Abstract
In April 2018, symptoms of phyllody, witches’ broom and little leaf were observed on eggplant on a farm located in Huiyang district, Guangdong province, China. Total DNA was extracted from the leaves of symptomatic plants and subjected to molecular detection using the phytoplasma universal primers R16mF2/mR1, P1/P7 and SecAfor1/SecArev3, which showed PCR products of the expected 1.4 kb, 1.8 kb and 0.8 kb fragments, respectively. The 16S rDNA sequence showed the highest similarity (100%) with ‘Celosia argentea’ phytoplasma strain Ca1 (GenBank accession KX426374), Crotalaria witches’ broom phytoplasma strain CrWB-Hnsy1 (GenBank accession EU650181) and pear decline phytoplasma (Taiwan II) isolate Heping (GenBank accession EF193157) from the 16SrII group. Phylogenetic analyses based on 16S rDNA and secA gene sequences confirmed the closer relationship of the eggplant phyllody phytoplasma (EPP) strain with the 16SrII-D subgroup. Virtual restriction fragment length polymorphism (RFLP) analysis of the 16S rDNA sequence indicated the eggplant phyllody phytoplasma belongs to subgroup D within the 16SrII group (16SrII-D). This is the first report of eggplant phyllody phytoplasma in China.
Résumé
En avril 2018, des plants d’aubergines affichant des symptômes de la phyllodie, du balai de sorcière et de la maladie de la petite feuille ont été collectées dans une ferme du district de Huiyang de la province du Guangdong, en Chine. L’ADN total extrait des feuilles des plants symptomatiques a été soumis à la détection moléculaire à l’aide des amorces universelles de phytoplasmes R16mF2/mR1, P1/P7 et SecAfor1/SecArev3, ce qui a engendré des produits de la taille attendue, soit des segments de 1,4 kb, 1,8 kb et 0,8 kb, respectivement. La séquence 16S de l’ADNr a affiché la plus forte similarité (100%) avec la souche Ca1 du phytoplasme de ‘Celosia argentea’ (obtention KX426374 de la GenBank), la souche CrWB-Hnsy1 du phytoplasme du balai de sorcière de la crotalaire (obtention EU650181 de la GenBank) et l’isolat du district de Heping du phytoplasme du dépérissement du poirier (Taiwan II) (obtention EF193157 de la GenBank) du groupe 16SrII. Les analyses phylogénétiques basées sur les séquences des gènes 16S de l’ADNr et secA ont confirmé la très étroite relation entre la souche du phytoplasme de la phyllodie de l’aubergine et le sous-groupe 16SrII-D. L’analyse virtuelle du polymorphisme de longueur des fragments de restriction (RFLP) de la séquence 16S de l’ADNr a indiqué que le phytoplasme de la phyllodie de l’aubergine appartenait au sous-groupe D du groupe 16SrII (16SrII-D). Il s’agit de la première mention du phytoplasme de la phyllodie de l’aubergine en Chine.
Introduction
Phytoplasmas are cell-wall-less prokaryotes that parasitize both plants and insects, with variable sizes ranging from 200–800 nm and a 680–1600 kb genome size (Bertaccini et al., Citation2014). Phytoplasmas can infect a wide range of plant species, including vegetables, fruit trees, ornamental plants, timber trees and shade trees (Liefting et al., Citation2004; Bertaccini et al., Citation2014). Phytoplasma diseases have been reported to be transmitted by grafting, leafhopper vectors and dodder (Weintraub & Beanland, Citation2006; Foissac & Wilson, Citation2010; Amaral Mello et al., Citation2011; Tohidi et al., Citation2015). Typical symptoms include witches’ broom, phyllody, virescence, little leaf, abnormal internode elongation and sterility. Previously, identification of phytoplasmas was based on indirect evidence such as electron microscopic observations, tetracycline treatment (Ishiie et al., Citation1967) and dodder transmission (Bertaccini, Citation2007; Hogenhout et al., Citation2008; Bertaccini et al., Citation2014). Over the last 20 years, PCR and nested-PCR amplification and sequencing of ribosomal rDNA and other housekeeping genes have provided direct identification of phytoplasmas (Lee et al., Citation1994, Citation1995).
Eggplant is an important vegetable crop cultivated worldwide. As the biggest producer and consumer of eggplants, China planted nearly one million hectares in 2017. Like many other vegetables, eggplant is threatened by abiotic and biotic stresses, and phytoplasma-associated diseases can cause yield losses of up to 40% (Mitra, Citation1993; Rao et al., Citation2011). Eggplants infected by phytoplasma show symptoms that include little leaf, witches’ broom, flower virescence, phyllody, giant calyxes and big bud (Rao et al., Citation2011; Rao & Kumar, Citation2017).
Phytoplasma-associated diseases have become a potential threat to eggplant cultivation worldwide. These diseases have been reported in Australia (Davis et al., Citation1997; Martin, Citation2010), Bangladesh (Siddique & Alam, Citation2001; Kelly et al., Citation2009), Japan (Okuda et al., Citation1997), India (Kumar et al., Citation2012, p. 2016), Russia (Ember et al., Citation2011), Iran (Tohidi et al., Citation2015), Oman (Al-Subhi et al., Citation2011), Egypt (Omar & Foissac, Citation2011), Turkey (Sertkaya et al., Citation2007) and Brazil (Boiteux et al., Citation1994; Amaral Mello et al., Citation2011; Rao & Kumar, Citation2017). However, phytoplasma-associated diseases in eggplant have not yet been reported in China. The objective of the present study was to identify the pathogen infecting symptomatic eggplants in China using molecular detection techniques and phylogenetic analyses.
Materials and methods
Sample collection and molecular identification
In April 2018, two eggplants exhibiting phyllody, little leaf and witches’ broom were collected from a farm located in Huiyang district, Guangdong province, China (23°02′35″N, 114°38′10″E) (). Total DNA was extracted from the leaves of two symptomatic and one asymptomatic eggplants using the CTAB (hexadecyltrimethyl-ammonium bromide) method (Doyle & Doyle, Citation1987). Two primer pairs for the 16S-23S ribosomal gene, R16mF2/mR1 (Gundersen & Lee, Citation1996) and P1/P7 (Deng & Hiruki, Citation1991; Smart et al., Citation1996), and an secA gene primer pair, SecAfor1/SecArev3 (Hodgetts et al., Citation2008), were used to perform PCR-based detection. DNA of the asymptomatic eggplant and a sample without a DNA template were used as negative controls. PCR products were then analysed by 1% agarose gel electrophoresis and visualized under UV light (Bio-Rad, USA). PCR products were cloned into pMD19T (Takara, Kusatsu, Japan) and sequenced by Sangon Biotech Co. (Shanghai, China). Virtual RFLP analysis was conducted using iPhyClassifier (http://www.ba.ars.usda.gov/data/mppl/) (Zhao et al., Citation2009) based on in silico digestion of 16S rDNA sequences.
Phylogenetic analysis
Sequence assembly and alignment were performed using DNAMAN software. The 16S rDNA sequences of different groups of phytoplasmas in eggplant were obtained by searching NCBI GenBank using the 16S rDNA sequence of eggplant phyllody phytoplasma strain isolated in China. Phylogenetic trees were constructed with MEGA7 software (Kumar et al., Citation2016b) using the maximum likelihood method based on 16S rDNA or secA gene sequences. To obtain support for the identified phylogenetic relationships, 1000 bootstrap replicates were performed.
Results and discussion
Molecular detection
To identify the pathogen infecting the leaves and flowers of eggplants in China, PCR was performed using phytoplasma 16S rDNA universal primers. PCR products of 1.4 kb and 1.8 kb in length were amplified from the total DNA of leaves from two symptomatic eggplants using the phytoplasma universal primers R16mF2/mR1 and P1/P7, respectively. In contrast, no PCR product was obtained from the negative controls. This indicated only the symptomatic eggplants had been infected by phytoplasma. Moreover, secA gene was also amplified using the specific primer pair SecAfor1/SecArev3. A 0.8-kb product was obtained from total DNA of leaves from the symptomatic eggplants, but no band was observed in the negative controls, confirming the PCR-based assay using 16S rDNA sequences.
Phylogenetic analysis
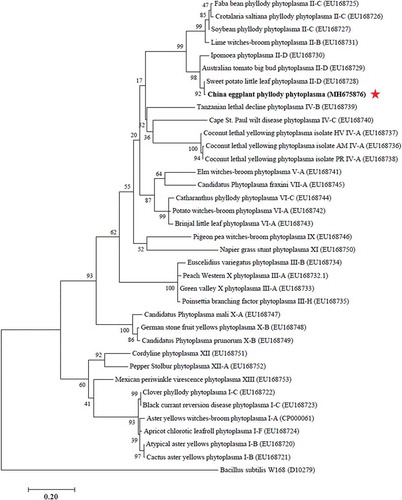
The 16S rDNA and secA gene sequences of EPP strain isolated in China were submitted to NCBI (GenBank accession numbers MH667642 and MH675876, respectively). The phylogenetic tree based on 16S rDNA sequence data show that China EPP clustered into the 16SrII-D subgroup, which includes eggplant big bud (EBB) phytoplasma (JX083377, JX441321, JX083373 and KT259050) found in Iran (Tohidi et al., Citation2015) and India (Kumar et al., Citation2012, Citation2016a), brinjal little leaf (BLL) phytoplasma (KX689253) found in India (Kumar et al., Citation2012), and EPP (HQ423156 and FN257482) found in Oman (Al-Subhi et al., Citation2011) and Egypt (Omar & Foissac, Citation2011) (). The phylogenetic tree based on secA sequences also places China EPP onto the same branch with Sweet potato little leaf phytoplasma (EU168728) within the 16SrII-D subgroup (). These results demonstrated that the EPP strain identified in China belongs to 16SrII-D subgroup of phytoplasma.
Fig. 2. Phylogenetic tree constructed from 16S rDNA sequence data by maximum likelihood. Thirty-eight 16S rDNA sequences deposited in GenBank were used, with Acholeplasma laidlawii used as the outgroup. The numbers on the branches are the bootstrap support obtained from 1000 replicates. The red star indicates the location of China EPP within the phylogenetic tree. BLL, brinjal little leaf. EBB, eggplant big bud. EPP, eggplant phyllody phytoplasma. EGC, eggplant giant calyx.
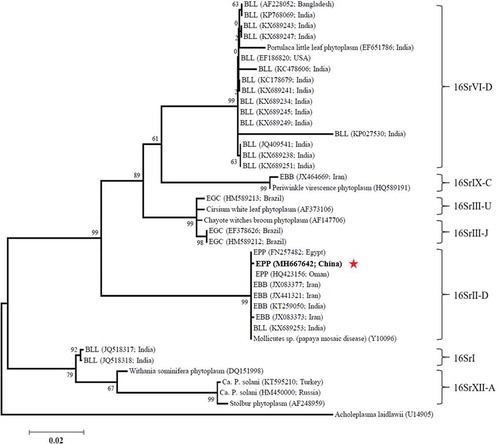
Fig. 3. Phylogenetic tree constructed from secA gene sequence data by maximum likelihood. Thirty-six secA sequences deposited in GenBank were used, with Bacillus subtilis W168 used as the outgroup. The numbers on the branches are the bootstrap support obtained from 1000 replicates. The red star indicates the location of China EPP within the phylogenetic tree.
Virtual RFLP analysis
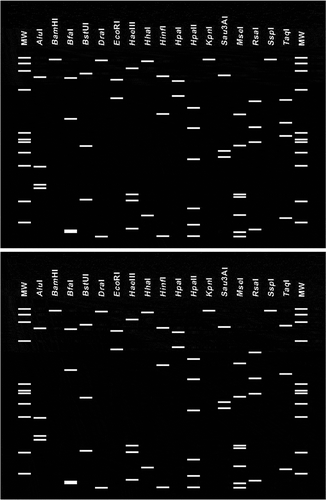
Virtual RFLP analysis based on 16S rDNA sequences indicated that EPP found in China belongs to the 16SrII-D subgroup and shares 99.9% similarity with the 16SrII-D subgroup reference strain (GenBank Y10096). The in silico RFLP profile obtained by iPhyClassifier had a similarity coefficient of 1.00 with the pattern from the 16SrII-D subgroup reference strain (GenBank Y10096) ().
Fig. 4. Virtual RFLP patterns from in silico digestion of the 16S rDNA sequence. The 16S rDNA sequence from Candidatus Phytoplasma australasia (GenBank Y10096) (bottom) was used as the reference strain for the 16SrII-D subgroup. Simulated digestions were performed with 17 endonucleases: AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, MboI, MseI, RsaI, SspI and TaqI. MW indicates the PhiX174DNA-HaeIII marker.
This is the first report of the 16SrII-D subgroup phytoplasma associated with eggplant phyllody in China. However, 16SrII-D subgroup phytoplasma in eggplant has already been reported in India (Yadav et al., Citation2016), Iran (Siampour et al., Citation2013), Oman (Al-Subhi et al., Citation2018) and Egypt (Omar & Foissac, Citation2011). Different groups of phytoplasma in eggplant have been identified worldwide (Rao & Kumar, Citation2017). The 16SrI, 16SrVI-D and 16SrIX-C subgroups are distributed mainly in Asia, including India, Bangladesh, Iran and Japan, while the 16SrIII-U and 16SrIII-J subgroups are only found in Brazil (Amaral Mello et al., Citation2011; Rao & Kumar, Citation2017).
This study extends current knowledge about the distribution of eggplant phytoplasma. Since China is the biggest eggplant producer worldwide, the identification of eggplant phyllody phytoplasma in south China urges growers to be aware of potential phytoplasma outbreaks.
Additional information
Funding
References
- Al-Subhi AM, Al-Saady NA, Khan AJ, Deadman ML. 2011. First report of a group 16SrII phytoplasma associated with witches’-broom of eggplant in Oman. Plant Dis. 95:360.
- Al-Subhi AM, Hogenhout SA, Al-Yahyai RA, Al-Sadi AM. 2018. Detection, identification, and molecular characterization of the 16SrII-D phytoplasmas infecting vegetable and field crops in Oman. Plant Dis. 102:576–588.
- Amaral Mello AP, Eckstein B, Flores D, Kreyci PF, Bedendo IP. 2011. Identification by computer-simulated RFLP of phytoplasmas associated with eggplant giant calyx representative of two subgroups, a lineage of 16SrIII-J and the new subgroup 16SrIII-U. Int J Syst Evol Microbiol. 61:1454–1461.
- Bertaccini A. 2007. Phytoplasmas: diversity, taxonomy, and epidemiology. Front Biosci. 12:673–689. [accessed 2007]. doi:10.2741/2092.
- Bertaccini A, Duduk B, Paltrinieri S, Contaldo N. 2014. Phytoplasmas and phytoplasma diseases: a severe threat to agriculture. Amer J Plant Sci. 05:1763–1788.
- Boiteux LS, Lima MI, Kitajima EW. 1994. Giant calyx: a disease of eggplant (Solanum melongena) associated with a mycoplasma-like organism in Brazil. Plant Pathol. 43:751–754.
- Davis RI, Schneider B, Gibb KS. 1997. Detection and differentiation of phytoplasmas in Australia. Aust J Agric Res. 48:535.
- Deng S, Hiruki C. 1991. Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J Microbiol Meth. 14:53–61.
- Doyle J, Doyle J. 1987. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 19:11–15.
- Ember I, Acs Z, Munyaneza JE, Crosslin JM, Kolber M. 2011. Survey and molecular detection of phytoplasmas associated with potato in Romania and southern Russia. Eur J Plant Pathol. 130:367–377.
- Foissac X, Wilson MR. 2010. Current and possible future distributions of phytoplasma diseases and their vectors. In: Weintraub P, Jones P, editors. Phytoplasmas: genomes, plant hosts, and vectors. Wallingford (UK): CABI; p. 309–324.
- Gundersen DE, Lee I-M. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Mediterr. 35:144–151.
- Hodgetts J, Boonham N, Mumford R, Harrison N, Dickinson M. 2008. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. Int J Syst Evol Microbiol. 58:1826–1837.
- Hogenhout SA, Oshima K, Ammar ED, Kakizawa S, Kingdom HN, Namba S. 2008. Phytoplasmas: bacteria that manipulate plants and insects. Mol Plant Pathol. 9:403–423.
- Ishiie T, Doi Y, Yora K, Asuyama H. 1967. Suppressive effects of antibiotics of tetracycline group on symptom development of mulberry dwarf disease. Jpn J Phytopathol. 33:267–275.
- Kelly PL, Arocha Y, Dider SZ. 2009. First report of a 16SrI, ‘Candidatus Phytoplasma asteris’ isolate affecting eggplant and Mikania sp. in Bangladesh. Plant Pathol. 58:789.
- Kumar J, Gunapati S, Singh S, Lalit A, Nc S, Tuli R. 2012. First report of a ‘Candidatus Phytoplasma asteris’ (16SrI group) associated with little leaf disease of Solanum melongena (brinjal) in India. New Dis Repts. 26:21.
- Kumar M, Katiyar A, Madhupriya RGP. 2016a. First report of association of Potato virus X and Potato virus Y and ‘Candidatus Phytoplasma trifolii’ in brinjal in India. Virus Dis. 27:207–208.
- Kumar S, Stecher G, Tamura K. 2016b. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee I-M, Gundersen DE, Hammond RW, Davis RE. 1994. Use of mycoplasmalike organism (MLO) group-specific oligonucleotide primers for nested-PCR assays to detect mixed-MLO infections in a single host plant. Phytopathology. 84:559–566.
- Lee I-M, Bertaccini A, Vibio M, Gundersen DE. 1995. Detection of multiple phytoplasmas in perennial fruit trees with decline symptoms in Italy. Phytopathology. 85:728–735.
- Liefting LW, Shaw ME, Kirkpatrick BC. 2004. Sequence analysis of two plasmids from the phytoplasma beet leafhopper-transmitted virescence agent. Microbiology. 150:1809–1817.
- Martin H. 2010. Eggplants. In: Persley DM, Cooke T, House S, editors. Diseases of vegetable crops in Australia. Australia: CSIRO Publishing; p. 139–142.
- Mitra DK. 1993. Little leaf, a serious disease of eggplant (Solanum melongena). In: Raychaudhuri SP, Teakle DS, editors. Management of plant diseases caused by fastidious prokaryotes. New Delhi (India): Associated Publishing Co; p. 73–78.
- Okuda S, Prince JP, Davis RE, Dally EL, Lee IM, Mogen B, Kato S. 1997. Two groups of phytoplasmas from Japan distinguished on the basis of amplification and restriction analysis of 16S rDNA. Plant Dis. 81:301–305.
- Omar AF, Foissac X. 2011. Occurrence and incidence of phytoplasmas of the 16SrII-D subgroup on solanaceous and cucurbit crops in Egypt. Eur J Plant Pathol. 133:353–360.
- Rao G, Mall S, Raj S, Snehi S. 2011. Review article: phytoplasma diseases affecting various plant species in India. Acta Phytopathol Entomol Hung. 46:59–99.
- Rao GP, Kumar M. 2017. World status of phytoplasma diseases associated with eggplant. Crop Prot. 96:22–29.
- Sertkaya G, Martini M, Musetti R, Osler R. 2007. Detection and molecular characterization of phytoplasmas infecting sesame and solanaceous crops in Turkey. Bull Insectol. 60:141–142.
- Siampour M, Izadpanah K, Galetto L, Salehi M, Marzachí C. 2013. Molecular characterization, phylogenetic comparison and serological relationship of the Imp protein of several ‘Candidatus Phytoplasma aurantifolia’ strains. Plant Pathol. 62:452–459.
- Siddique A, Alam KR. 2001. Electron microscopy and molecular characterization of phytoplasmas associated with little leaf disease of brinjal (Solanum melongena L.) and Periwinkle (Catharanthus roseus) in Bangladesh. J Phytopathol. 149:237–244.
- Smart CD, Schneider B, Blomquist CL, Guerra LJ, Harrison NA, Ahrens U, Lorenz KH, Seemuller E, Kirkpatrick BC. 1996. Phytoplasma-specific PCR primers based on sequences of the 16S-23S rRNA spacer region. Appl Environ Microbiol. 62:2988–2993.
- Tohidi Z, Salehi M, Ghasemi S, Khanchezar A, Shahamiri SM. 2015. Association of a 16SrIX-C phytoplasma with eggplant phyllody in Iran. Crop Protect. 4:247–256.
- Weintraub PG, Beanland L. 2006. Insect vectors of phytoplasmas. Annu Rev Entomol. 51:91–111.
- Yadav V, Mahadevakumar S, Tejaswini GS, Shilpa N, Sreenivasa MY, Amruthavalli C, Janardhana GR. 2016. First report of 16SrII-D phytoplasma associated with eggplant big bud (Solanum melongena L.) in India. Plant Dis. 100:517.
- Zhao Y, Wei W, Lee IM, Shao J, Suo X, Davis RE. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int J Syst Evol Microbiol. 59:2582–2593.

