Abstract
Okra (Abelmoschus esculentus (L.) Moench) is an economically important vegetable crop that is widely cultivated in the tropics for its nutritional properties. There are concerns, though, that in China, commercial okra production is at risk from an emerging leaf blight disease, that has been recently observed in Fujian Province. Fungi were isolated from diseased okra leaves collected in 2016 and 89 fungal isolates were tentatively identified as Choanephora cucurbitarum based on cultural and sporangia characters. DNA sequences of the internal transcribed spacer (ITS) region and the D1/D2 region of the large subunit (LSU) of the rRNA gene from two representative isolates (C6 and C7) were 99% identical to those of C. cucurbitarum obtained from NCBI. Phylogenetic analysis based on the ITS sequences showed that C6 and C7 clustered with C. cucurbitarum. All 89 fungal isolates were found to be pathogenic to okra leaves. We provide the first confirmation that, based on morphological, pathogenicity and phylogenetic analyses, the causal agent of leaf blight of okra in China was C. cucurbitarum.
Résumé
L’okra (Abelmoschus esculentus [L.] Moench) est une importante culture légumière sur le plan économique et nutritionnel, produite à grande échelle sous les tropiques. On craint par contre qu’en Chine la production commerciale soit menacée par l’émergence d’une brûlure helminthosporienne qui a, depuis peu, été observée dans la province du Fujian.Des champignons ont été isolés à partir de feuilles d’okra infectées, collectées en 2016, et, en se basant sur les caractères culturaux et sur ceux des sporanges, 89 isolats fongiques ont été provisoirement identifiés en tant que Choanephora cucurbitarum. Les séquences de la région de l’espaceur transcrit interne (ITS) de l’ADN et de la région D1/D2 de la grande sous-unité du gène de l’ARNr de deux isolats représentatifs (C6 et C7) étaient identiques à 99% à ceux de C. cucurbitarum obtenus du NCBI. L’analyse phylogénétique basée sur les séquences de l’ITS a montré que C6 et C7 formaient une grappe avec C. cucurbitarum. Les 89 isolats se sont avérés pathogènes à l’égard des feuilles d’okra. Nous sommes les premiers à confirmer que, en se basant sur les analyses morphologiques, phylogénétiques et de pathogénicité, l’agent causal de la brûlure helminthosporienne chez l’okra en Chine est C. cucurbitarum.
Introduction
Okra (Abelmoschus esculentus (L.) Moench) is a warm, rainy season crop in China that is popular for its medicinal and nutritional properties (Li et al., Citation2016). Under optimum growth conditions, the pods grow rapidly and are ready for harvest in about 60 days after planting (Ray et al., Citation2016). Immature pods (3–7-day-old fresh fruit) may be used to produce a health drink or may be consumed directly (Kumar et al., Citation2013).
Choanephora cucurbitarum (Berk. and Rav.) Thaxt. is a fungal plant pathogen that causes flower rot and leaf blights on a wide range of crops (Johnson et al., Citation2014; Park et al., Citation2015). Warm, humid conditions, especially at 25°C and 70–90% relative humidity (RH), promote infection where mycelium and monosporous sporangiola are produced on lesions (Barnett & Lilly, Citation1955). Water-soaked necrotic lesions on leaves and leaf blight caused by this fungus have been recorded on Withania somnifera (Indian ginseng) and Lablab purpureus (Hyacinth bean) (Das et al., Citation2017). Choanephora curcurbitarum is an emerging pathogen and has been recorded from Egypt and Korea as blossom and pod blight of okra (Hussein & Ziedan, Citation2013) and has caused 15% and 20–30% disease incidence in cowpea and pepper, respectively, in India (Chaudhary & Vishwa, Citation1999).
Leaf blights have caused significant losses in okra crops elsewhere in Fujian and necrotic lesions were observed for the first time on okra leaves in Zhangzhou, Fujian Province, China in June 2016; no lesions were observed on flowers and fruits. Previously, only leaf curl disease has been reported on okra in China (Xie et al., Citation2010). Since okra crops in the region may be at risk from this new disease, the main objective of the study was to isolate and identify the causal agent of this disease on okra, using morphology and molecular analysis, as well as pathogenicity tests.
Materials and methods
Isolate collection
Disease surveys on okra seedlings were conducted from April to August, 2016, in Zhangzhou, Fujian Province, China and in total, about 95 plants (6 plants per field on average from 16 fields) with typical water-soaked necrotic lesions were randomly collected for analysis. Diseased okra leaves from the sampled plants were placed in plastic bags and stored at 28°C for 24 h prior to analysis. The diseased leaves were surface sterilized with 1% NaOCl solution for 1 min, and then rinsed with sterile distilled water three times. The surface sterilized leaves were incubated on potato dextrose agar (PDA) at 28°C in the dark. After 3 days, fungal colonies that grew from the tissues were sub-cultured on freshly prepared PDA, and pure cultures were obtained using single spore isolation (Kirk, Citation1984).
Pathogenicity assay
Pathogenicity tests of all the isolates obtained from the diseased leaves were conducted on attached (in planta) leaves. The isolates were grown on PDA at 28°C in the dark for 13–14 days. The conidia were harvested by flooding the Petri dish with sterile distilled water and scraping the colony surface. The resulting conidial suspension was adjusted to 104 conidial mL−1 with sterile distilled water with the aid of a hemocytometer. Two leaves on each of three sterile distilled water washed healthy okra plants for each fungal isolate were inoculated with 2 μL of the conidial suspension using a pipette (Eppendorf, Germany), while two leaves of three healthy okra plants were inoculated with double distilled water as a control treatment. The okra plants were arranged as a completely randomized design, in a sterilized growth chamber set at 28ºC, 80% RH and a 12 h:12 h light-dark cycle. At 3–4 days after inoculation, the inoculated okra leaves were photographed and some of the leaves were harvested to isolate fungi from developing lesions to determine if these fungi were similar to those used for inoculation, to fulfil Koch’s postulates. Pathogenicity of fungal isolates obtained from inoculated leaves was determined by inoculating healthy leaves. The rest of the leaves were allowed to incubate up to 26 days after inoculation to monitor lesion development.
Isolate characterization
Eighty-nine purified isolates were identified according to the descriptions of Campbell & Johnson (Citation2013). Briefly, the shape and size of sporangia, sporangiola and sporangiospores, as well as size and shape of zygospores were measured using an Olympus BX51 microscope (Olympus Corporation, Tokyo, Japan). Means were calculated from 15–20 structures of each diagnostic character, per isolate.
Mycelial cultures of two isolates, C6 and C7, were grown in submerged liquid PDA culture at 28°C in the dark. After 5 days, the mycelial mats of C6 and C7 were dried between sterile filter papers and ground to a fine powder in liquid nitrogen. Total genomic DNA was extracted using a DNA mini kit (51 304, Qiagen, GmbH, Hilden, Germany) and DNA concentration was quantified and verified using a NanoDrop spectrophotometer and 1% agarose gel electrophoresis, respectively. The extracted genomic DNA samples were diluted using sterilized distilled water to a final concentration of 50 ng μL−1 and stored at −80°C prior to rDNA amplification. PCR amplification of rDNA-ITS was done using ITS1/ITS4 primers (Vaghefi et al., Citation2012), and the D1/D2 region of the large subunit (LSU) of rDNA was amplified using LSU-F(GCGGAGGAAAAGAAAATAACAAT)/LSU-R(GTCATTTAGAGTCATTAAGCCAACA) primers. The LSU primers were designed based on GenBank accession number JN939195. DNA samples were mixed with ITS1/ITS4 primers for the amplification of rDNA-ITS, and DNA samples were mixed with LSU-F/LSU-R primers for the amplification of LSU. The amplification reaction contained 100 ng genomic DNA (2 μL) as template, 5 μL of 10 × PCR buffer (100 mM Tris-HCl pH 8.3, 500 mM KCl and 0.1% gelatin), 1.5 mM MgCl2, 10 mM deoxyribonucleotide triphosphate (dNTPs), 15 μM primers (ITS1 and ITS4 or LSU-F and LSU-R), and 0.75 units of Taq DNA Polymerase (TaKaRa, China) in a total volume of 50 μL. The thermal cycling programme was run on a BIO-RAD PCR System S1000 TM Thermal Cycler using the following conditions: 5 min initial denaturation at 95°C, followed by 30 cycles of 30 s denaturation at 95°C, 30 s annealing at 55°C, 50 s extension at 72°C, and a final 5 min extension at 72°C. The recovered PCR products were purified with a Gel Extraction Kit (DP208, Tiangen, Beijing, China) and cloned into the pMD 19-T simple vector (D104A, Takara, Dalian, China). Sequences were determined by a commercial sequencing service (Lifetech, Shanghai, China).
Phylogenetic analysis
The C6 and C7 ITS rDNA sequences were compared with seven published sequences for Choanephora species, and eight isolates of five different species as outgroups. The C6 and C7 LSU sequences were compared with three published sequences for Choanephora species, and five different species as outgroups. For phylogenetic reconstruction, all the sequences other than those for C6 and C7 were obtained from NCBI. A neighbour-joining phylogenetic tree was constructed using the maximum composite likelihood method, by using MEGA7 with bootstrapped values of 1000 replicates (Kumar et al., Citation2016). The phylogenetic tree was drawn to scale, with branch lengths in the same units as those of the inferred evolutionary distances.
Results
Fungal symptoms
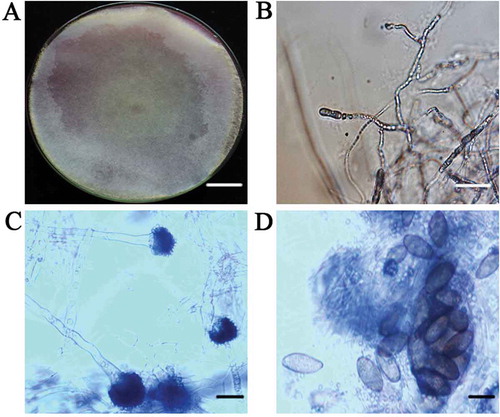
Approximately 5–10% of the plants in each field inspected had leaf blight symptoms, and 89 isolates were obtained from the 95 diseased leaf samples. All 89 isolates were similar in morphology. The isolates grew to 90 mm diameter within 3–5 days on PDA and had abundant aerial mycelia that secreted a scarlet pigment (). The 35–78 μm diameter sporangia possessed columellae and were brown-black, sub-globose and asperate (). The broadly ellipsoid to ovoid, non-branching, aseptate sporangiophores measured 8–15 μm × 18–29 μm, and were brown to pale brown, smooth-walled, and hyaline with nodding sporangia (). We found no evidence of zygospores. From the morphological characteristics, the fungal cultures were tentatively identified as C. cucurbitarum (Campbell & Johnson, Citation2013). Isolates C6 and C7, which were used for the molecular identification, had the same morphological characteristics, but were isolated from different fields.
Pathogenicity
Symptoms first appeared on inoculated leaves at 4 days after inoculation as small, 2–5 mm diameter, water-soaked necrotic lesions (). The necrotic area increased to encompass the whole leaf within 18–26 days after inoculation (). There was no evidence of lesions on the control leaves. Similar results were observed on leaves that were treated with the conidia of the fungi isolated from the inoculated leaves.
Fig. 1 (Colour online) (a) Typical necrotic symptom on naturally infected okra leaves. Scale bar = 1 cm. (b) Okra leaf exhibiting necrotic lesion 4 days after inoculation with a conidial suspension of Choanephora cucurbitarum isolate C7. Scale bar = 1 cm.
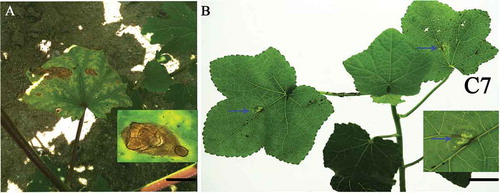
Fig. 2 (Colour online) (a) Purified colony of C. cucurbitarum isolate C7 grown on potato dextrose agar at 28°C with a 12-h photoperiod for 10 days (top of colony). Scale bar = 1 cm. (b) Mycelia of isolate C7. Scale bar = 10 µm. (c) Sporangiophore of isolate C7 bearing sporangiola. Scale bar = 20 µm. (d) Sporangiola of isolate C7. Scale bar = 50 µm.
Molecular identification and phylogenetic analysis
The ITS sequences from C6 and C7 were 615 bp in length and identical. The C6 (GenBank KY080446) and C7 (GenBank KY080447) sequences showed 99% homology to the C. cucurbitarum strain CBS 674.93 isolated from China (GenBank: JN943006). The LSU sequences of isolates C6 (GenBank: MF977740) and C7 (GenBank: MF977741) were 693 bp in size identical, and 99% homologous to those of the C. cucurbitarum strain CBS 674.93 (GenBank: JN939195). Phylogenetic analysis based on the ITS sequences revealed that isolates C6 and C7 clustered with other C. cucurbitarum isolates, and also formed their own separate clade within the C. cucurbitarum isolate sequence (). Phylogenetic analysis based on the LSU sequences revealed that isolates C6 and C7 clustered with the other C. cucurbitarum isolate (), and also formed their own separate clade within the C. cucurbitarum isolate sequence, similar to the sequence data for the ITS.
Discussion
The fungus causing leaf blight of okra in China was identified as C. cucurbitarum based on our morphological, pathogenicity and phylogenetic analysis. Choanephora cucurbitarum has previously been reported to be a causal pathogen for fruit and flower rots and leaf blights in a range of crops, including pepper, millet, squash, rice, pumpkin, pea, sorghum and bean (Hussein & Ziedan, Citation2013; Johnson et al., Citation2014), but there is no report of okra leaf blight of okra caused by C. cucurbitarum. Previous studies have suggested that Choanephora is a weak pathogen that mostly affects flowers and young fruits and occasionally leaves. The fungus also affects spent flowers and other senescent plant material (Park et al., Citation2015). Choanephora leaf blight has been observed in a number of crops, including Hyacinth bean in West Bengal, India, with a disease incidence of 5–20% (Das et al., Citation2017), soybeans in Louisiana, USA (Holcomb, Citation2003) and amaranth (Amaranthus spp.), cucumber (Cucumis sativus) and cowpea (Vigna unguiculata) (Johnson et al., Citation2014). In China, the first report of C. cucurbitarum was from peas (Pisum sativum) and cucumber (Chang & Yang, Citation1984), and it has since spread to other crops, such as Moringa oleifera, causing significant yield losses (He et al., Citation2017).
Molecular identification based on the ITS and LSU regions showed that there was a 99% sequence similarity among isolates C6, C7 and C. cucurbitarum (CBS 674.93) from NCBI. Phylogenetic analysis based on the ITS sequences showed that C6, C7 and CBS 674.93 (JN943006) were clustered together (), and C6, C7 and CBS 674.93 (China) were clustered together based on the LSU (). However, C6 and C7 did form their own separate clade within the C. cucurbitarum cluster. Previously, C. cucurbitarum was reported to cause leaf and stem disease of ice plant (Mesembryanthemum crystallinum), and flower and fruit disease of okra (Park et al., Citation2015; Das et al., Citation2017). In our study, we only observed lesions on leaves of naturally infected plants, with no lesions observed on flowers and fruits.
Fig. 3 Phylogenetic tree constructed by the neighbour-joining method comparing the ITS region of Choanephora cucurbitarum isolates C6 and C7 with other sequences of isolates of 3 Choanephora species and 8 isolates of 5 species used as outgroups. GenBank accession numbers are shown next to each species. The bar indicates nucleotide substitutions per site. Numbers of bootstrap support values ≥ 50% based on 1000 replicates.
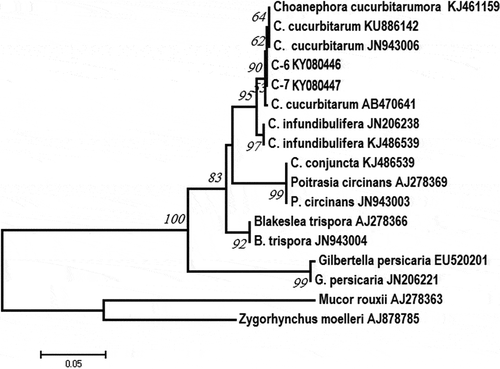
Fig. 4 Phylogenetic tree constructed by the neighbour-joining method comparing the LSU region of isolates C6 and C7 with sequences of 3 isolates of 2 Choanephora species and 6 isolates of 5 species used as outgroups. GenBank accession numbers are shown next to each species. The bar indicates nucleotide substitutions per site. Numbers of bootstrap support values ≥ 50% based on 1000 replicates.
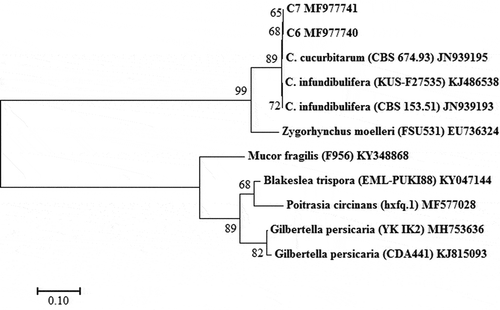
Choanephora includes only two species (i.e. C. cucurbitarum and C. infundibulifera (C. infundibuliferaf. infundibulifera)), which belong to the family Choanephoraceae, Order Mucorales and Phylum Zygomycota, according to Kirk’s taxonomic criteria (Kirk, Citation1984). Previous studies have shown the two species can cause leaf necrosis (Park et al., Citation2015; Das et al., Citation2017; Siddhartha et al., Citation2017), but they can be distinguished by the shape and the presence of striations on sporangiolum (Kirk, Citation1984). In our study, phylogenetic analysis based on the LSU and ITS sequences revealed that C. infundiburifera is a closely related species of C. cucurbitarum, but the C. infundiburifera sequences (JN206238, KJ486539, KJ486538 and JN939193) did not cluster with the C. cucurbitarum sequences (, ).
This is the first evidence of C. cucurbitarum causing leaf blight on okra in China, to our knowledge. Further studies are in progress in okra nurseries to monitor the epidemic and severity of leaf blight caused by C. cucurbitarum. In addition, disease control strategies, including the use of fungicides, are being evaluated. Results of this research will help to better understand diseases threatening C. cucurbitarum susceptible crops and develop effective control strategies to improve crop production.
Acknowledgements
This work was supported by the Natural Science Foundation of Fujian Province (Grant No. 2018J01043); Provincial Public Project in Fujian Province (Grant No. 2018R1024-3); Science and Technology Innovation Foundation of FAAS (20170524); Hundred Young Talents of FAAS (Grant No. YC2015-3).
Additional information
Funding
References
- Barnett HL, Lilly VG. 1955. The effects of humidity, temperature and carbon dioxide on sporulation of Choanephora cucurbitarum. Mycologia. 47:26–29.
- Campbell CK, Johnson EM. 2013. Identification of pathogenic fungi. Hoboken, NJ: John Wiley & Sons.
- Chang CW, Yang HC. 1984. Zygospore formation of Choanephora cucurbitarum [isolated from Pisum sativum and Cucumis sativus]. Trans Mycol Soc Japan. 25:67–74.
- Chaudhary RG, Vishwa D. 1999. A note on the seedling blight of pigeonpea caused by Choanephora cucurbitarum (Berk. and Rav.) Thaxt. Ann Plant Prot Sci. 7:107–109.
- Das S, Dutta S, Mandal B. 2017. First report of Choanephora cucurbitarum, causing leaf blight of hyacinth bean in India. J Plant Pathol. 99:541.
- He Y, Chen X, Huang M, Gao X, Zhang Z, Zhang C, Zhang Y, Li F. 2017. First report of seed pod rot of Moringa oleifera caused by Choanephora cucurbitarum in China. Plant Dis. 101:1824.
- Holcomb GE. 2003. First report of petunia blight caused by Choanephora cucurbitarum in the United States. Plant Dis. 87:751.
- Hussein E, Ziedan E. 2013. First report of pod blight of okra caused by Choanephora cucurbitarum in Egypt. J Agri Tech. 9:135–140.
- Johnson UE, Ishoro AP, Effiong US, Aniediabasi M, Ntui OE, Johnson UI. 2014. Determination of pathogenicity of Choanephora cucurbitarum (Berkeley and Ravenel) Thaxt, amongst commonly cultivated vegetables in Calabar, Cross River state, Nigeria. J Phytopathol. 3:55–61.
- Kirk PM. 1984. A monograph of the Choanephoraceae. Mycological Papers. 152:1–61.
- Kumar DS, Tony DE, Kumar AP, Kumar KA, Rao DBS, Nadendla R. 2013. A review on: Abelmoschus Esculentus (Okra). Int Res J Pharm App Sci. 3:129–132.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li BJ, Guo MY, Chai AL. 2016. First report of Fusarium solani causing fusarium root rot on okra (Abelmoschus esculentus) in China. Plant Dis. 100:526.
- Park JH, Cho SE, Choi IY, Shin HD. 2015. First report of Choanephora rot of Okra caused by Choanephora cucurbitarum in Korea. J Phytopathol. 163:503–506.
- Ray S, Saha SK, Raychaudhuri U, Chakraborty R. 2016. Preparation of okra-incorporated dhokla and subsequent analysis of nutrition, antioxidant, color, moisture and sensory profile. J Food Meas Charact. 11:639–650.
- Siddhartha D, Subrata D, Bholanath M. 2017. First report on blossom and leaf blight of aubergine (Solanum melongena L.) caused by Choanephora infundibulifera (Currey) Sacc., in India. J Mycol Plant Pathol. 47:69–73.
- Vaghefi N, Pethybridge SJ, Ford R, Nicolas ME, Crous PW, Taylor PW. 2012. Stagonosporopsis spp. associated with ray blight disease of Asteraceae. Australas Plant Pathol. 41:675–686.
- Xie K, Cai JH, Hu DM, Wei X, Jia Q, Qin BX, Liu YL. 2010. First report of okra leaf curl disease in China. J Plant Pathol. 92:S109.
