Abstract
Camellia sinesis (tea) is an economically important crop in China, especially in Fujiang Province. Leaf necrosis symptoms were observed on C. sinensis in Fujian Province, China, during 2015. Initial symptoms were small brown, irregular lesions on young and mature leaves, which become necrotic with a brown margin. Isolations from diseased leaves yielded several pycnidial-forming fungi, including two Lasiodiplodia species which were characterized based on morphology and combined analysis of the internal transcribed spacer (ITS) and translation elongation factor 1-α (tef) gene sequences. The isolates obtained from symptomatic leaves from C. sinensis were identified as Lasiodiplodia theobromae and L. pseudotheobromae. Pathogenicity of both species was demonstrated and symptoms which developed on inoculated C. sinensis leaves were similar to those observed on diseased plants in the field. This is the first report of L. theobromae and L. pseudotheobromae causing leaf necrosis on Camellia sinensis in China.
Résumé
Camellia sinesis (thé) est une culture économiquement importante en Chine, principalement dans la province du Fujian. En 2015, des symptômes de la nécrose des feuilles ont été observés sur C. sinesis dans la province du Fujian. Les symptômes initiaux consistaient en de petites lésions brunes de forme irrégulière se développant sur les feuilles, jeunes et matures, qui par la suite se caractérisaient par une nécrose aux marges brunes. Des isolements faits à partir de feuilles mortes ont engendré différents champignons produisant des pycnides, y compris deux espèces de Lasiodiplodia qui ont été caractérisées en se basant sur leur morphologie et sur l’analyse combinée de l’espaceur transcrit interne (ITS) et des séquences du gène du facteur d’élongation de la transcription 1-α (tef). Les isolats obtenus des feuilles symptomatiques de C. sinesis ont été identifiés en tant que Lasiodiplodia theobromae et L. pseudotheobromae. La pathogénicité des deux espèces a été démontrée et les symptômes qui se sont développés sur les feuilles de C. sinesis inoculées étaient analogues à ceux observés sur les plants infectés trouvés au champ. Il s’agit de la première mention de L. theobromae et de L. pseudotheobromae causant la nécrose des feuilles chez Camellia sinesis en Chine.
Introduction
Camellia sinensis (L.) Kuntze (Theaceae), commonly known as tea, is a major economic crop in China (Maharachchikumbura et al., Citation2013), which was one of the earliest countries to begin tea cultivation. In Fujian province, tea was cultivated on 242 900 hectares in 2014, making it the 5th largest tea growing region in China (China Agriculture Research System Statistical Data 2014). The raw materials for tea products (buds and leaves) are affected by a number of diseases caused by fungi, viruses and bacteria. Diseases caused by fungi can result in reduction of the quantity and quality of the tea leaves. Species of Colletotrichum, Pestalotiopsis, Diaporthe and Botryosphaeria are known to cause leaf necrosis of this crop (Maharachchikumbura et al., Citation2013; Jayawardena et al., Citation2015; Liu et al., Citation2015; Gao et al., Citation2016). Studies relating Botryosphaerious taxa with pathogenicity of C. sinensis in China have not been extensively conducted. This work is part of a larger study identifying fungal taxa associated with C. sinensis in China causing leaf necrosis.
Diseased leaves of C. sinensis cultivar ‘Purple Rose’, which is commonly grown in Fujian Province, were observed in June 2015. The symptoms initially began as small brown, irregular lesions on young and mature leaves, which became necrotic, 2–4 cm diameter with a brown margin. Symptoms mainly occured on the upper surface of the leaves. The objective of this study was to identify the causal agent of this leaf necrosis based on morphology, molecular and pathogenicity studies.
Materials and methods
Sample collection and isolation of fungi
Diseased leaves were collected from 20 infected plants from six fields in Zhangzhou, Fujian, China in June 2015. Small pieces of leaves from the margins of the necrotic areas were surface-sterilized in 75% alcohol for 2 min, 5% NaClO for 1 min, rinsed twice in sterilized distilled water and incubated on potato dextrose agar (PDA) at 25°C for 5 days. From the fungal cultures growing from the diseased leaf pieces, a single spore isolation technique was followed to obtain pure cultures (Chomnunti et al., Citation2014). Single germinating conidia were transferred to fresh PDA dishes and incubated at 28°C. Microscopic examination of morphological characteristics of the cultures was conducted using an Axio Imager Z2 photographic light microscope (Carl Zeiss Microscopy, Germany). Seventeen isolates (JZB313001–JZB313017) were assessed for colony characteristics, which appeared to separate the isolates into two different groups – Group A and B. Five isolates of each group (Group A: JZB313005, JZB313006, JZB313007, JZB313009, JZB3130015; Group B: JZB313001, JZB313002, JZB313003, JZB313004, JZB3130010) were examined for shape and colour of conidia, and the length and width of 40 conidia were measured from each culture.
DNA extraction, amplification and phylogeny
Mycelium was harvested from actively growing 7-day-old, PDA cultures of seven isolates; three from Group A and four from Group B. Total genomic DNA was extracted according to the protocol of Philips et al. (Citation2013). ITS and tef1-α gene regions were amplified using primer pairs ITS1/ITS4 (White et al., Citation1990), and 728F/986R (Carbone & Kohn, Citation1999), respectively. PCR was performed in a BIORAD1000TM Thermal Cycle in a total volume of 25 µL. The PCR mixtures contained 0.3 µL TaKaRa Ex-Taq DNA polymerase, 12.5 µL of 2× PCR buffer (200 mM Tris HCl (pH 8.4), 500 mM KCl supplied with 1mL of 50 mM MgCl2) with 2.5 µL of dNTPs, 1 µL of each primer, 9.2 µL of double-distilled water and 100–500 ng of DNA template. The thermal cycling program followed Philips et al. (Citation2013). The PCR products were verified by staining with ethidium bromide on 1.2% agarose electrophoresis gels and purified according to the manufacturer’s instructions of a Qiagen purification kit (Qiagen, USA). DNA sequencing of the genes were conducted by Sunbiotech Company, Beijing, China. The DNA sequences generated in this study were deposited in GenBank and the alignments in TreeBase (S19576) (www.treebase.org/treebase/index.html).
The systemic placement of taxa was based on the phylogeny of the combined analysis of the ITS and tef gene data. DNAStar V.5.1 and SeqMan V.5.00 were used to obtain consensus sequences from sequences generated from forward and reverse primers. Phylogenetic trees were constructed based on the initial blast results obtained from NCBI blast tool (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). A combined dataset of two gene regions were aligned using Clustal X1.81 (Thompson et al., Citation1997). The sequences were further aligned using default settings of MAFFT v.7 (Katoh & Toh, Citation2008; http://mafft.cbrc.jp/alignment/server/) and manually adjusted using BioEdit V.7.0.9.0 (Hall, Citation1999), where necessary. A maximum parsimony analysis (MP) was also performed using PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford, Citation2002). Ambiguously aligned regions were excluded and gaps were treated as missing data. Trees were inferred using the heuristic search option with Tree Bisection Reconnection (TBR) branch swapping and 1000 random sequence additions. Maxtrees were set at 5000, branches of zero length were collapsed and all multiple parsimonious trees were saved. Tree Length (TL), Consistency Index (CI), Retention Index (RI), Rescaled Consistency Index (RC) and Homoplasy Index (HI) were calculated for trees generated under different optimality criteria. The robustness of the most parsimonious trees was evaluated by 1000 bootstrap replications resulting from maximum parsimony analysis (Hillis & Bull, Citation1993). The Kishino–Hasegawa tests (Kishino & Hasegawa, Citation1989) were performed in order to determine whether the trees inferred under different optimality criteria, were significantly different. The fungal strains that were used are listed in . In addition, Bayesian analyses (BA) were performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck, Citation2003). Suitable models were first selected using models of nucleotide substitution for each gene, using Mr. Modeltest (Nylander, Citation2004). SYM+G model was selected for the combined gene analysis. The analyses of four Markov chain Monte Carlo (MCMC) chains were run from random trees for 100 000 000 generations and sampled every 1000 generations. The first 2000 trees, representing the burn-in phase of the analyses, were discarded and the remaining 8000 trees used for calculating posterior probabilities (PP) in the majority rule consensus tree. Lasiodiplodia citricola was used as the outgroup taxon (). Bayesian inference and maximum parsimony produced nearly identical topologies (Bayes trees not shown).
Table 1. GenBank accession numbers for ITS and tef1-α sequence data for Lasiodiplodia species used in this study. The sequence data for isolates JZB313001 to JZB313007 were obtained in this study and the sequence data for the other isolates were obtained from Phillips et al. (Citation2013).
Pathogenicity studies
Pathogenicity tests were conducted on attached and detached healthy leaves of C. sinensis cultivar ‘Purple Rose’, using fungal strains JZB313005 (a group A isolate) and JZB31001 (a group B isolate). Cultures used for the pathogenicity tests were grown on PDA for 7 days. Spore suspensions of 1 × 106 conidia mL−1 were prepared according to Yan et al. (Citation2013). Pathogenicity of the strains were assessed by presence or absence of symptoms of leaf necrosis.
Attached leaf inoculations were conducted on 18 tea plants planted in plastic pots for each fungal strain. Six attached healthy leaves per tea plant were selected. Wounds (1 cm) were made using a sterilized blade on the upper surface of three selected leaves. Colonized PDA agar discs (5 mm diameter) were placed on wounded and non-wounded leaves of 8 tea plants and wrapped with parafilm (BEMIS, USA). Wounded and non-wounded leaves were inoculated with sterile PDA plugs on one tea plant as the control. Conidial suspension was sprayed on eight tea plants with wounded and non-wounded leaves and one tea plant was sprayed with sterile water as a control.
Detached leaf inoculation consisted of 16 leaves, including two controls per isolate. Eight leaves were inoculated per inoculation method; three wounded inoculation leaves, three non-wounded inoculation leaves, one wounded control leaf and one non-wounded control leaf. Leaves were surface sterilized by immersion in 75% ethanol for 30 s and 1% NaClO for 30 s. One cm wounds were made by a sterilized blade on the upper surface of leaves (three leaves per inoculation method). Colonized PDA agar discs (5 mm diam) were placed on wounded and non-wounded leaves and wrapped with parafilm (BEMIS, USA). Wounded and non-wounded leaves inoculated with sterile PDA plugs were used as the control. The conidial suspension was sprayed on wounded and non-wounded leaves. Wounded and non-wounded control leaves were sprayed with sterile water.
Inoculated plants and leaves were incubated in a moist chamber at ± 95% relative humidity and at 28°C until symptoms appeared. Isolations were made from lesions that developed on inoculated leaves, followed by fungal identification using morphological charcateristics and phylogenetic analysis to fulfil Koch’s postulates.
Results and discussion
Isolation of fungi
Seventeen isolates were obtained from 30 diseased samples collected from the field. The cultures were deposited in Beijing Academy of Agriculture and Forestry Sciences culture collection as isolates JZB313001–JZB313017.
Pathogen identification
Cultures of the isolates grew to 8.0 cm diam. on PDA after 7 days. All of them had white-grey fluffy mycelia (,). The reverse of the isolates was black and became dark brown-black with age (,). Conidiomata were produced after 10 days on PDA. Based on culture characteristics these isolates were separated into two groups; Group A with seven isolates and group B with 10 isolates. Conidiomata were not clearly observed in group A, but in group B, dark brown, simple to aggregate, frequently setose, ostiolate conidiomata were clearly observed. Group A isolates had hyaline, cylindrical, rounded end aseptate paraphyses with a mean length of 56 µm. Group B isolates had hyaline, cylindrical, rounded end and septate paraphyses with a mean length of 54 µm. Conidiophores were not observed in group A, while in group B, conidiophores were hyaline, cylindrical and simple. Both groups had hyaline thin-walled, smooth conidiogenous cells. However, the conidiogenous cells in group A, had cylindrical, slightly swollen bases and group B isolates had conidiogenous cells of cylindical to sub-obpyriform shape (). Conidia of both groups were initially hyaline and aseptate, becoming 1-septate and dark brown some time after release from the conidiomata. However, there were several differences in the conidia among the two groups. Conidia of group A isolates were ellipsoidal with rounded apexes and bases with L/W = 1.7 (22–30 × 14–17 (x = 27 × 16, n = 40); ). Conidia of group B isolates were subovoid to ellipsoid-ovoid with broadly rounded apexes tapering to truncate bases with L/W = 1.9 (22–31 × 13–17 (x = 26.5 × 14.3); ). From these morphological characteristics, we concluded that we had isolated species belonging to the genus Lasiodiplodia.
Fig. 1 (Colour online) a, b, c, Natural symptoms of the leaf necrosis caused by Lasiodiplodia species on tea (Camellia sinensis ‘Purple Rose’). d, Conidia of Lasiodiplodia pseudotheobromae (isolate JZB313005). e, Upper view of a 7-day-old colony of L. pseudotheobromae on PDA. f, Reverse view of a 7-day-old colony of L. pseudotheobromae on PDA. g, Conidiogenous cells of Lasiodiplodia theobromae (isolate JZB313001). h, Conidia of L. theobromae. i, Upper view of a 7-day-old colony of L. theobromae on PDA. j, Reverse view of a 7-day-old colony of L. theobromae on PDA. Scale bars: d, g, h = 10 μm.

Phylogenetic analysis
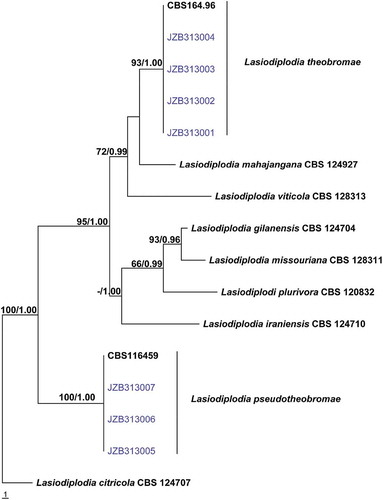
The combined gene alignment for Lasiodiplodia comprised 16 isolates with 808 characters including gaps. Parsimony analysis indicated that 732 characters were constant, 36 variable characters were parsimony-uninformative and 40 characters parsimony-informative. The Kishino–Hasegawa (KH) test showed that the trees (4) generated from parsimonious analysis did not differ significantly (TL = 102, CI = 0.814, RI = 0.865, RC = 0.704, HI = 0.186).
Four representative isolates of L. theobromae and three representative isolates of L. pseudotheobromae obtained in this study clustered together with their ex-type strain respectively with high bootstrap and posterior probability values (). Based on both morphology and phylogenetic analyses the Group A isolates were identified as L. theobromae and the Group B isolates as L. pseudotheobromae.
Fig. 2 (Colour online) Phylogram generated from parsimony analysis based on combined ITS and tef sequence data of 16 isolates of Lasiodiplodia. The sequence data for isolates JZB313001–JZB313007 were obtained in this study and sequence data for the other isolates were obtained from Phillips et al. (Citation2013). Parsimony bootstrap support values above 50% and Bayesian posterior probabilities greater than 0.95 are indicated above the nodes. The ex-type and voucher strains are in bold. The tree is rooted with Lasiodiplodia citricola CBS 124 707.
Pathogenicity studies
Brown necrotic lesions appeared after 10 days on wounded and non-wounded leaves in both detached and attached inoculated leaf assays, while the control leaves remained healthy for both of the isolates studied (). The initial lesions for isolate JZB313001, which was identified as L. theobromae, was observed after 3 days while the initial lesions for isolate JZB313005, which was identified as L. pseudotheobromae, were observed after 5 days. Lesions on the wounded leaves were larger than those on the non-wounded leaves. The symptoms on the inoculated plants were similar to those observed in the field. The experiments were conducted three times with similar results. Fungi isolated from lesions developing on the inoculated leaves were found to be identical to the original isolates used for inoculation based on morphology and phylogenetic analyses. The results revealed that L. pseudotheobromae and L. theobromae were the causal agents of the leaf necrosis.
Fig. 3 (Colour online) Pathogenicity results. a, d, Control b, Necrotic lesions obtained after 7 days of inoculation by Lasiodiplodia pseudotheobromae (isolate JZB313005) in the detached leaf tests c, Necrotic lesions obtained after 7 days of inoculation by Lasiodiplodia theobromae (isolate JZB313001) in the detached leaf tests e, Necrotic lesions obtained after 21 days of inoculation by L. pseudotheobromae (isolate JZB313005) in the attached leaf tests f, Necrotic lesions obtained after 21 days of inoculation by Lasiodiplodia theobromae (isolate JZB313001) in the attached leaf tests.

Lasiodiplodia is a genus of hemibiotrophic phytopathogenic fungi (Tudzynski & Sharon, Citation2003). Recent developments in the phylogeny have established the genus Lasiodiplodia within the family Botryosphaeriaceae (Philips et al., Citation2013). This genus was introduced by Ellis & Everhart (Citation1894) to accommodate the type species Lasiodiplodia tubericola Ellis & Everh., but they did not provide a description for the species. Therefore, Griffin & Maublanc (Citation1909) considered L. theobromae as the type species of this genus (Phillips et al., Citation2013). At present, this genus comprises of 31 species (Dissanayake et al., Citation2016) and the placement of this genus is well-established (Phillips et al., Citation2013; Slippers et al., Citation2013). Species differentiation within this genus using morphological characters has been shown not to be reliable due to the overlapping morphological characters between species (Phillips et al., Citation2013). However, the use of conidial (especially dimensions) and paraphyses morphology can be used in species differentiation (Phillips et al., Citation2013). Species of this genus can be accurately identified from combined ITS and tef1-α sequence data (Slippers et al., Citation2013). Some of the species of this genus are important pathogens causing canker, fruit rots and leaf spots on economically important crops such as Carica papaya, Citrus sinensis, Mangifera indica and Vitis vinifera. Lasiodiplodia pseudotheobromae is known as a pathogen on Vitis vinifera in China (Dissanayake et al., Citation2015; Liu et al., Citation2015). Lasiodiplodia theobromae has a cosmopolitan distribution and was formerly considered to be a cryptic species (Alves et al., Citation2008; Phillips et al., Citation2013). Lasiodiplodia pseudotheobromae and L. theobromae are pathogens on many host plant families, including Actinidiaceae, Aloaceae, Amaryllidaceae, Anacardiaceae, Annonaceae, Araceae, Arecaceae, Asteraceae, Bromeliaceae, Combretaceae, Euphorbiaceae, Fabaceae, Malvaceae, Moraceae, Myrtaceae, Rosaceae, Rutaceae, Sapindaceae, Theaceae, Thymelaceace and Vitaceae, worldwide (Sakalidis et al., Citation2011; Philips et al., Citation2013; Deng et al., Citation2015; Dissanayake et al., Citation2015, Citation2016; Zhou et al., Citation2015). These species have also been recorded as endophytes of many plant families (Chethana et al., Citation2016b; Sakalidis et al., Citation2011).
To the best of our knowledge, this study represents the first report of both L. pseudotheobroame and L. theobromae occurring as pathogens on Camellia sinensis. Botryosphaeria dothidea (Botryosphaeriaceae) has been reported to cause leaf necrosis of tea in China (Jayawardena et al., Citation2015). Lasiodiplodia theobromae has been reported to cause leaf spot of tea oil Camellia (C. olifera) in China (Zhu et al., Citation2014). Pathogenicity testing in this study provides confirmation that these two Lasiodiplodia species are pathogenic to tea leaves.
Correct species identification of plant pathogenic genera is important to implement proper disease control strategies (Chethana et al., Citation2016a). Research on new records of fungal species in hosts can provide useful information in understanding the interactions between hosts and fungi as well as their geographic distribution. Lasiodiplodia species can switch their lifestyle from endophytic to pathogenic, which is influenced by both internal and external environmental factors (Sakalidis et al., Citation2011). Incidence of this disease needs more attention from tea growers. These results can provide plant pathologists and breeders with a reference as well as insight for the prevention and control of this disease.
Acknowledgements
The research was funded by CARS-23, KJCX20140402. Any opinions, finding, conclusions or recommendations expressed in this publication are those of the authors, and there is no conflict of interest to declare.
Additional information
Funding
References
- Alves A, Crous PW, Correia A, Phillips AJL. 2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 28:1–13.
- Carbone I, Kohn L. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 91:553–556.
- Chethana KWT, Li XH, Zhang W, Hyde KD, Yan JY. 2016b. Trail of decryption of molecular research on Botryosphaeriaceae in woody plants. Phytopathol Mediterr. 55:147–171.
- Chethana KWT, Phillips AJL, Zhang W, Chen Z, Hao YY, Hyde KD, Li XH, Yan JY. 2016a. Mycosphere Essay 5: is it important to name species of Botryosphaeriaceae? Mycosphere. 7:870–882.
- Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, et al. 2014. The sooty moulds. Fungal Divers. 66:1–36.
- Deng TJ, Li QL, Chen XL, Huang SP, Guo TX, Mo JY, Hsiang T, Wei JM. 2015. First report of Lasiodiplodia theobromae associated with stem canker of Cassia fistula in Guangxi, South China. Plant Dis. 99:288.
- Dissanayake AJ, Philips AJL, Li XH, Hyde KD. 2016. Botryosphaeriaceae: current status of genera and species. Mycosphere. 7:1001–1073.
- Dissanayake AJ, Zhang W, Liu M, Chukeatirote E, Yan JY, Li XH, Hyde KD. 2015. Lasiodiplodia pseudotheobromae causes pedicel and peduncle discolouration of grapes in China. Australas Plant Dis Notes. 10:article 21.
- Ellis JB, Everhart BM. 1894. New species of fungi from various localities. Proc Acad Natural Sci Philadelphia. 46:322–386.
- Gao Y, Liu F, Cai L. 2016. Unravelling Diaporthe species associated with Camellia. Syst Biodivers. 14:102–117.
- Griffin WM, Maublanc A. 1909. Sur une maladie du cacaoyer. Bulletin De La Société Mycologique De France. 25:51–58.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 42:artcle182.
- Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock WAM, Dissanayake AJ, Glocking SL, Goonasekara ID, et al. 2014. One stop shop: backbones trees for important phytopathogenic genera: I. Fungal Divers. 67:21–125.
- Index Fungorum. 2018. [ accessed 2018 Aug 16]. http://www.indexfungorum.org/names/names.asp
- Jayawardena RS, Li HL, Hyde KD, Li XH, Xu W, Yan JY. 2015. First report of Botrosphaeria dothidea casuing leaf necrosis of Camellia sinensis in Fujian Province, China. Plant Dis. 100:854.
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9:276–285.
- Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data. J Mol Evol. 29:170–179.
- Liu F, Weir BS, Damm U, Crous PW, Wang Y, Liu B, Wang M, Zhang M, Cai L. 2015. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia. 35:63–86.
- Maharachchikumbura SSN, Chukeatirote E, Guo LD, Crous PW, Mckenzie EHC, Hyde KD. 2013. Pestalotiopsis species associated with Camellia sinensis (tea). Mycotaxon. 123:47–61.
- Nylander JAA. 2004. MrModeltest 2.0. Program distributed author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW. 2013. The Botryosphaeriaceae: genera and species known from culture. Stud Mycol. 76:51–167.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572.
- Sakalidis ML, Hardy GES, Burgess TI. 2011. Endophytes as potential pathogens of the baobab species Adansonia gregorii: a focus on the Botryosphaeriaceae. Fungal Ecol. 4:1–14.
- Slippers B, Boissin E, Phillips AJL, Groenewald JZ, Lombard L, Wingfield MJ, Postma A, Burgess T, Crous PW. 2013. Phylogenetic lineages in the Botryosphaeriales: A systematic and evolutionary framework. Stud Mycol. 76:31–49.
- Swofford DL. 2002. PAUP* 4.0: phylogenetic analysis using parsimony (* and other methods). Sunderland: Sinauer Associates.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882.
- Tudzynski P, Sharon A. 2003. Fungal pathogenicity genes. Appl Microbiol Biot. 3:187–212.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York (USA): Academic; p. 315–322.
- Yan JY, Xie Y, Zhang W, Wang Y, Liu JK, Hyde KD, Robert C, Zhang GZ, Wang ZY, Yao SW, et al. 2013. Species of Botryosphaeriaceae involved in grapevine dieback in China. Fungal Divers. 61:221–236.
- Zhou Y, Gong G, Cui Y, Zhang D, Chang X, Hu R, Liu N, Sun X. 2015. Identification of Botryosphaeriaceae species causing kiwifruit rot in Sichuan Province, China. Plant Dis. 99:699–708.
- Zhu H, Niu XQ, Song WW, Yu FY, Tang QH, Qin WQ, Chen LQ. 2014. First report of leaf spot of tea oil Camellia (Camellia oleifera) caused by Lasiodiplodia theobromae in China. Plant Dis. 98:10.
