Abstract
Leaf blight of sunflower caused by Alternaria spp. occurs frequently in the Beijing municipality of China. A total of 482 Alternaria isolates were obtained from sunflower leaves with typical dark fleck symptoms sampled from 13 different districts in the Beijing municipality in 2014. Morphological observations and sequence analyses of the rDNA-ITS region and the histone 3 gene revealed that two Alternaria species, A. tenuissima (86.5% frequency) and A. alternata (13.5% frequency), were associated with the disease. Four isolates of A. tenuissima and six of A. alternata were pathogenic to sunflower leaves with disease incidence and disease index ranging from 50−100% and 16.7–33%, respectively. This is the first comprehensive examination of the involvement of Alternaria species in leaf blight of sunflower of Beijing, China.
Résumé
La brûlure helminthosporienne du tournesol, causée par Alternaria spp., apparaît fréquemment à Beijing, en Chine. En tout, 482 isolats d’Alternaria ont été obtenus de feuilles de tournesol affichant les taches noires typiques, échantillonnées dans 13 districts de Beijing en 2014. Les observations morphologiques et les analyses de séquences de la région de l’ITS de l’ADNr et du gène de l’histone 3 ont révélé que deux espèces d’Alternaria, A. tenuissima (fréquence de 86.5%) et A. alternata (fréquence de 13.5%), étaient associées à la maladie. Quatre isolats d’A. tenuissima et six d’A. alternata étaient pathogènes à l’égard des feuilles de tournesol, ayant une incidence de la maladie et un indice de la maladie variant de 50 à 100% et de 16.7 à 33%, respectivement. Il s’agit du premier examen approfondi de l’implication des espèces d’Alternaria dans la brûlure helminthosporienne chez le tournesol à Beijing.
Introduction
Sunflower (Helianthus annuus L.) is the fourth largest oil crop in China, with a cultivation area of approximately 1 million ha, and total production of 2 million tons annually. Sunflower leaf blight caused by Alternaria spp. is one of the most destructive and widely occurring diseases in the world, including China (Hansford, Citation1943; Qi, Citation1966; Tubaki & Nishihara, Citation1969; Allen et al., Citation1981; Kong et al., Citation1995). The pathogenic fungi can invade the discs, stems and leaves of sunflower, causing round brown lesions, leaf drop and even death of the plants. The disease can also result in premature defoliation under conditions of warm temperature and high relative humidity (Prathuangwong et al., Citation1991). Sunflower leaf blight can reduce the seed yield by 28–81% and oil content by 19–34% (Balasubrahmanyam & Kolte, Citation1980; Bai, Citation1986). Qi (Citation1966) reported for the first time that sunflower leaf blight occurred in China. In the early 1970s, the sunflower planting area in China rapidly increased, and leaf blight disease became a major problem due to the lack of disease-resistant sunflower varieties. It has been a serious threat to China’s sunflower production for many years (Yu et al., Citation1996). Nine Alternaria species have been reported to cause leaf blight on sunflower worldwide (Wang et al., Citation2014). However, the population structure of Alternaria species associated with sunflower leaf blight in Beijing has not been thoroughly studied. Thus, the objectives of the present study were to identify the Alternaria species associated with leaf blight on sunflower in Beijing, China using morphological and molecular methods as well as pathogenicity tests.
Materials and methods
Isolation and characterization of the pathogen
Sunflower leaves with dark fleck symptoms () were sampled from 170 fields from 13 different districts in Beijing, China. Pieces of leaf tissue (5 × 5 mm) were cut from the margins of lesions with a sterile scalpel, surface-disinfected with 0.3% sodium hypochlorite for 2 min and 70% ethanol for 40 s, and then rinsed three times with sterile distilled water. The leaf pieces were incubated in Petri dishes containing potato dextrose agar (PDA) amended with streptomycin sulphate (50 mg·L−1) at 25°C in the dark for 7 days. A total of 482 putative Alternaria isolates were obtained and subcultured onto PDA and pure cultures were obtained using single spore technique (). Colony characteristics and sporulation patterns of 10 isolates representing different morphologies from different regions were examined as described by Simmons (Citation2007) on potato carrot agar (PCA) dishes.
Table 1. Geographic origins and number of Alternaria isolates obtained from infected sunflower leaves collected in Beijing, China.
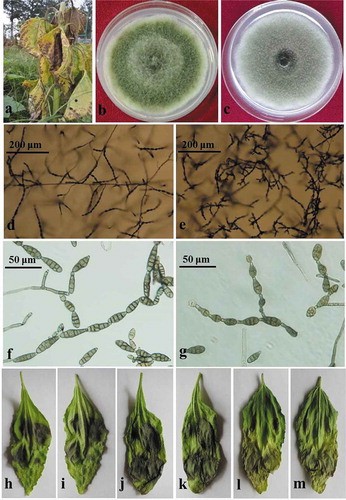
Fig. 1 (Colour online) Symptoms of sunflower leaf blight and morphologies of the causal fungi, Alternaria tenuissima and A. alternata. (a), Necrotic flecks and chlorotic lesions on diseased sunflower leaves. (b, d, f), Colonies, sporulation patterns and conidia of the representative isolates of Alternaria tenuissima. (c, e, g), Colonies, sporulation patterns and conidia of the representative isolates of A. alternata. (h–j), Pathogenicity of the representative isolates of A. tenuissima on detached sunflower leaves. (k–m), Pathogenicity of the representative isolates of A. alternata on detached sunflower leaves.
Pathogenicity tests
A total of 10 Alternaria isolates (four tentative A. tenuissima, six tentative A. alternata) from different regions were selected and tested for their pathogenicity on sunflower leaves of cv. ‘Gankui No. 2ʹ. Spore suspensions of the Alternaria isolates were prepared by flooding 7-day-old cultures on PDA with sterile distilled water, gently scratching the colony surface and collecting the liquid. After filtering through four layers of sterile cheesecloth, the spore suspension was adjusted to 106 spores·mL−1 with the aid of a hemocytometer. A 20 μL drop of spore suspension of the Alternaria isolates were inoculated individually on each side of the main vein of the upper leaf surface of fully expanded detached leaves from 30-day-old plants (two points per leaf) (Pryor & Michailides, Citation2002). Control leaves were inoculated similarly with sterile distilled water. For each isolate tested, 15 fully expanded sunflower leaves were inoculated and placed in Petri dishes resting in five plastic boxes (19 × 14 × 5 cm (length × width × height)). The plastic boxes were lined with two layers of blotting paper moistened with sterile distilled water, covered to maintain high humidity, and incubated in a growth chamber at 25°C and 90% RH with a 12 h daily fluorescent light photoperiod (4000 lux) for 7 days. Symptoms on the inoculated leaves were observed and compared with those which occurred on naturally infected leaves in the field. Disease severity (DS) was scored using the standard scale of Santha et al. (Citation2009): 0 = no symptoms on leaf; 1 = small, circular, scattered, brown spots on leaves covering 1% or less of the leaf area; 3 = spots enlarging, dark brown in colour, covering 1–10% of the leaf area; 5 = spots enlarging, dark brown in colour, target like appearance covering 11–25% of leaf area; 7 = spots dark brown, coalescing with target like appearance covering 26–50% of leaf area; 9 = spots uniformly dark brown covering 51% or more of leaf area. Disease index (DI) was calculated by the formula described by Bensassi et al. (Citation2009): DI = [100× ∑ (n× corresponding DS)]/(N × 9), where n is the number of inoculation points corresponding to each disease rating, N is the number of total inoculation points. Isolations were made from lesions that developed on inoculated sunflower leaves, and the fungi obtained were identified using morphological and molecular techniques to determine if the lesions were caused by the same fungus used to inoculate sunflower leaves, thereby fulfilling Koch’s postulates.
Molecular identification
All 482 Alternaria isolates were cultured on PDA dishes at 25°C in darkness for 7 days. Approximately 20 mg (fresh weight) of mycelia of each isolate was harvested by scratching the colony surface with an inoculating loop, and ground in liquid nitrogen. Genomic DNA was extracted using the revised method of Adachi et al. (Citation1993). The rDNA-ITS region, including ITS1, ITS2 and the 5.8S ribosomal DNA, were amplified using primer set ITS1 (5ʹ-TCC GTA GGT GAA CCT GCG G-3ʹ) and ITS4 (5ʹ-TCC TCC GCT TAT TGA TAT GC-3ʹ) at the following thermal profile: initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 40 s, annealing at 58°C for 40 s, extension at 72°C for 1 min; final extension at 72°C for 10 min (White et al., Citation1990). The partial coding sequence of the histone 3 gene was amplified using primer set H3-1a (5ʹ-ACT AAG AG ACC GCC CGC AGG-3ʹ) and H3-1b (5ʹ-GCG GGC GAG CTG GAT GTC CTT-3ʹ) using the following thermal cycling profile: initial denaturation at 96°C for 2 min; 30 cycles of denaturation at 96°C for 15 s, annealing at 55°C for 30 s, extension at 75°C for 35 s; final extension at 72°C for 2 min (Glass & Donaldson, Citation1995). PCR amplification was done in a 25 μL volume containing 10.5 μL ddH2O, 12.5 μL Premix Ex Taq (v. 2.0, TaKaRa; containing 0.625 U Taq DNA polymerase, 200 μM dNTP and 2 mM MgCl2), 0.5 μL each of the primers (10 μM) and 1 μL DNA template (100 μg·mL−1).
The PCR amplification products were purified with AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Hangzhou, China) and sent to Beijing Sunbiotech Co. Ltd (Beijing, China) for sequencing. The obtained sequences were analysed using BLAST on the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). Sequence alignment was done using Clustal X 1.83 version program (Thompson et al., Citation1994) with manual adjustment. Phylogenetic analysis was carried out using the neighbour-joining method of MEGA 5 version (http://www.Megasoftware.net/) (Udayashankar et al., Citation2012). Alternaria infectoria was used as an outgroup. The rDNA-ITS region or histone 3 gene of the A. tenuissima and A. alternata strains retrieved from GenBank were also used as references in phylogeny analysis.
Results and discussion
Isolation and characterization of the pathogen
A total of 482 Alternaria isolates were obtained from the infected leaf samples (). The Alternaria isolates were tentatively classified into either A. tenuissima (Nees) Wiltshire or A. alternata (Fr.) Keissl groups according to the morphological characters on PDA and PCA. The Alternaria isolates classified as A. tenuissima produced greyish-green to olive brown colonies with aerial mycelia on top of the colony on PDA (). Sporulation patterns showed the primary chains contained up to 12 conidia in length with one or two lateral branches forming occasionally on PCA (). Conidiophores were short and arose singly. Conidia were typically ovoid to obclavate (). The isolates classified as A. alternata produced dark grey, dense colonies (). Sporulation patterns of this species had primary chains of 8−12 conidia in length with secondary and occasionally tertiary chains arising from apical and median conidia on PCA (). Conidiophores were single or fasciculate, straight or bent. Conidia were obpyriform to ellipsoid ().
Pathogencity test
The sunflower leaves developed nearly round or oval, dark brown to black lesions after inoculation with the 10 Alternaria isolates, which were similar to those observed on naturally infected leaf samples in sunflower fields (–). All control leaves inoculated with sterile distilled water remained asymptomatic. Fungi isolated from lesions on inoculated leaves had similar morphological and molecular characteristics as the fungal isolates used for inoculation (four isolates of A. tenuissima, six isolates of A. alternata), fulfilling Koch’s postulates. There was no statistical difference between the two species or among isolates from different geographic origins for disease index ().
Table 2. Disease incidence and disease index of four isolates of Alternaria tenuissima and six isolates of A. alternata on detached sunflower leaves.
Molecular identification
The PCR amplifications of the ITS1-ITS2 regions of rDNA produced fragments of 570 bp from all the 482 Alternaria isolates. Sequence analyses of the partial coding sequences of the histone 3 gene indicated the Alternaria isolates could be divided into two species, A. tenuissima (546 bp) and A. alternata (440 bp). With regard to all the 482 isolates, the GenBank accession numbers of their ITS1-ITS2 sequences were from KR866865−KR867346, whilst those of the histone 3 gene were from KR866383−KR866864. Phylogenetic analyses were conducted based on the sequences of the ITS1–ITS2 regions of rDNA and the partial coding sequences of the histone 3 gene of the four A. tenuissima isolates and six A. alternata isolates used in the pathogenicity tests. Phylogenetic analysis of the partial coding sequence of the histone 3 gene divided all the tested Alternaria isolates into two distinct clades, A. tenuissima and A. alternata (), but all isolates were grouped in one clade using the ITS sequence data ().
Fig. 2 Phylogenetic tree based on the partial coding sequences of histone 3 genes of four A. tenuissima isolates, six A. alternata isolates and three reference sequences retrieved from GenBank. The tree was constructed by neighbour-joining using the Kimura two-parameter distance method. Bootstrap values (in percentage) above 70 from 1000 pseudo-replicates are shown for major lineages within the tree. The marker denotes a measurement of relative phylogenetic distance. Alternaria infectoria was used as an outgroup.
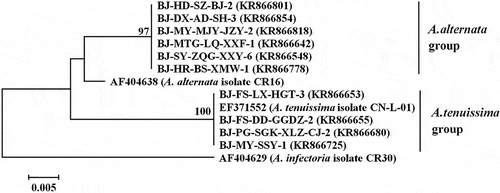
Fig. 3 Phylogenetic tree based on the sequences of the ITS region of rDNA of four A. tenuissima isolates, six A. alternata isolates, and three reference sequences retrieved from GenBank. The tree was constructed by neighbour-joining using the Kimura two-parameter distance method. Bootstrap values (in percentage) above 70 from 1000 pseudo-replicates are shown for major lineages within the tree. The marker denotes a measurement of relative phylogenetic distance. Alternaria infectoria was used as an outgroup.

Different culture media, such as PCA, DRYES, MEA, PDA and DG18 have been used to evaluate the morphological characteristics to discriminate pathogenic Alternaria species (Andersen & Thrane, Citation1996; Andersen et al., Citation2001, Citation2005). However, Pryor & Michailides (Citation2002) showed that morphological traits were insufficient to clearly distinguish among some small-spored Alternaria species. It is reported that the analysis of the sequences of rDNA-ITS regions is not sufficiently variable to estimate a phylogeny among the small-spored Alternaria species (Kusaba & Tsuge, Citation1995) as was shown in this study. The use of EF-1α, CAL, CHS and THN sequences are also insufficient to delineate species (Roberts et al., Citation2000; Pryor & Michailides, Citation2002; Hong et al., Citation2006; Andrew et al., Citation2009). DNA sequence analysis based on the partial coding sequences of the histone 3 gene has proven to be effective to differentiate the small-spored Alternaria species (Wang et al., Citation2014; Zheng et al., Citation2015; Zhao et al., Citation2016a, Citation2016b, Citation2018). Kang et al. (Citation2002) also found the partial coding sequence of the histone 3 gene was better able to distinguish Alternaria spp. than that of the ITS region of rDNA.
Population structure of Alternaria species in this study was dominated by A. tenuissima, whereas A. helianthi was considered as the main pathogen of sunflower leaf blight in China 20 years ago (Yu et al., Citation1996). Possible reasons for this change may relate to infection sources from diseased plant debris in soil and infected sunflower seeds. Alternaria tenuissima is highly adaptable to different environments, and has been identified as the causal agent of many diseases on other plants, including wheat (Bensassi et al., Citation2009), peanut (Banuett & Herskowitz, Citation1994), potato (Zheng et al., Citation2015; Zhao et al., Citation2018), muskmelon (Zhao et al., Citation2016a) and watermelon (Zhao et al., Citation2016b). All these plants were often rotated with sunflower in Beijing, which would increase the risk of sunflower plants becoming infected by A. tenuissima. Alternaria alternata has also been reported as the main cause of leaf blight of Zanthoxylum piperitum (Yang et al., Citation2013), cherry fruit (Zhao & Liu, Citation2012), potato (Zheng et al., Citation2015; Zhao et al., Citation2018), muskmelon (Zhao et al., Citation2016a) and watermelon (Zhao et al., Citation2016b) in China. In addition, A. tenuissima and A. alternata have been found to infect the seeds of sunflower (Dawar & Ghaffar, Citation1991; Viswanathan, Citation1996), which was a main cause of sunflower plant infection. According to the present results, Alternaria tenuissima and A. alternata should be treated as major target pathogens when making decisions on sunflower management strategies in Beijing, China.
Additional information
Funding
References
- Adachi Y, Watanabe H, Tanabe K, Doke N, Nishimura S, Tsuge T. 1993. Nuclear ribosomal DNA as a probe for genetic variability in the Japanese pear pathotype of Alternaria alternata. Appl Environ Microb. 59:3197–3205.
- Allen SJ, Kochman JK, Brown JF. 1981. Losses in sunflower yield caused by Alternaria helianthi in Southern Queensland. Aust J Exp Agr. 21:98–100.
- Andersen B, Hansen ME, Smedsgaard J. 2005. Automated and unbiased image analyses as tools in phenotypic classification of small-spored Alternaria spp. Phytopathology. 95:1021–1029.
- Andersen B, Krøger E, Roberts RG. 2001. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol Res. 105:291–299.
- Andersen B, Thrane U. 1996. Differentiation of Alternaria infectoria and Alternaria alternata based on morphology, metabolite profiles, and cultural characteristics. Can J Microbiol. 42:685–689.
- Andrew M, Peever TL, Pryor BM. 2009. An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia. 101:95–109.
- Bai JA. 1986. Sunflower disease prevention and problems. Chinese J Oil Crop Sci. 4:9–17. In Chinese.
- Balasubrahmanyam N, Kolte SJ. 1980. Effect of different intensities of Alternaria blight on yield and oil content of sunflower. J Agr Sci. 94:749–751.
- Banuett F, Herskowitz I. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Gene Dev. 8:1367–1378.
- Bensassi F, Zid M, Rhouma A. 2009. First report of Alternaria species associated with black point of wheat in Tunisia. Ann Microbiol. 59:465–467.
- Dawar S, Ghaffar A. 1991. Detection of the seed-borne mycoflora of sunflower. Pak J Bot. 23:173–178.
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb. 61:1323–1330.
- Hansford CG. 1943. Contributions towards the fungus flora of Uganda.—v. fungi imperfecti. Proc Linn Soc London. 155:34–67.
- Hong SG, Maccaroni M, Figuli PJ. 2006. Polyphasic classification of Alternaria isolated from hazelnut and walnut fruit in Europe. Mycol Res. 110:1290–1300.
- Kang JC, Crous PW, Mchau GR, Serdani M, Song SM. 2002. Phylogenetic analysis of Alternaria spp. associated with apple core rot and citrus black rot in South Africa. Mycol Res. 106:1151–1162.
- Kong GA, Kochman JK, Brown JF. 1995. A greenhouse assay to screen sunflower for resistance to Alternaria helianthi. Ann Appl Biol. 127:463–478.
- Kusaba M, Tsuge T. 1995. Phylogeny of Alternaria fungi known to produce host-specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Curr Genet. 28:491–498.
- Prathuangwong S, Kao SW, Sommartya T, Sinchaisri P. 1991. Role of four Alternaria spp. causing leaf and stem blight of sunflower in Thailand and their chemical controls. Kasetsart J. 25:112–124.
- Pryor BM, Michailides TJ. 2002. Morphological, pathogenic, and molecular characterization of Alternaria isolates associated with Alternaria late blight of pistachio. Phytopathology. 92:406–416.
- Qi PK. 1966. Fungal diseases of cultivated plants in Jilin Province, China. Beijing: Science Press. In Chinese.
- Roberts RG, Reymond ST, Andersen B. 2000. RAPD fragment pattern analysis and morphological segregation of small-spored Alternaria species and species groups. Mycol Res. 104:151–160.
- Santha LPM, Sujatha M, Chander RS. 2009. Analysis of cultural and genetic diversity in Alternaria helianthi and determination of pathogenic variability using wild Helianthus species. J Phytopathol. 157:609–617.
- Simmons EG. 2007. Alternaria: an identification manual. CBS Biodiversity Series 6. Utrecht (the Netherlands): CBS Fungal Biodiversity Centre.
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Tubaki K, Nishihara N. 1969. Alternaria helianthi (Hansf.) comb. nov. Trans Brit Mycol Soc. 53:147–149.
- Udayashankar A, Nayaka SC, Archana B, Anjana G, Niranjana S, Mortensen CN, Lund OS, Prakash H. 2012. Specific PCR-based detection of Alternaria helianthi: the cause of blight and leaf spot in sunflower. Arch Microbiol. 194:923.
- Viswanathan R. 1996. Seed mycoflora composition in sunflower. Ind Phytopathol. 49:286–289.
- Wang TY, Zhao J, Sun P, Wu XH. 2014. Characterization of Alternaria species associated with leaf blight of sunflower in China. Eur J Plant Pathol. 140:301–315.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protoc. 18:315–322.
- Yang WX, Liu F, Zhang N. 2013. First report of Alternaria alternata causing blight on Zanthoxylum piperitum in China. Plant Dis. 97:840.
- Yu L, Zhang LH, Li C, Guan CJ. 1996. Alternaria helianthi on sunflower was identified and compared its forms with its closed species. J Jilin Agr Univ. 18:22–24. In Chinese.
- Zhao J, Bao SW, Ma GP, Wu XH. 2016a. Characterization of Alternaria species associated with muskmelon foliar diseases in Beijing municipality of China. J Gen Plant Pathol. 82:29–32.
- Zhao J, Bao SW, Ma GP, Wu XH. 2016b. Characterization of Alternaria species associated with watermelon leaf blight in Beijing municipality of China. J Plant Pathol. 98:135–138.
- Zhao J, Ma GP, Liu YY, Wu XH. 2018. Alternaria species infecting potato in southern China. Can J Plant Pathol. 40:312–317.
- Zhao YZ, Liu ZH. 2012. First report of black spot disease caused by Alternaria alternata on cherry fruits in China. Eur J Plant Pathol. 140:301–315.
- Zheng HH, Zhao J, Wang TY, Wu XH. 2015. Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathol. 64:425–433.
