Abstract
A total of 15 out of 26 local and imported plant samples from British Columbia nurseries were obtained, and following DNA extraction, PCR was conducted to amplify the fungal ribosomal ITS region which was sequenced using High Throughput Sequencing methods. Bioinformatics analysis of these sequences revealed the presence of Heterobasidion irregulare and H. occidentale. A total of 529 Heterobasidion Illumina DNA sequence reads were found in the 13/15 successfully PCR amplified plant samples, with H. irregulare comprising 86% of the Heterobasidion DNA reads and was more commonly found (13/15 samples) than H. occidentale (10/15 samples). Previously, H. irregulare has been mentioned only once before as being present in BC. The presence of Heterobasidion DNA in nearly all successfully PCR amplified samples from plants grown locally or recently imported, is interpreted as the result of local spore dispersal, likely originating from nearby infected trees. This raises the question whether asymptomatic nursery plants can participate in the spread of H. irregulare in BC and elsewhere in Canada.
Résumé
En tout, 15 échantillons de végétaux locaux et importés de pépinières de Colombie-Britannique sur 26 ont été obtenus et, après en avoir extrait l’ADN, la PCR a été utilisée pour amplifier la région de l’ITS de l’ADN ribosomique fongique, qui a été séquencée par séquençage à haut débit. L’analyse bio-informatique de ces séquences a révélé la présence d’Heterobasidion irregulare et d’H. occidentale. En tout, 529 lectures de séquences d’ADN d’Heterobasidion Illumina ont été trouvées dans les 13 échantillons de végétaux sur 15 amplifiés avec succès par PCR, H. irregulare représentant 86% des lectures d’ADN d’Heterobasidion et étant plus couramment trouvé (13 échantillons sur 15) qu’H. occidentale (10 échantillons sur 15). Jusque-là, H. irregulare avait été recensé qu’une seule fois en Colombie-Britannique. L’occurrence d’ADN d’Heterobasidion dans presque tous les échantillons de végétaux cultivés localement ou récemment importés, amplifiés avec succès par PCR, est interprétée comme résultant d’une dispersion locale de spores qui, probablement, provient d’arbres infectés croissant à proximité. Cela soulève la question de savoir si les végétaux asymptomatiques produits en pépinières peuvent contribuer à la dispersion d’H. irregulare en Colombie-Britannique et ailleurs au Canada.
Introduction
Heterobasidion species are important basidiomycete pathogens of coniferous trees throughout the temperate regions of the world, and they are particularly damaging in plantations (Garbelotto and Gonthier Citation2013). In North America, there are currently two species: Heterobasidion occidentale which is found on the west coast from Mexico, Canada to southern Alaska, extending eastward as far as Colorado (Garbelotto and Gonthier Citation2013), and H. irregulare which is found throughout North America from Mexico-Cuba to the mixed-wood forests of Canada, with the exception of British Columbia.
In Canada, H. irregulare has been reported from red pine (P. resinosa) plantations in southern regions of Ontario (Punter Citation1970) and Quebec (Laflamme and Blais Citation1995), causing high mortality on mature trees left after thinning. Heterobasidion occidentale found only in British Columbia is mainly reported as a butt rot (Morrison and Johnson Citation1999) on Douglas-fir (Pseudotsuga menziesii), western hemlock (Tsuga heterophylla) amabilis fir (Abies amabilis) and Sitka spruce (Picea sitchensis).
Heterobasidion irregulare has also been reported as an introduced invasive species in Italy, associated with movement of infected wood crates by US troops during World War II (Gonthier et al. Citation2004). As an introduced species in Europe, its host range was different from the one observed in North America where it is associated only with pines. In Italy, the spread of H. irregulare is not limited to pine woodlands but also to pure oak forests (Gonthier et al. Citation2012) and its spores are present irrespective of vegetation type. It has higher saprobic capacity, higher fruiting body production and higher sporulation rate (Giordano et al. Citation2014), all factors that could fasten its spread and increase its invasiveness. Due to these factors and its potential threat to European pine trees, it was added to the European and Mediterranean Plant Protection Organization (EPPO) A2 List of pests recommended for regulation as quarantine pests since 2015 (EPPO Citation2017).
Heterobasidion irregulare presence in Italian oak stands seems to be only as a saprobe, producing large amount of sporulating fruiting bodies that lead to further infections. The H. irregulare EPPO (Citation2015a) Mini Data Sheet states ‘The possible role of non-host plants carrying saprophytic populations of the fungus remains to be clarified’. Are there other introduction and dissemination pathways for Heterobasidion species? The EPPO Pest Risk Analysis on H. irregulare (EPPO Citation2015b) rates the role of ‘Plant for Planting’ as a pathway for the spread of this pathogen as ‘low’ for likelihood of entry but ‘high’ for uncertainty associated with the evaluation of this pathway. More information about the possible role on ‘Plant for Planting’ on Heterobasidion dissemination is required.
The objective of this study was to develop a High Throughput Sequencing (HTS) method coupled with a bioinformatics pipeline to detect new invasive forest pathogens from asymptomatic live nursery plant material samples. Here, we describe the discovery of the presence of H. irregulare DNA in British Columbia, along with the presence of H. occidentale on live nursery plants.
Materials and methods
DNA extraction
In 2015, a total of 26 live plants from southwestern British Columbia nurseries were sampled, 11 grown locally and 15 recently imported () and used for this study. Samples, consisting of stem or ramets with leaves, were collected, shipped with icepacks and kept frozen at −20°C at arrival until extractions. Stems and foliage were separated, thin-sliced using a scalpel and ground in a mortar with liquid nitrogen. About 50 mg of stem and foliage tissues were sampled to be ground again using a Christison M3 Mixermill with a tungsten bead, twice for 2 min at 30 hertz without extraction buffer and then once for 1 min at 30 hertz with the extraction buffer. Extraction was done with the Plant DNeasy mini kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions. One microlitre of the eluate was used as genomic DNA (gDNA) template for PCR.
Table 1. Asymptomatic live plant samples collected in 2015 in British Columbia.
Amplicon preparation for illumina sequencing
The eluted DNA (gDNA) was used as template for Polymerase Chain Reaction (PCR). In order to obtain a comprehensive data set of all fungal DNA present in our samples, a High Throughput Sequencing method based on Illumina Miseq sequencing system was used. DNA amplification, primer constructs, purification and sequencing was done as described in Bérubé et al. (Citation2018). Primers ITS1F and ITS7G were used to amplify the ITS regions of the ribosomal DNA fragment (ITS1-5.8S) for metabarcoding of fungi present in the plant samples.
Each of the 15 samples were tagged with differing indexes and then pooled in equimolar amount of 4 ng DNA per sample with DNA from samples of another experiment to fill up an Illumina run. Final quantification of pool, verification of primer artifact removal and amplicon quality check were done with the Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). Pooled DNA samples were sent to the Next-Generation Sequencing Platform, Genomics Centre, CHU de Québec- Université Laval Research Centre, Quebec City, QC, Canada, which performed paired-end 300 bp sequencing using MiSeq Reagent Kit v3 (600-cycles) through an Illumina MiSeq system.
Bioinformatic analysis: villumina reads treatment and clusterization
A bioinformatics treatment of HTS DNA sequences was executed to create Operational Taxonomic Units (OTUs), our proxies for fungal species (Huse et al. Citation2010; Kunin et al. Citation2010). Sequence analysis was done as described in Bérubé et al. (Citation2018). The sequences set was then organized into clusters with USEARCH 64 bit v8.0.1623 with a sequence similarity threshold of 97% to agglomerate reads and form the OTUs, the most abundant sequences type serving as cluster seeds. As no single similarity threshold will accurately reflect the species level throughout the fungal kingdom, a 3% dissimilarity cutoff was selected as a compromise in order to avoid overestimating fungal diversity versus masking rare OTUs and putative new emerging fungal pathogens (Nilsson et al. Citation2008; Huse et al. Citation2010; Schoch et al. Citation2012). Representative sequences, which are the most frequent sequence in each OTU, were extracted and then screened against Genbank nt database using local BLAST. Twenty sequence reads from each OTU of interest were aligned to correct the 0.8% sequencing errors (Quail et al. Citation2012) associated with the Illumina platform and produce consensus sequences used for building a phylogenetic tree with MEGA v.7.0.26 (Tamura et al. Citation2013) as described in Bérubé et al. (Citation2018). Output Excel files were then organized alphabetically by Latin names and then parsed for plant pathogens of interest and those on the quarantine species list of Canada and other industrialized countries.
Results
Fungal DNA from 15 of the 26 plants samples was successfully PCR amplified and their DNA sent for Illumina sequencing. A total of 828,690 clean Illumina DNA reads were obtained after quality control from those 15 plants samples, in a Illumina run containing 10.9 million reads when including all other samples (not from BC) in the run. Cluster analysis, with a sequence similarity threshold of 97% gave 2,430 fungal OTUs, our proxies for fungal species, from 765,144 reads (singletons not counted).
After bioinformatics analysis of DNA sequences was obtained, we found the presence of H. irregulare and H. occidentale DNA. A total of 529 DNA Heterobasidion Illumina sequence reads were found on the 13/15 successfully PCR amplified plant samples (), with H. irregulare comprising 86% of the Heterobasidion DNA reads and more commonly found (13/15 samples) than H. occidentale (10/15 samples). Heterobasidion DNA was found to be present on Rhododendron yakushimanum, Hydrangea macrophylla, Buxus sempervirens, Hydrangea paniculata, Rhododendron ‘PJM Elite’, and Buxus ‘Green velvet’ ( and ). All those plants samples are considered non-host plants for Heterobasidion species.
Table 2. Putative identification of the 20 most common fungi and species of the genus Heterobasidion present on BC nursery plants collections. Number of DNA reads, plant sample number (1–15) and origin (L: locally grown plants, IM: imported plants grown in BC nurseries) are given for 15 plants samples.
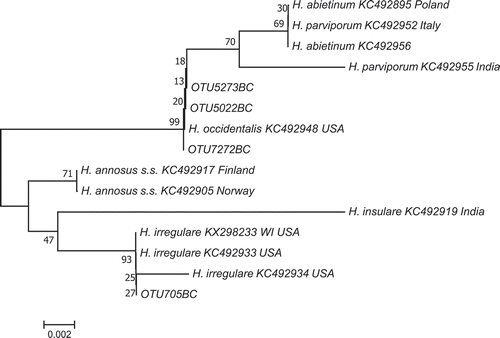
Phylogenetic analysis showed that fungal OTU 705 containing 456 reads clustered () with North American H. irregulare isolates, whereas OTUs 5022, 5273 and 7272, containing respectively 29, 28 and 16 reads, clustered with American H. occidentale. The 97% similarity threshold used to cluster reads often forms multiple OTUs as read length, Illumina-sequencing error and natural sequence variability may add-up, leading to creation of separate OTUs. Once corrected to obtain a consensus sequence, OTUs 5022, 5273 and 7272 had exactly the same sequence. All consensus DNA sequences from this study are deposited in Genbank (accession numbers: OTU705BC, MK188851; OTU5022BC, MK188853; OTU5273BC, MK188854; OTU7272BC, MK188855).
Fig. 1 Phylogenetic relationship among Heterobasidion DNA reads consensus sequence isolated from nursery plants based on Neighbour-joining analysis on the ITS1 partial 5.8s ribosomal sequence data. Bootstrap percentages are indicated at nodes (10,000 replicates), and branch length is proportional to the number of changes indicated. GenBank accession numbers are indicated after species names.
Discussion
Two recent reports (Ginns Citation2017; Shamoun et al. Citation2017) mention H. irregulare on tree stumps in BC, but this species is common in Oregon (Garbelotto and Gonthier Citation2013). Our study is the first to show the presence of Heterobasidion DNA presumably originating from spores on BC nursery plant material. However, we have not cultured isolates from spores or fruiting bodies, or observed symptoms on nursery plants.
The presence of H. irregulare as 86% of the Heterobasidion DNA reads and on nearly all plants samples is surprising as H. occidentale was, until recently, the only species currently known to be present in BC, commonly and widely found in forests. As it is difficult to differentiate those species in the field, the presence of H. irregulare may have gone unnoticed for a long time. In eastern Canada, only H. irregulare is known to exist but H. annosus senso stricto has also been detected recently using HTS methods (Tremblay et al. Citation2018). The H. irregulare spore counts are presently low in Canada (Bérubé et al. Citation2017) and they are expected to rise as number of infected plantation increases. Its newly discovered presence in BC could eventually lead to damage in pine plantations as those observed in eastern Canada.
Heterobasidion DNA presence on nearly all successfully PCR amplified plant samples grown locally or recently imported is interpreted as the result of local spore dispersal originating from nearby infected trees. The origin of the Heterobasidion DNA, whether from spores on the plant leaf surface, or an endophytic life stage in non-coniferous nursery plant material, or if the spores detected are viable, is unknown. Petrini et al. (Citation1990) demonstrated that Spiniger meineckellus (the asexual spore state of Heterobasidion) is present inside fructicose lichen tissues. This raises the question whether asymptomatic nursery plants can participate in the dissemination and spread of H. irregulare in BC and elsewhere in Canada. Plating of surface-sterilized foliage from nursery plants on agar media would answer this question.
It is important to mention that read counts in HTS methods are considered semi-quantitative (Amend et al. Citation2010), without inferring strong quantitative relationships between read count and spore count. Read counts only give an indication of the order of magnitude of spore presence, meaning H. irregulare may not actually be dominant in BC air spore samples. If precise indication of spore load for one specific fungal species is required, quantitative PCR would become the tool of choice. Further tree samplings are needed to locate possibly infected trees and collect fruiting bodies for species identification.
The detection of H. irregulare presence demonstrates that the monitoring of emerging forest pathogens using HTS is an efficient method that is not limited in its scope by the number and availability of DNA probes targeting specific fungal species (Tremblay et al. Citation2018). A typical Illumina run yields thousands of fungal species that informs in depth of potential pathogen species present, but potentially also new pathogens overlooked in surveys that may be identified by their DNA sequence.
References
- Amend AS, Seifert KA, Bruns TD. 2010. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol. 19:5555–5565.
- Bérubé JA, Gagné PN, Ponchart JP, Tremblay ED, Bilodeau GJ. 2018. Detection of Diplodia corticola spores in Ontario and Québec based on HighThroughput Sequencing (HTS) methods. Can J Plant Pathol. In press.
- Bérubé JA, Potvin A, Stewart D. 2017. Importance of local and long-distance Heterobasidion irregulare aerial basidiospore dispersal for future infection centres in thinned red pine plantations in Quebec. For Chron. 93:241–245.
- EPPO. 2015a. Mini data sheet on Heterobasidion irregulare (Panel review date 2015-03). https://gd.eppo.int/taxon/HETEIR/documents.
- EPPO. 2015b. Pest risk analysis for Heterobasidion irregulare. Paris: EPPO. http://www.eppo.int/QUARANTINE/Pest_Risk_Analysis/PRA_intro.htm.
- EPPO. 2017. EPPO A2 list of pests recommended for regulation as quarantine pests (version 2017-09). http://www.eppo.int/QUARANTINE/listA2.htm
- Garbelotto M, Gonthier P. 2013. Biology, epidemiology, and control of Heterobasidion species worldwide. Ann Rev Phytopathol. 51:39–59.
- Ginns J 2017. Polypores of British Columbia. Prov. B.C., Victoria, B.C. Tech. Rep. 104. www.for.gov.bc.ca/hfd/pubs/Docs/Tr/TR104.htm
- Gonthier G, Lione G, Capretti P, Garbelotto M. 2014. The saprobic and fruiting abilities of the exotic forest pathogen Heterobasidion irregulare may explain its invasiveness. Biol Invasions. 16:803–814.
- Gonthier P, Lione G, Giordano L, Garbelotto M. 2012. The American forest pathogen Heterobasidion irregulare colonizes unexpected habitats after its introduction in Italy. Ecol Applications. 22:2135–2143.
- Gonthier P, Warner R, Nicolotti G, Mazzaglia A, Garbelotto M. 2004. Pathogen introduction as a collateral effect of military activity. Mycol Res. 108:468–470.
- Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 12:1889–1898.
- Kunin V, Engelbrekston A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 12:118–123.
- Laflamme G, Blais R. 1995. Détection du Heterobasidion annosum au Québec. Phytoprotection. 76:39–43.
- Morrison DJ, Johnson ALS. 1999. Incidence of Heterobasidion annosum in precommercial thinning stumps in coastal British Columbia. Eur J Forest Pathol. 29:1–16.
- Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. 2008. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinform. 4:193–201.
- Petrini O, Hake U, Dreyfuss MM. 1990. An analysis of fungal communities isolated from fruticose lichens. In: Mycologia. p. 444–451.
- Punter D. 1970. Fomes annosus in eastern Canada. In: Toussoun TA, Bega RV, Nelson PE, editors. Root diseases and soil-borne pathogens. Berkeley: University of California Press; p. 156–170.
- Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific biosciences and illumina MiSeq sequencers. BMC Genomics. 13:341.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Lévesque CA, Chen W, Consortium FB. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci. U.S.A. 109:6241–6246.
- Shamoun SF, Hammett C, Kassatenko I, Sumampong G, Li X. 2017. Development of species-specific PCR primers for rapid detection and identification of North American Heterobasidion species. Can J Plant Pathol. 40:158.doi:https://doi.org/10.1080/07060661.2017.1416041
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evolution. 30:2725–2729.
- Tremblay ED, Duceppe MO, Bérubé JA, Kimoto T, Lemieux C, Bilodeau GJ. 2018. Screening for exotic forest pathogens to increase survey capacity using metagenomics. Phytopathology. 108:1509–1521. doi:10.1094/PHYTO-02-18-0028-R
