Abstract
A baseline for genetic diversity of Ralstonia solanacearum species complex (RSSC) causing bacterial wilt of field and greenhouse grown Solanum lycopersicum (tomato) in Louisiana was established based on biovar, phylotype and phylogenetic analysis of the egl gene sequence. RSCC strains were characterized as Biovars 1, 3 and 6 and belonged to phylotypes I and II. Phylotypes were subdivided into sequevars based on differences in the nucleotide sequence of the egl gene. Phylotype II strains clustered with closely related North American strains in sequevar 7 and phylotype I strains clustered with Asian strains in sequevar 14. All three RSCC strains isolated from greenhouse grown tomato were phylotype II sequevar 7 and were identical to strain AW1 from tomato in Alabama (AL). While all RSCC strains were pathogenic on tomato ‘Roma’, only six induced a hypersensitive reponse (HR) on Nicotiana tabacum. Four of the HR positive strains were also pathogenic on pepper ‘Yolo Wonder’. This is the first study to characterize RSSC strains causing bacterial wilt of tomato in Louisiana and confirms that strains originating from Asia are present in North America. Genetic characterization of RSCC strains currently present in Louisiana is the first step towards the development of strain-specific management practices, including the identification of strain-specific host resistance.
Résumé
Une base de référence, en ce qui a trait à la diversité génétique du complexe d’espèces Ralstonia solanacearum (CERs) causant la flétrissure bactérienne chez la tomate cultivée en champ et en serre en Louisiane, a été établie en se basant sur le biovar, le phylotype et l’analyse phylogénétique de la séquence du gène egl. Les souches de CERs ont été caractérisées en tant que Biovars 1, 3 et 6 appartenant aux phylotypes I et II. En se basant sur les différences qui existent dans la séquence de nucléotides du gène egl, les phylotypes ont été divisés en séquevars. Les souches du phylotype II se sont groupées avec les souches nord-américaines étroitement apparentées dans le séquevar 7, et les souches du phylotype I se sont groupées avec les souches asiatiques dans le séquevar 14. Les trois souches de CERs isolées à partir de tomates cultivées en serre étaient du phylotype II séquevar 7, et étaient identiques à la souche AW1 de la tomate cultivée en Alabama. Tandis que toutes les souches de CERs étaient pathogènes à l’égard de la tomate ‘Roma’, seulement six ont induit une réaction d’hypersensibilité (RH) chez Nicotiana tabacum. Quatre des souches provoquant une RH étaient également pathogènes à l’égard du poivron ‘Yolo Wonder’. Cette étude est la première à caractériser les souches de CERs causant la flétrissure bactérienne de la tomate en Louisiane et confirme que des souches issues d’Asie sont établies en Amérique du Nord. La caractérisation génétique des souches de CERs actuellement établies en Louisiane est la première étape qui concoure à développer des stratégies de gestion propres à ces souches, y compris la détermination de la résistance de l’hôte spécifique de la souche.
Introduction
In south-eastern Louisiana (LA), USA, the majority of fresh market tomatoes are produced on small mixed-crop farms, with an average farm size of 0.6 ha (1.4 acres). Bacterial wilt, caused by Ralstonia solanacearum species complex (RSSC) (Gillings & Fahy, Citation1994), is a major limiting factor to small-scale tomato production in LA and other southern states and is one of the most challenging diseases of tomato to manage. Producers in LA report annual losses due to bacterial wilt, with plant loss ranging from 10–50% depending on the season. The RSSC consists of a heterogeneous group of widely geographically distributed but related strains (Fegan & Prior, Citation2005). Although RSSC strains share common traits, groups within the species have different characteristics, such as host range, optimal temperature to cause disease, and differential utilization of carbohydrates (Hayward, Citation1964). Historically, the population of RSSC worldwide has been classified using a dichotomous system consisting of race and biovar (Hayward, Citation1964; French et al., Citation1995). However, because host range of RSCC strains are broad and can overlap, and biovar tests tend to be inconsistent and for the most part do not correspond to phylogenetically coherent groups (Champoiseau et al., Citation2009), this system has been mostly replaced by a hierarchical classification system that groups strains of RSSC into phylotypes and sequevars (Fegan & Prior, Citation2005; Prior & Fegan, Citation2005). However, in developing countries where bacterial wilt is a major constraint of tomato and eggplant production and where molecular tools are not economically feasible or readily accessible, biovar is the primary method of characterizing RSCC strains of horticulture food crops into infrasubspecies groups.
Phylotypes, which correspond to the geographic origin of strains, are defined as monophyletic lineages based on phylogenetic analysis of 16S–23S internal transcribed spacer (ITS) region sequence data (Fegan & Prior, Citation2005). To date, strains of RSCC have been divided into four phylotypes (I–IV). Phylotype I strains originate from Asia, phylotype II strains from the Americas, phylotype III strains from Africa and Mayotte Island (Indian Ocean) and phylotype IV strains from Australia, Indonesia and Japan. Phylotype II strains have two recognizable subclusters, IIA and IIB (Fegan & Prior, Citation2005; Prior & Fegan, Citation2005; Castillo & Greenberg, Citation2007). Over the past 25 years, the bacterium has been reclassified several times. Most recently, Safini et al. (Citation2014) and Prior et al. (Citation2016) proposed the division of RSCC into three species – R. pseudosolanacearum (phylotypes I and III), R. solanacearum (phylotypes IIA and IIB) and R. syzgii (phylotype IV). However, in this study, we have used RSCC or R. solanacearum when referring to the strains used.
Phylotypes can be further subdivided into a variable and additive number of sequevars, based on a 1% difference in the nucleotide sequence of the egl gene (Fegan & Prior, Citation2005). The egl gene has provided robust phylogenies for the majority of RSSC strains characterized, although exceptions have occurred (Lewis Ivey et al., Citation2007; Hong et al., Citation2012). When first described, 23 sequevars were identified based on egl sequences from 140 RSCC strains (Fegan & Prior, Citation2005). Since these first reports, the number of sequevars has increased dramatically to at least 57.
In the southeastern USA (Georgia (GA), South Carolina (SC) and North Carolina (NC)), RSSC strains isolated from solanaceous crops are classified as race 1, biovars 1 or 3, phylotype II and sequevar 7 (Ji et al., Citation2007; Hong et al., Citation2012). Strains from Florida (FL) are more diverse than those from GA, SC and NC, belonging to phylotypes II and I and at least four different sequevars (4, 5/6, 7, 14 and an undefined sequevar) (Hong et al., Citation2012; Weibel et al., Citation2016). Although bacterial wilt of tomato was first reported in LA in 1913 (Edgerton & Moreland, Citation1913), and RSSC is endemic in the state, strains of RSSC from LA have not been characterized. Thus the objective of this study was to characterize the phylotype, endoglucanase (egl) sequevar and biovar of isolates in small-scale tomato production regions of LA.
Materials and methods
Sample collection, bacterial isolation and isolate storage
Tomato plants with bacterial wilt symptoms were collected from small-scale production fields and greenhouses in three major tomato production parishes (East Baton Rouge, Livingston and St. Helena) in south-eastern LA in 2015. In the field and laboratory, tomato stem tissue was tested for bacterial streaming by suspending them in clean water. Ralstonia solanacearum-specific immunoflow strip tests (Agdia Inc., Elkhart, IN) were also used according to the manufacturer’s instructions to confirm the presence of the bacterial wilt pathogen. Symptomatic stem tissue (water soaking and brownish-black vascular discolouration) was surface-disinfected in 70% ethanol and 10-fold serial dilutions of macerated stem tissue in 10 mM potassium phosphate buffer (pH 7.0) were plated onto Kelman’s tetrazolium chloride (TZC) (Kelman, Citation1954) medium. The plates were incubated at 28°C for 3–5 days. Pure cultures from 48–72 h single colony streaks of each isolate were stored in nutrient broth (NB) containing 15% glycerol at −80°C for long-term storage. Completion of Koch’s postulates was conducted on four representative strains as previously described (Madrid Jimenez et al., Citation2016). All laboratory practices were conducted according to US Department of Agriculture (USDA)–Animal and Plant Health Inspection Service (APHIS) permit P526P-15–04705 conditions. Presumptive RSCC isolates are listed in .
Table 1. Genotypic and phenotypic diversity of Ralstonia solanacearum species complex strains from Louisiana.
Pathogenicity tests on tomato and pepper
Pathogenicity tests were performed using the RSSC susceptible tomato ‘Roma VF’ and the RSSC susceptible pepper ‘Yolo Wonder’. A strain of RSCC (WLS-2) isolated from a tomato plant with bacterial wilt symptoms in 2009 () (unpublished) served as the positive control. Seedlings without RSSC inoculum served as negative controls. Roots of 4-week-old tomato or pepper seedlings were wounded by making small cuts through the seedling mix with a sterile razor blade. Five plants per RSSC strain were drenched with 50 mL of inoculum (~108 CFU mL−1). Plants were grown under BSL-2 greenhouse conditions as described by Madrid Jimenez et al. (Citation2016). Run-off inoculum was collected in closed containers and autoclaved to kill all viable RSCC cells. Strains that resulted in wilting symptoms, vascular discolouration and were positive for bacterial streaming after 21 days were considered pathogenic.
Hypersensitive response (HR) test on tobacco
All 11 strains () were tested to determine if they could induce a hypersensitive response (HR) on tobacco (Nicotiana tabacum) (Klement & Goodman, Citation1967). Each RSCC strain was suspended into sterile deionized water (SDW) to an OD600 = 0.1–0.15 (~108 CFU mL−1) and ~0.5 mL of each suspension was infiltrated into the intercellular space of fully expanded tobacco leaves using a 3-mL hypodermic syringe (without a needle). Strain MLI37-14 (Xanthomonas vesicatoria) was used as the positive control and sterile deionized water (SDW) as the non-inoculated control. Each strain was inoculated twice on the same leaf. Inoculated plants were grown under standard greenhouse conditions (~30°C and 80% relative humidity) with no supplemental lighting. Infiltrated areas were assessed for collapsed tissue and desiccation at the site of infiltration 24, 48 and 72 h after inoculation. Pathogenicity and hypersensitive tests were repeated once.
DNA extraction and identification of isolates
Ralstonia solanacearum strains were grown on TZC medium for 3 days at 28°C. Total genomic DNA was extracted using the Cetyl Trimethyl Ammonium Bromide (CTAB) method (modified from Zhang et al., Citation1998). DNA was quantified spectrophotometrically at 260 nm, diluted to 50 ng μL−1 and stored at 4ºC for further studies. For whole cell extractions, RSCC strains were grown on TZC medium for 3 days at 28°C and a loopful of bacterial cells were suspended in 200 µL of SDW. The whole cell extractions were incubated at −20ºC for 24 h, thawed at room temperature, and then stored at −20ºC. The taxonomic identity of RSCC was verified with RSCC-specific primers 759/760 (Opina et al., Citation1997) as previously described (Lewis Ivey et al., Citation2007). Race 3-specific PCR was performed using the primer pair 630/631 (5′ ATA CAG AAT TCG ACC GGC AC-3′ and 5′ ATT CAC ATG CAA TTC GCC TAC-3′) (Fegan et al., Citation1998). PCR reactions contained 12 µL Master Mix (GoTaq DNA polymerase at 100 units mL−1, 400 μM dNTP, and 3 mM MgCl2; Promega Corporation, Madison, WI), 1 µL of each primer (0.6 µM), 10 µL DNAse-free water and 1 µL template (DNA). Template DNA for RSCC reference strain UW551 was provided by C. Allen, University of Wisconsin-Madison (Madison, WI) and served as the positive control. PCR was performed in a C-1000 Touch thermocycler (BioRad Laboratories, Foster, CA) using the following programme: 96°C for 5 min; 30 cycles of 94°C for 15 s, 60°C for 30 s and 72°C 30 s; followed by 72°C for 10 min. Amplified PCR products were separated by horizontal gel electrophoresis in 2% agarose gels and run in TBE buffer (pH 8.3) for 1 h at 120 V. DNA was visualized under UV light with GelRed Nucleic acid (Phenix Research Product, Candler, NC) staining and photographed using a UVP ChemiDoc-It2 Imager (UVP LLC, Upland, CA) system with VisionWorksLS Image Acquisition and Analysis Software (Upland, CA). R3b2-specific PCR was repeated once.
Biovar determination
Biovar designation of each strain was determined as previously described by French et al. (Citation1993) and Subedi (Citation2015). Sugar/alcohol solutions (10%) were prepared by dissolving cellobiose, lactose, maltose, dulcitol, mannitol or sorbitol in SDW. Ten millilitres of each sugar/alcohol solution was mixed with 90 mL of basal medium (BM; pH 7) and 150 μL of each mixture was dispensed into five individual wells in a sterile 96-well plate. Control wells contained BM only. A droplet (7 μL) of 48-h-old culture suspension (~108 CFU mL−1) of each RSCC isolate was added to each well and incubated at 28°C for 5 days and ambient room temperature for an additional 2 days. Water served as the non-inoculated control. Internal reference strains were not used due to APHIS permit P526P-15–04705 condition restrictions. Isolates capable of oxidizing the sugar/alcohol turned the BM from green to yellow. Biovar was scored using published biovar type profiles (He et al., Citation1983; French et al., Citation1993; Hayward, Citation1994; Xue et al., Citation2011). The plates were scored on days 3, 5 and 7. The biovar test was repeated twice.
Phylotype identification
Phylotype specific primers (Fegan & Prior, Citation2005) were used in combination with a species-specific reverse primer (Fegan & Prior, Citation2005) and the species-specific primers 759/760 (Opina et al., Citation1997). Template DNA for RSCC reference strains GMI1000 and K60 (CFBP2047) representing phylotypes I and II respectively were provided by C. Allen, University of Wisconsin-Madison (Madison, WI) and served as controls. PCR assays were performed as previously described (Lewis Ivey et al., Citation2007). PCR product was separated by horizontal gel electrophoresis in 1.5% agarose with TBE buffer at 50 V for 2 h. DNA was visualized and photographed as described above. Phylotype-specific-multiplex PCR was repeated twice.
Sequevar identification
The egl gene was amplified from RSCC strains using primers EndoF (5′ ATGCATGCCGCTGGTCGCCGC-3′) and EndoR (5′ GCGTTGCCCGGCACGAACACC-3′) using previously described PCR assays (Fegan et al., Citation1998). Amplified PCR product was separated by horizontal gel electrophoresis in 1.5% agarose gels and run in TBE buffer (pH 8.3) for 70 min at 100 V. DNA was visualized and photographed as described above. PCR products were purified using an ExoSAP-IT PCR Cleanup kit (Affymetrix ThermoFisher Scientific, Waltham, MA) before sequencing. Sequencing of the forward and reverse amplicons was performed for six of the 11 strains with a two-step cycle sequencing programme by Macrogen Corp. (Rockville, MD). Sanger sequencing was conducted using the 3130 Genetic Analyzer (Applied Biosystems, Forest City, CA) by Macrogen Corp. (Rockville, MD). Sequence reads were edited, trimmed, and consensus sequences automatically assembled using Geneious 8.1 (Kearse et al., Citation2012). A multiple sequence alignment (MSA) was estimated using the on-line MAFFT version 7 with the iterative refinement method G-INS-I with 200PAM/k = 2 scoring matrix (Katoh & Standley, Citation2013). Sequence data were compared to sequences published in GenBank using the Basic Local Alignment Search Tool (BLAST) service. Reference sequences () used in the analysis were selected from previously published reports (Ji et al., Citation2007; Lewis Ivey et al., Citation2007; Lebeau et al., Citation2011; Hong et al., Citation2012) with strains deposited in GenBank. A maximum likelihood (ML) topology for the egl MSA was inferred in GARLI v. 2.01 (Zwickl, Citation2006) using the best-fit model of nucleotide substitution selected using Akaike’s Information Criterion corrected for small sample sizes (AICc) in jModelTest2 v. 2.1.6. (Posada, Citation2008; Darriba et al., Citation2012). The ML topology is the best tree from 20 replicate searches. Independent searches were terminated after 10 000 generations without an improvement in the likelihood score greater than 0.01 log-likelihood units. Node support was estimated in a bootstrap analysis with 1008 pseudo-replicates, with the tree with the highest likelihood after two replicate searches used to represent each bootstrap pseudo-replicate dataset. Support values were mapped to the ML tree using the program SumTrees 3.3.1, which utilizes the DendroPy Phylogenetic Computing Library version 3.12.0 (Sukumaran & Holder, Citation2010). Bootstrap support values ≥70% were considered robust and are represented in the phylogenetic tree.
Table 2. List of Ralstonia solanacearum reference species complex strains used in the phylogenetic analysis.
Results
Isolates, pathogenicity and HR
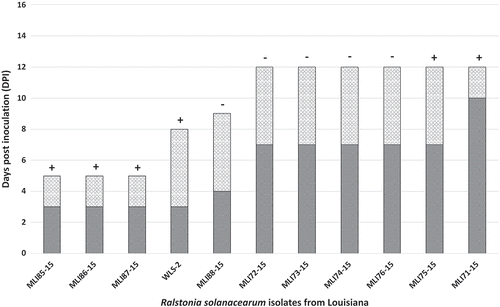
A total of 11 isolates were confirmed to be RSCC (). Seven of the isolates were from Livingston parish, three from Saint Helena parish and one from East Baton Rouge parish. The three isolates from Saint Helena parish were isolated from greenhouse produced tomatoes. Six isolates, all from field grown tomato (MLI71–15, MLI72-15, MLI73-15, MLI74–15, MLI75-15, MLI76-15) caused the development of wilt symptoms on ‘Roma VF’ tomato within 7–10 DPI and plants were dead 12 DPI (). Of these, only isolates MLI71-15 and MLI75-15 induced a HR in tobacco. Field isolate MLI88-15 did not induce a HR but caused wilting symptoms within 4 DPI and the plants were dead by 9 DPI. Field isolate WLS-2 induced a HR, caused wilting symptoms within 3 DPI and plants were dead by 8 DPI. The three isolates from greenhouse grown tomatoes (MLI85-15, MLI86-15 and MLI87-15) developed symptoms within 3 DPI and were dead within 5 DPI. On pepper, only isolates MLI85-15, MLI86-15, MLI71-15 and WLS-2 developed wilt symptoms (data not shown). MLI85-15 and MLI86-15 first induced wilt symptoms on pepper within 7 DPI and MLI71-15 and WLS-2 within 10 DPI (data not shown). Non-inoculated control plants (tomato and pepper) did not develop symptoms nor did the water control induce a HR on tobacco. Plants inoculated with Xanthomonas vesicatoria strain MLI37-14, which served as the HR test positive control, induced a HR on tobacco leaves within 24 h.
Fig. 1 Pathogenicity of Ralstonia solanacearum species complex (RSCC) strains isolated from tomato in Louisiana on susceptible tomato ‘Roma VF’ and hypersensitive response (HR) on Nicotiana tabacum (tobacco). The dark grey bars indicate the number of days post inoculation that wilt symptoms were first observed and the light grey bars indicate the number of days post inoculation that the plants died from RSCC infection. The + symbol above each bar indicates a positive HR 24 h after infiltration into tobacco and – symbol indicates no HR within 72 h post infiltration.
Biovar determination
Biovars 1, 3 and 6 were found in LA (). Biovar 1 was only found in St. Helena parish. Biovar 3 was present in Livingston and biovar 6 in Livingston and East Baton Rouge parishes. The majority (57%) of RSCC strains from Livingston parish were biovar 6.
Molecular characterization
All of the 11 field isolates (strains) generated a 280-bp RSCC species-complex-specific amplicon with the PCR primers 759/760 (data not shown). None of the RSCC strains from LA produced the expected 304 bp R3Bv2 specific amplicon (data not shown). Reference strain UW551 produced a single 304 bp product and no product was produced when water was used as template.
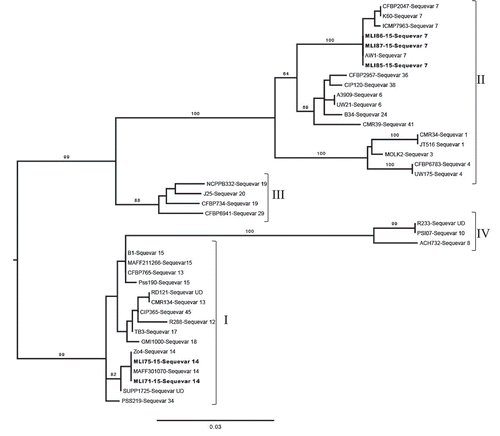
Eight (73%) of the strains generated a ~370-bp amplicon and were identified as phylotype II (). The other three strains (27%) generated a 144-bp phylotype I-specific amplicon and were identified as phylotype I. A partial fragment (750-bp) of the egl gene was amplified from six (MLI71-15, MLI75-15, MLI85-15–MLI88-15) of the 11 RSCC strains tested (Table 3). No PCR amplicon was produced from MLI72-15, MLI73-15, MLI 74-15, MLI76-15, MLI77-15 or WLS-2. The six strains that generated a partial egl fragment were sequenced and compared with 36 reference strains of RSCC representing the known extent of phylotype diversity and 22 different sequevars. The sequence quality of strain MLI88-15 was consistently poor, however, based on a partial egl nucleotide sequence, this sequence was 98% similar to reference strain AW1 (accession: KC188082.1) isolated from tomato in AL (data not shown); MLI88-15 was not included in the final analysis and was not assigned a sequevar. Ralstonia solanacearum strains MLI85-15, MLI86-15 and MLI87-15, isolated from greenhouse tomatoes, were nested within a subclade of phylotype II that includes all sequevar 7 strains and AW1 originally isolated from tomato in AL (). The closely related sequevar 7 strains include CFBP2047 (K60) from the USA and the Kenyan strain ICMP7963. Strains MLI75-15 and MLI71-15, both isolated from Livingston parish but from different fields, formed a monophyletic group with the phylotype I reference strain MAAF301 070 originally isolated from tomato in Japan and Zo4 isolated from ginger in the Philippines. The egl sequences for MLI71-15, MLI75-15, MLI85-15, MLI86-15 and MLI87-15 were published in GenBank ().
Fig. 2 Maximum likelihood phylogenetic tree based on the partial (750-bp) endoglucanase gene sequences of five Ralstonia solanacearum species complex from Louisiana (MLI71-15, MLI75-15, MLI85-15, MLI86-15 and MLI87-15 shown in bold text) and 36 reference strains previously deposited in GenBank. Bootstrap support values ≥70 are shown above the branches. The scale bar represents an estimated three nucleotide substitutions per 100 nucleotides. The midpoint of the path between the two most distant taxa on the tree was used to root the phylogeny. Strains within each of the four phylotypes (I, II, II and IV) are shown in brackets. <>
Discussion
To establish a baseline for the diversity of Ralstonia solanacearum (RSCC) causing bacterial wilt of tomato on small-scale farms in Louisiana (LA), RSCC isolates were characterized using biovar and the hierarchical classification system as described by Hayward (Citation1994) and Fegan & Prior (Citation2005), respectively. In this study, we observed more genetic and phenotypic diversity among the strains confirmed to be RSCC than expected from a small sample size; although none were characterized as the select agent (regulated plant pest) Ralstonia solanacearum race 3 biovar 2. Five of the 11 strains did not induce HR on tobacco but were pathogenic on ‘Roma VF’ tomato. These results are consistent with other studies that characterized RSCC strains from the southern USA including FL, NC, SC and GA (Robertson et al., Citation2004; Hong et al., Citation2012). Variation in the ability of RSCC strains from LA to utilize various carbohydrates was also observed. Strains belonged to Biovars 1, 3 and 6. In the USA, only biovars 1, 2 and 3 have been reported. Since the original four biovars were proposed by Hayward (Citation1964) the number of biotypes has expanded to accommodate outlier strains (He et al., Citation1983; French et al., Citation1993; Hayward, Citation1994). Biovar 6 was recently described by Xue et al. (Citation2011) and Gaitonde & Ramesh (Citation2014) and contains strains that use all the disaccharides and oxidize all the hexose alcohols, except dulcitol. While many of the traits used to determine biovar are probably associated with only one gene and may easily mutate, biovar type is thought to be relatively stable in culture and nature (Denny, Citation2006). Over a span of two years, the biovar 6 strains in this study consistently utilized sorbitol and not dulcitol (data not shown). This is the first report of Biovar 6 in the USA.
All the LA Biovar 1 strains were characterized as phylotype II based on phylotype-specific multiplex PCR and Biovar 3 strains were characterized as phylotype I. Biovar 3 strains, which cause significant losses to solanaceous crops, are widely distributed throughout Asia, Australia, Africa, Europe and South and Central America (Hayward, Citation1964, Citation1994) and were reported for the first time in FL in 2007 (Ji et al., Citation2007). Initial observations indicated that the Biovar 1 strains originally isolated from greenhouse tomato (MLI85-15, MLI86-15 and MLI87-15) were more aggressive than all of the other strains on tomato ‘Roma VF’. Although this was not supported statistically (χ2 = 3.5, P = 0.174), these three strains killed the tomato plants within 5 days compared to between 8 and 12 days by the other strains. Our results are similar to those reported by Prior et al. (Citation1990) in which biovar 1 strains were more aggressive than biovar 3 strains on tomato ‘Capitan’. Lopes et al. (Citation1994) also indicated that biovar 1 strains were more aggressive than biovar 3 in six tomato cultivars. In contradiction to our study, Biovar 3 strains isolated from pepper in FL were more aggressive on tomato, tobacco and pepper cultivars than biovar 1 (Hong et al., Citation2012). Given our small sample size, identifying a clear link between biovar and aggressiveness is unrealistic. Further studies including a broad collection of RSCC strains isolated from tomato or different crops are needed in order to identify differential biochemical characteristics of RSCC strains (biovars) and any correlation with aggressiveness.
For over 40 years, the majority of strains collected from the south-eastern USA have been classified as phylotype II sequevar 7, and this group is assumed to be endemic in the region. Phylogenetic analysis using egl sequences grouped the three strains isolated from greenhouse tomato in LA into phylotype II sequevar 7 with the USA strains AW1 and CFBP2047 (also referred to as K60), as well as a strain from Kenya (ICMP7963). The remaining two Louisianan strains (MLI71-15 and MLI75-15) from fields in Livingston parish, grouped with phylotype I strains from the Philippines (Zo4) and Japan (MAFF 301 070) and probably originated from Asia. The presence of Asian strains in LA is not unique to the south-eastern USA. Phylotype I strains were first reported in 2007 in FL causing an outbreak of bacterial wilt in pepper and these strains clustered with diverse hosts in Asia belonging to sequevar 13 (Ji et al., Citation2007). Unfortunately, the sequences of these strains are not yet published in GenBank and we could not include them in our phylogenetic analysis. In this study, the egl sequences of MLI71-15 and MLI75-15 were grouped with Zo4 and MAFF 301070, which belong to sequevar 14 and were isolated from ginger (Xu et al., Citation2009) and tomato (Liu et al., Citation2009), respectively. In addition to the Philippines and Japan, sequevar 14 strains have been reported in China (Xu et al., Citation2009; Liu et al., Citation2017), Guatemala (Sanchez Perez et al., Citation2008; Weibel et al., Citation2016), French Guiana (Deberdt et al., Citation2014), Indonesia (Waki et al., Citation2013) and the USA (Weibel et al., Citation2016). Because this is the first study to characterize RSCC strains in LA, we cannot say with certainty that the phylotype I strains are recent introductions to the state. In this study, five of the 11 strains did not produce an egl amplicon. The inability to amplify egl from species of Ralstonia has been previously reported (Castillo & Greenberg, Citation2007; Lewis Ivey et al., Citation2007). While the absence of egl in Ralstonia can reduce virulence, egl mutants can still cause disease in tomatoes (Roberts et al., Citation1988). Given the complexity of the species complex, further studies to address the apparent absence of egl in some strains and the use of egl alone to identify sequevar is warranted.
A high level of molecular diversity was found amongst the LA strains characterized in this study. Despite the small sample size, we established baseline data for genetic diversity of RSCC in LA and contributed to the scientific knowledge of the national distribution of RSCC on tomato. Following the new nomenclature proposed by Safini et al. (Citation2014) and Prior et al. (Citation2016), strains of R. pseudosolanacearum and R. solanacearum are present in LA production systems. Genetic characterization of RSCC strains currently present in LA is the first step in the development of strain-specific management practices and these data will be used in future studies to screen for strain-specific host resistance. To better determine the origin of RSCC strains from LA, a larger collection of strains and further characterization using more advanced gene analysis tools, such as multilocus sequence analysis, is needed.
Acknowledgements
We thank C. Allen, University of Wisconsin, Madison for kindly providing DNA of representative phylotype and biovar 3 race 2 strains.
Additional information
Funding
References
- Castillo JA, Greenberg JT. 2007. Evolutionary dynamics of Ralstonia solanacearum. Appl Environ Microbiol. 73:1225–1238.
- Champoiseau PG, Jones JB, Allen C. 2009. Ralstonia solanacearum race 3 biovar 2 causes tropical losses and temperate anxieties. Plant Health Prog. doi:10.10.1094/PHP-2009-0313-01-RV
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Deberdt P, Guyot J, Coranson-Beaudu R, Launay J, Noreskal M, Rivière P, Vigné F, Laplace D, Lebreton L, Wicker E. 2014. Diversity of Ralstonia solanacearum in French Guiana expands knowledge of the “emerging ecotype”. Phytopathology. 104:586–596.
- Denny T. 2006. Plant-Associated Bacteria: plant pathogenic Ralstonia species. Dordrecht (The Netherlands): Springer.
- Edgerton CW, Moreland CC 1913. Diseases of the tomato in Louisiana. LSU Agricultural Experiment Station Reports 590.
- Fegan M, Prior P. 2005. Bacterial wilt: the disease and the Ralstonia solanacearum Species Complex. St. Paul (MN): American Phytopathological Society.
- Fegan M, Taghavi M, Sly LI, Hayward AC. 1998. Bacterial wilt disease: molecular and ecological aspects. New York (NY): Springer-Verlag.
- French EB, Gutarra L, Aley P, Elphinstone J. 1995. Culture media for Ralstonia solanacearum isolation, identification and maintenance. Phytopathology. 30:126–130.
- French ER, Aley P, Torres E, Nydegger U. 1993. Diversity of Pseudomonas solanacearum in Peru and Brazil. ACIAR Proceedings. 45:70–77.
- Gaitonde S, Ramesh R. 2014. Genetic characterization of Ralstonia solanacearum infecting eggplant Solanum melongena L. from Goa and Western region of India. Int J Curr Sci. 12:128–139.
- Gillings M, Fahy P. 1994. Bacterial Wilt: the disease and its causative agent, Pseudomonas solanacearum. Wallingford (UK): CAB International.
- Hayward AC. 1964. Characteristics of Pseudomonas solanacearum. J Appl Bacteriol. 27:265–277.
- Hayward AC. 1994. Bacterial Wilt: the disease and its causative agent, Pseudomonas solanacearum. Wallingford (UK): CAB International.
- He LY, Sequeira L, Kelman A. 1983. Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 67:1357–1361.
- Hong JC, Norman DJ, Reed DL, Momol MT, Jones JB. 2012. Diversity among Ralstonia solanacearum strains isolated from the Southeastern United States. Phytopathology. 102:924–936.
- Ji P, Allen C, Sanchez-Perez A, Yao J, Elphinstone JG, Jones JB, Momol MT. 2007. New diversity of Ralstonia solanacearum strains associated with vegetable and ornamental crops in Florida. Plant Dis. 91:195–203.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kelman A. 1954. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology. 44:693–695.
- Klement Z, Goodman RN. 1967. The hypersensitive reaction to infection by bacterial plant pathogens. Annu Rev Phytopathol. 5:17–44.
- Lebeau A, Daunay MC, Frary A, Palloix A, Wang JF, Dintinger J, Chiroleu F, Wicker E, Prior P. 2011. Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology. 101:154–165.
- Lewis Ivey ML, McSpadden Gardner BB, Opina N, Miller SA. 2007. Diversity and geographic distribution of Ralstonia solanacearum from eggplant in the Philippines. Phytopathology. 97:1467–1475.
- Liu Y, Kanda A, Yano K, Kiba A, Hikichi Y, Aino M, Kawaguchi A, Mizoguchi S, Nakaho K, Shiomi H, et al. 2009. Molecular typing of Japanese strains of Ralstonia solanacearum in relation to the ability to induce a hypersensitive reaction in tobacco. J Gen Plant Pathol. 75:369–380.
- Liu Y, Wu D, Liu Q, Zhang S, Tang Y, Jiang G, Li S, Ding W. 2017. The sequevar distribution of Ralstonia solanacearum in tobacco-growing zones of China is structured by elevation. Plant Pathol. 147:541.
- Lopes CA, Quezado-Soares AM, de Melo PE. 1994. Differential resistance of tomato cultigens to biovars I and III of Pseudomonas solanacearum. Plant Dis. 78:1091–1094.
- Madrid Jimenez A, Sidhu JK, Lewis Ivey ML. 2016. Bacterial wilt of tomato (Solanum lycopersicum) in Louisiana is caused by phylotype II, biovar 1 and 3 strains of Ralstonia solanacearum. Plant Dis. 100:1775.
- Mahbou Somo Toukam G, Cellier G, Wicker E, Guilbaud C, Kahane R, Allen C, Prior P. 2009. Broad diversity of Ralstonia solanacearum strains in Cameroon. Plant Dis. 93:1123–1130.
- Opina N, Tavner F, Hollway G, Wang JF, Li TH, Maghirang R, Fegan M, Hayward AC, Krishnapillai V, Hong WF, et al. 1997. A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solancearum (formerly Pseudomonas solancearum). Asia Pac J Mol Biol Biotechnol. 5:19–30.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Poussier S, Prior P, Luisetti J, Hayward C, Fegan M. 2000. Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst Appl Microbiol. 23:479–486.
- Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B, Allen C. 2016. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics. 17:90.
- Prior P, Fegan M. 2005. Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta Hortic. 695:127–136.
- Prior P, Steva H, Cadet P. 1990. Aggressiveness of strains of Pseudomonas solanacearum from the French West Indies (Martinique and Guadeloupe) on tomato. Plant Dis. 74:962–965.
- Roberts DP, Denny TP, Schell MA. 1988. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J Bacteriol. 170:1445–1451.
- Robertson AE, Wechter WP, Denny TP, Fortnum BA, Kluepfel DA. 2004. Relationship between avirulence gene (avrA) diversity in Ralstonia solanacearum and bacterial wilt incidence. Mol Plant-Microbe Interact. 17:1376–1384.
- Safini I, Cleenwerck I, De Vos P, Fegan M, Sly L, Kappler U. 2014. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. IJSEM. 64:3087–3103.
- Sanchez Perez A, Mejia L, Fegan M, Allen C. 2008. Diversity and distribution of Ralstonia solanacearum strains in Guatemala and rare occurrence of tomato fruit infection. Plant Pathol. 57:320–331.
- Stulberg M, Huang Q. 2015. A TaqMan-based multiplex qPCR assay and DNA extraction method for phylotype IIB sequevars 1&2 (select agent) strains of Ralstonia solanacearum. PLoS One. 10:e0139637.
- Subedi N 2015. Characterization and management of Ralstonia solanacearum populations in South Asia [ dissertation]. Columbus (OH): The Ohio State University.
- Sukumaran J, Holder MT. 2010. DendroPy: A Python library for phylogenetic computing. Bioinformatics. 26:1569–1571.
- Waki T, Horita M, Kurose D, Mulya K, Tsuchiya K. 2013. Genetic diversity of Zingiberaceae plant isolates of Ralstonia solanacearum in the Asia-Pacific region. Jarq. 47(3):283–294.
- Weibel J, Tran TM, Bocsanczy AM, Daughtrey M, Norman DJ, Mejia L, Allen C. 2016. A Ralstonia solanacearum strain from Guatemala infects diverse flower crops, including new asymptomatic hosts Vinca and Sutera, and causes symptoms in geranium, mandevilla vine, and new host African daisy (Osteospermum). Plant Health Prog. 17:114–121.
- Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Guilbaud C, Fegan, M Prior P. 2007. Ralstonia solanacearum strains from martinique (French West Indies) exhibiting a new pathogenic potential. Appl Environ Microbiol. 73:6790–6801.
- Wicker E, Lefeuvre P, de Cambiaire JC, Lemaire C, Poussier S, Prior P. 2012. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 6:961–974.
- Xu J, Pan ZC, Prior P, Xu JS, Zhang Z, Zhang H, Zhang LQ, He LY, Feng J. 2009. Genetic diversity of Ralstonia solanacearum strains from China. Eur J Plant Pathol. 125:641–653.
- Xue QY, Yin YN, Yang W, Heuer H, Prior P, Guo JH, Smalla K. 2011. Genetic diversity of Ralstonia solanacearum strains from China assessed by PCR-based fingerprints to unravel host plant- and site-dependent distribution patterns. FEMS Microbiol Ecol. 75:507–519.
- Zhang D, Ghislain M, Huamán Z, Golmirzaie A, Hijmans R. 1998. RAPD variation in sweetpotato [Ipomoea batatas (L.) Lam.] cultivars from South America and Papua New Guinea. Genet Resour Crop Evol. 45:271–277.
- Zwickl DJ 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [dissertation]. Austin (TX): The University of Texas at Austin.
