2012. Effects of the mutation of selected genes of Cotton leaf curl Kokhran virus on infectivity, symptoms and the maintenance of Cotton leaf curl Multan betasatellite. Virus Res. 169:107–116.
- Kamberoglu MA, Caliskan AF, Desbize C. 2016. Current status of some cucurbit viruses in cukurova region (Adana and mersin provinces) of Turkey and molecular characterization of zucchini yellow mosaic virus isolates. Rom Biotechnol Lett. 21:11709–11719.
- Khan AJ, Akhtar S, Briddon RW, Ammara U, Al-Matrooshi AM, Mansoor S. 2012. Complete nucleotide sequence of Watermelon chlorotic stunt virus originating from Oman. Viruses. 4:1169–1181.
- Kheyr-Pour A, Bananej K, Dafalla GA, Caciagli P, Noris E, Ahoonmanesh A, Lecoq H, Gronenborn B. 2000. Watermelon chlorotic stunt virus from the Sudan and Iran: sequence comparisons and identification of a whitefly-transmission determinant. Phytopathology. 90:629–635.
- Kumar S, Stecher G, Tamura K, Garcia-Alonso L, Alemán A, García-García F, Rodriguez JA, Daub JT, Muntané G, Rueda A, et al. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lapidot M, Gelbart D, Gal-On A, Sela N, Anfoka G, Haj AF, Abou-Jawada Y, Sobh H, Mazyad H, Aboul-Ata -A-A, et al. 2014. Frequent migration of introduced cucurbit-infecting begomoviruses among Middle Eastern countries. Virol J. 11:181.
- Mowat WP, Dawson S, Juntti N, Teppema JS, UytdeHaag FG, Osterhaus AD. 1987. Detection and identification of plant viruses by ELISA using crude sap extracts and unfractionated antisera. J Virol Methods. 15:233–247.
- Muhire BM, Varsani A, Martin DP, Kuhn JH. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 9:e108277.
- Nawaz-ul-Rehman MS, Fauquet CM. 2009. Evolution of geminiviruses and their satellites. FEBS Lett. 583:1825–1832.
- Ragab AI 2013. Genetic variability of the whitefly Bemisia tabaci and its secondary endosymbionts in the Arabian Peninsula [master’s thesis]. Thuwal (Saudi Arabia): King Abdullah University of Science and Technology (KAUST).
- Samsatly J, Sobh H, Jawhari M, Najjar C, Haidar A, Abou-Jawdah Y 2012. First report of Watermelon chlorotic stunt virus in cucurbits in Lebanon. Plant Dis. 96:1703.
- Sattar MN 2012. Diversity and interactions of begomoviruses and their associated DNA-satellites [PhD Thesis]. Uppsala (Sweden): Swedish University of Agricultural Sciences.
- Sattar MN, Iqbal Z, Tahir MN, Ullah S. 2017. The prediction of a new CLCuD epidemic in the old world. Front Microbiol. 8:631.
- Sattar MN, Kvarnheden A, Saeed M, Briddon RW. 2013. Cotton leaf curl disease - an emerging threat to cotton production worldwide. J Gen Virol. 94:695–710.
- Tahir MN, Amin I, Briddon RW, Mansoor S, Martin DP. 2011. The merging of two dynasties-identification of an African cotton leaf curl disease-associated begomovirus with cotton in Pakistan. PLoS One. 6:e20366.
- Walkey DGA, Alhubaishi AA, Webb MJW. 1990. Plant virus diseases in the Yemen Arab Republic. Trop Pest Manag. 36:195–206.
- Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A; Ictv Report Consortium. 2017. ICTV virus taxonomy profile: geminiviridae. J Gen Virol. 98:131–133.

