Abstract
Mint (Mentha sp.; family: Lamiaceae) plants showing yellowing and stunting symptoms from Salalah, Dhofar region, Oman were shown to be infected by a monopartite begomovirus (family: Geminiviridae). Cloning and sequencing of restriction endonuclease digested rolling-circle amplified viral DNA identified a begomovirus associated with the symptomatic mint plants. Further analysis of complete sequences showed the virus to be a variant of the ‘Oman’ strain of Chili leaf curl virus (ChLCV-OM), a virus prevalent in a number of crops across Oman. However, the ChLCV-OM identified in mint has a recombination pattern distinct from all other ChLCV isolates. Additionally, PCR amplification and sequencing showed the mint plants to also harbour the betasatellite, Tomato leaf curl betasatellite (ToLCB). This is the first identification of ChLCV in association with ToLCB infecting mint. The distinct recombination pattern of ChLCV-OM from mint suggests that the host is epidemiologically important in the spread of ChLCV-OM to other crops in Oman.
Résumé
Des plants de menthe (Mentha sp.; famille: Lamiaceae) de Salalah, de la région du Dhofar au Sultanat d’Oman, affichant des symptômes de jaunissement et de rabougrissement, ont été infectés par un bégomovirus monopartite (famille: Geminiviridae). Le clonage et le séquençage d’un ADN viral amplifié en cercle roulant digéré par une endonucléase de restriction ont permis d’identifier un bégomovirus associé aux plants de menthe symptomatiques. Une analyze plus poussée des séquences complètes a montré que le virus était une variante de la souche ‘Oman’ du virus de la frisolée du piment (ChLCV-OM), un virus répandu dans un certain nombre de cultures partout au sultanat. Toutefois, le ChLCV-OM identifié chez la menthe possède un schéma de recombinaison distinct de tous les autres isolats de ChLCV. De plus, l’amplification par PCR et le séquençage ont montré que les plants de menthe hébergent également le ß-satellite de la frisolée de la tomate (ToLCB). C’est la première fois que le ChLCV est identifié de concert avec le ToLCB quant à l’infection chez la menthe. Le schéma de recombinaison distinct du ChLCV-OM chez la menthe suggère que l’hôte est important sur le plan épidémiologique en ce qui a trait à la dissémination de ChLCV-OM dans d’autres cultures au Sultanat d’Oman.
Introduction
Plant-infecting viruses with circular single-stranded DNA (ssDNA) genomes known as Geminiviruses belong to the genus Begomovirus (family Geminiviridae) (Zerbini et al., Citation2017). The genus Begomovirus is the leading genus of the family Geminiviridae of circular ssDNA viruses encompassing many of the economically important geminiviruses and are transmitted by the whitefly (Bemisia tabaci) species complex (Zerbini et al., Citation2017). The genomes of the begomoviruses consist of either two ~2.7 kb genomic components (bipartite), referred to as DNA-A and DNA-B, or a single ~2.7 kb genomic component (monopartite) homologous to the DNA-A component of bipartite begomoviruses. The majority of begomoviruses native to the New World (NW) are bipartite; only a single monopartite begomovirus has so far been reported from the NW (Melgarejo et al., Citation2013; Sánchez-Campos et al., Citation2013). On the contrary, in the Old World (OW), the majority of the begomoviruses characterized are monopartite (Zerbini et al., Citation2017). The DNA-A component of begomoviruses from the OW encodes six genes of which one, the V2 gene (also known as the precoat protein), is absent from begomoviruses originating from the NW. Two of the overlapping genes being transcribed from the virion sense strand encode for coat protein (V1) and precoat protein (V2), while four other overlapping genes belong to the complementary sense strand and encode for the replication-associated protein (Rep), transcriptional activator protein (TrAP), replication enhancer (REn) and C4 protein. The DNA-B component encodes for one gene in the complementary sense known as movement protein (MP), and the other one in the virion sense, the nuclear shuttle protein (NSP). Both MP and NSP are required for cell-to-cell movement as well as for the export of replicated viral DNA from the nucleus to the cytoplasm, respectively.
Another important feature of the OW begomoviruses is that a majority of the monopartite begomoviruses are found associated with a class of ssDNA satellite molecules known as betasatellites, while these satellite molecules are absent in NW viruses (Zhou, Citation2013). Sequence analyses revealed three important characteristic features of betasatellites: (1) DNA segment that is highly conserved among the betasatellites referred to as satellite conserved region (SCR); (2) adenine (A) rich region; and (3) a gene on the complementary sense strand encoding for a protein known as βC1 (Briddon et al., Citation2003). Betasatellites play an important role in determining the host range and the satellite encoded βC1 protein is also a major symptom severity determinant (Briddon et al., Citation2001; Saunders et al., Citation2004).
Mint (Mentha sp.; family; Lamiaceae) is an important crop that has been cultivated for both food and medicinal purposes for thousands of years and is affected by a number of plant pathogens. During a survey in December 2016 in the vicinity of Salalah (Dhofar region, Oman), leaf yellowing and stunting symptoms were observed on mint plants (). The plants showed typical symptoms associated with begomovirus infection and the observation was further supported by the presence of higher numbers of an insect vector (B. tabaci) on the plants. The objective of this study was to characterize the putative begomovirus associated with virus-like symptoms on mint.
Materials and methods
Leaf samples were collected from the three symptomatic, as well as two non-symptomatic, mint plants in a mixed cropping field where tomato and chili pepper were the other crops. Total nucleic acids were extracted from the leaf samples using a modified CTAB method (Porebski et al., Citation1997). Nucleic acids were used as a template in polymerase chain reaction (PCR) along with the following set of primers for the detection of begomoviruses (FD-CP-382 5′-CTSARCTTCGACAGCCCXTA-3′; RD-CP-1038 5′-TGMGTACAXGCCATATACAA-3′). The nucleic acid samples were also used as a template for rolling circle amplification (RCA) reaction, to amplify all circular DNA molecules, using TempliPhi 100 Amplification System (GE Healthcare). As high molecular weight products were produced only from symptomatic plant samples, they were subsequently digested using a number of restriction enzymes to identify the release of unit length (~2.7 kb) fragments. Restriction with the endonuclease HindIII resulted in ~2.7 kb fragments which were gel purified and cloned into Hind III digested pUC19 plasmid vector. The betasatellite-specific primers Sat101/Sat102 (Shahid et al., Citation2017a) were used in the PCR reactions and the products were cloned in pTZ257R/T vector system (Thermo Fisher Scientific). Potentially full-length virus and betasatellite clones were sequenced in both orientations at Macrogen Inc. (Seoul, South Korea).
Three potentially full-length clones (MV2, MV3 and MV4) from each symptomatic plant were obtained from Hind III digested RCA products. The sequences for all three clones were found to be 2,763 nt in length and further analysis revealed that the sequences were identical. On the basis of 100% similarity, the complete genome sequence of only one isolate (MV3) was deposited in the nucleotide sequence database under the accession number MG566078. Further analysis of the deposited sequence showed it to have an organization typical of monopartite begomoviruses (or DNA-A component of bipartite begomoviruses native to the OW).
Results and discussion
The PCR product for the detection of begomoviruses of the expected ~700 nt was obtained from symptomatic plant leaf samples while no amplicons were obtained from DNA isolated from non-symptomatic plants, confirming the presence of begomovirus in symptomatic plants. The PCR reactions using primer pairs for the detection of begomovirus DNA-B component (Rojas et al., Citation1993) and/or alphasatellites (Bull et al., Citation2003) did not yield any products. An initial screen of the full length sequence (MV3) against the GenBank nucleotide sequence database using BLASTn (Altschul et al., Citation1990; run online http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed this to have a high level of sequence identity to the isolates of Chili leaf curl virus (ChLCV). ‘Species Demarcation Tool’ (SDT; Muhire et al., Citation2014) analysis of all the available ChLCV sequences and the other selected begomovirus sequences showed MV3 to have nucleotide sequence identity in the range of 90% to isolates of the ‘Oman’ strain of ChLCV (ChLCV-OM), with the highest (94.9%) similarity to the two isolates reported from tomato, originating from Oman (JN604496, JN604497). To all the other isolates of ChLCV as well as other begomoviruses, the nucleotide sequence identity was less than 88.6%. Based on the criteria set for species/strain demarcation of begomoviruses, the ChLCV isolate from mint is a variant of ChLCV-OM. This is supported by the phylogenetic analysis (Neighbour-joining method) that showed MV3 to cluster with other ChLCV-OM isolates and was distinct from isolates of the other ChLCV strains ().
Fig. 2. Phylogenetic analysis of the sequences using available sequences in the GenBank database. (a) The dendrograms based on alignments of the complete nucleotide sequences of the genomes of selected monopartite begomoviruses and (b) betasatellite components. Vertical branches are arbitrary; horizontal branches are proportional to calculated mutation distance. Values at nodes of the Neighbour-joining phylogenetic tree indicate the percentage boot strap values (1000 replicates). The begomovirus phylogenetic tree was arbitrarily rooted on the sequence of Tomato pseudo-curly top virus (X84735) and the betasatellite tree on Ageratum yellow vein Singapore alphasatellite (FJ956707) as outgroup. The GenBank database accession numbers are indicated in each case.
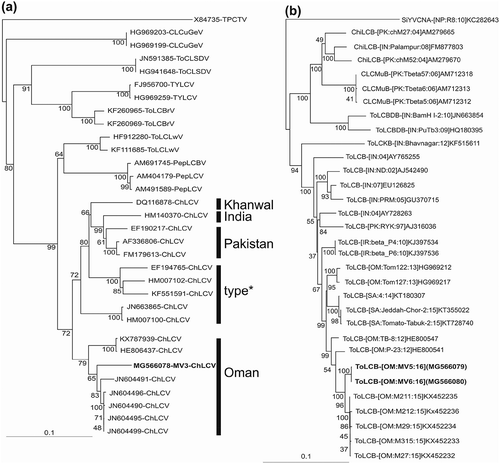
The RDP analysis of all available ChLCV sequences showed MV3 to have a recombination pattern distinct from all other ChLCV isolates, including other ChLCV-OM isolates. The analysis showed MV3 to be an intraspecific recombinant having ChLCV [IN: Pat: Chi: 08] (HM007117) as major parent, with a fragment (coordinates 21 to 2255 of MV3) of ChLCV-OM [OM: TH3: Chili: 11] (JN604498). This was supported by seven of the nine detection algorithms implemented in RDP with average P-values between 2.39 × 10−41 and 1.656 × 10−5. The possible recombination event might have occurred outside (South Asian countries) of Oman and this ChLCV was subsequently introduced to Oman, or it might have occurred in Oman and was identified later. Two clones (MV5 and MV6), from different plant samples, were obtained as PCR products using primer pair Sat101/Sat102 (Shahid et al., Citation2017a). The sequences of MV5 and MV6 were found to be 1367 and 1368 nt in length, respectively, and were deposited in the GenBank nucleotide database under the accession numbers MG566079 and MG566080, respectively. The sequences encompassed features typical of betasatellites, including a single gene in the complementary sense (known as βC1), encoding a predicted product of 118 amino acids, a region of sequence well conserved among all betasatellites (known as the satellite conserved region), and an adenine rich region (Briddon et al., Citation2003).
The species demarcation tool (SDT) analysis showed MV5 and MV6 to have 99.5% nucleotide sequence identity with each other and the highest nucleotide identity (98.1–98.2%) to the sequences of five ToLCB isolates (KX452232–KX452236) recently obtained from Phaseolus vulgaris originating from Oman (Shahid et al., Citation2017b). This shows that the ChLCV-OM isolate infecting Mentha is associated with ToLCB. The phylogenetic analysis revealed that betasatellites obtained here cluster with other isolates of ToLCB from the Arabian Peninsula, but are distinct from isolates originating from South Asia ().
Mentha spp. have been shown to host a number of distinct viruses, including begomoviruses. In India, mint has been shown to be infected with Chili leaf curl India virus and Ageratum yellow vein betasatellite (Saeed et al., Citation2014), Chili leaf curl India virus and Andrographis yellow vein leaf curl betasatellite (Saeed et al., Citation2017), Tomato leaf curl virus and Cotton leaf curl Multan betasatellite (Borah et al., Citation2010), Tomato leaf curl Pakistan virus (Samad et al., Citation2009) and possibly also Pedilanthus leaf curl virus (based on partial sequence available in the GenBank database). This is the first report of a Mentha spp. infected by ChLCV and ToLCB. The varying recombination patterns between the ChLCV-OM isolates characterized here and the ChLCV-OM infecting crop plants in Oman suggest that possibly Mentha is not an alternate host for the virus infecting other crops, although ChLCV-OM has so far been identified in tomato, pepper and watermelon (Khan et al., Citation2013; Shahid et al., Citation2017a). The genetic ‘fingerprint’ of ChLCV-OM infecting mint obtained here should allow the epidemiology of the virus to be studied in the future – specifically to investigate the movement of the virus between different crops and weeds that may harbour the virus in the off-season.
Acknowledgements
We are thankful to the farmers and technicians for their help in sample collection for this research work.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Borah B, Cheema G, Gill C, Dasgupta I. 2010. A geminivirus-satellite complex is associated with leaf deformity of Mentha (Mint) plants in Punjab. Indian J Virol. 21:103–109.
- Briddon R, Mansoor S, Bedford I, Pinner M, Saunders K, Stanley J, Zafar Y, Malik K, Markham P. 2001. Identification of DNA components required for induction of cotton leaf curl disease. Virology. 285:234–243.
- Briddon RW, Bull SE, Amin I, Idris AM, Mansoor S, Bedford ID, Dhawan P, Rishi N, Siwatch SS, Abdel-Salam AM. 2003. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology. 312:106–121.
- Bull S, Briddon R, Markham P. 2003. Universal primers for the PCR-mediated amplification of DNA 1: a satellite-like molecule associated with begomovirus-DNA β complexes. Mol Biotechnol. 23:83–86.
- Khan AJ, Akhtar S, Al-Zaidi AM, Singh AK, Briddon RW. 2013. Genetic diversity and distribution of a distinct strain of Chili leaf curl virus and associated betasatellite infecting tomato and pepper in Oman. Virus Res. 177:87–97.
- Melgarejo TA, Kon T, Rojas MR, Paz-Carrasco L, Zerbini FM, Gilbertson RL. 2013. Characterization of a New World monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J Virol. 87:5397–5413.
- Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 9:e108277. doi:10.1371/journal.pone.0108277.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15:8–15.
- Rojas MR, Gilbertson R, Maxwell D. 1993. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 77:340–347.
- Saeed S, Khan A, Kumar B, Ajayakumar P, Samad A. 2014. First report of Chilli leaf curl India virus infecting Mentha spicata (Neera) in India. Plant Dis. 98:164.
- Saeed ST, Kumar B, Shasany A, Samad A. 2017. Molecular identification of Chilli leaf curl India virus along with betasatellite molecule causing leaf curl disease of menthol mint (Mentha arvensis var. Kosi) in India. J Gen Plant Pathol. 83:333–336.
- Samad A, Gupta MK, Shasany A, Ajayakumar P, Alam M. 2009. Begomovirus related to Tomato leaf curl Pakistan virus newly reported in Mentha crops in India. Plant Pathol. 58:404.
- Sánchez-Campos S, Martínez-Ayala A, Márquez-Martín B, Aragón-Caballero L, Navas-Castillo J, Moriones E. 2013. Fulfilling Koch’s postulates confirms the monopartite nature of Tomato leaf deformation virus: a begomovirus native to the New World. Virus Res. 173:286–293.
- Saunders K, Norman A, Gucciardo S, Stanley J. 2004. The DNA β satellite component associated with ageratum yellow vein disease encodes an essential pathogenicity protein (βC1). Virology. 324:37–47.
- Shahid M, Al-Sadi A, Briddon R. 2017a. First report of Chilli leaf curl virus and Tomato leaf curl betasatellite infecting watermelon (Citrullus lanatus) in Oman. Plant Dis. 101:1063.
- Shahid MS, Briddon RW, Al‐Sadi AM. 2017b. Identification of Mungbean yellow mosaic Indian virus associated with Tomato leaf curl betasatellite infecting Phaseolus vulgaris in Oman. J Phytopathol. 165:204–211.
- Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A. 2017. ICTV’ virus taxonomy profile: geminiviridae. J Gen Virol. 98:131–133.
- Zhou X. 2013. Advances in understanding begomovirus satellites. Annu Rev Phytopathol. 51:357–381.

