Abstract
Outbreaks of light leaf spot (Pyrenopeziza brassicae), a disease new to North America, occurred along with the diseases blackleg (Leptosphaeria spp.) and white leaf spot (Mycosphaerella capsellae) on Brassica crops in western Oregon. Disease symptoms in seed fields planted in the fall of 2013 were observed in the spring of 2014. Field surveys were conducted for disease during 2014 and 2015 to determine the geographic range of light leaf spot, blackleg, and white leaf spot within the Willamette Valley of western Oregon. Infected seed crops included turnip and oilseed rape in Benton, Lane, Linn, Marion, Polk, and Yamhill counties. New protocols that are more sensitive for detection of light leaf spot were developed for the pathogen population in the Pacific Northwest of the USA. Pyrenopeziza brassicae was detected when samples also contained L. maculans or M. capsellae. The test did not react with a range of other fungi evaluated. Sensitivity of the PCR protocol reported is robust at 10 pg of genomic DNA. Standard and real-time PCR screenings developed in this research increase sensitivity for detection of P. brassicae in affected Brassica hosts, which is critical for timely diagnosis and management of this newly-introduced disease in Brassica crops in the Pacific Northwestern region of the USA.
Résumé
Des éclosions de cylindrosporiose (Pyrenopeziza brassicae), une maladie nouvelle en Amérique du Nord, se sont produites parallèlement à la manifestation de la jambe noire (Leptosphaeria spp.) et la tache blanche (Mycosphaerella capsellae) dans les cultures de Brassica dans l’ouest de l’Oregon. Les symptômes de la maladie dans les champs semés à l’automne 2013 sont apparus au printemps 2014. Des études sur la maladie ont été menées en 2014 et 2015 sur le terrain pour déterminer la répartition géographique de la cylindrosporiose, de la jambe noire et de la tache blanche dans la vallée de la Willamette, dans l’ouest de l’Oregon. Dans les comtés de Benton, Lane, Linn, Marion, Polk et Yamhill, les cultures semencières infectées incluaient le navet et le colza oléagineux. De nouveaux protocoles plus sensibles visant la détection de la cylindrosporiose ont été développés en fonction de la population de l’agent pathogène dans le nord-ouest des États-Unis. Pyrenopeziza brassicae a été détecté lorsque les échantillons contenaient aussi L. maculans ou M. capsellae. Le test n’a pas réagi à une gamme d’autres champignons évalués. La sensibilité rapportée du protocole de la PCR est robuste à 10 pg d’ADN génomique. Les criblages par PCR standard et en temps réel développés pour cette recherche ont accru la sensibilité pour la détection de P. brassicae chez les hôtes du genre Brassica, ce qui est crucial pour un diagnostic et une gestion propices de cette maladie nouvellement introduite dans les cultures de Brassica dans le nord-ouest des États-Unis.
Introduction
Light leaf spot caused by Pyrenopeziza brassicae B. Sutton & Rawl. (asexual stage Cylindrosporium concentricum Grev.) (McCartney and Doughty Citation2007; Farr and Rossman Citation2019) had not been previously reported in North America until it was discovered in seed fields of turnip and winter oilseed rape during March 2014 in western Oregon (Ocamb et al. Citation2015). This disease was subsequently detected during 2016 in Washington state on mustard cover crops and the weed, birdsrape mustard (Carmody et al. Citation2016; Carmody Citation2017). The sexual stage of the light leaf spot fungus has not yet been found in the Pacific Northwest of the USA and examinations of the mating types have failed to show successful reproduction, although apothecia of P. brassicae developed when European isolates were crossed in studies conducted by Carmody (Citation2017). The recent light leaf spot outbreaks in western Oregon co-occurred with blackleg (Plenodomus lingam (Tode) Höhn). [syn. Leptosphaeria maculans (Desm.) Ces. & De Not.]; asexual stage formerly Phoma lingam [(Tode: Fr.) (Desm.) (de Gruyter et al. Citation2013; Farr and Rossman Citation2019)] and white leaf spot (Mycosphaerella capsellae A.J. Inman & Sivan.; asexual stage: Pseudocercosporella capsellae (Ellis & Everh.) Deighton) (Fitt et al. Citation2006; Inman and Fitt Citation2007; Koike et al. Citation2007; Rimmer and van der Berg Citation2007).
Light leaf spot is a significant economic problem on oilseed rape in various parts of the world, with seed losses during 2014 estimated at more than £140M ($171 million USD) in the United Kingdom. Losses due to blackleg have historically been economically damaging for producers of oilseed rape in the UK, Australia, and North America (Fitt et al. Citation2006) while white leaf spot has been reported to cause yield losses in Australia on oilseed rape when susceptible varieties are planted (Gunasinghe et al. Citation2016). Light leaf spot losses were estimated to be greater than those due to blackleg in six years out of 10 in UK oilseed rape surveys conducted from 2005 through 2014 (CropMonitor Citation2016). Losses in oilseed rape due to light leaf spot result from stand die-out, reduced pod numbers, premature pod ripening, or an overall growth reduction with less severe infections (McCartney and Doughty Citation2007). Oilseed rape and cauliflower are reported to be more susceptible to light leaf spot than Brussels sprouts or kale (McCartney and Doughty Citation2007).
The pathogens that incite blackleg and white leaf spot are readily discerned when the fungi produce their characteristic anamorphic spore stage on infected plant tissues after incubation in moist chambers at 5°C for 3 to 7 days. Pyrenopeziza brassicae requires a relatively long time to confirm in symptomatic plants – a minimum of 10 days of incubation at 5°C is usually needed before conidia are observed on symptomatic plant samples from western Oregon. Due to the slow growth rate of P. brassicae, the time required to isolate this fungus from infected plant tissues and identify via sequencing can exceed six weeks.
A PCR-based method for the identification of European P. brassicae isolates was developed using primers that targeted unique genomic sequences downstream of mating type genes (Foster et al. Citation1999). Subsequently, this protocol was modified to a two-step, nested PCR to increase sensitivity, improving its utility as a diagnostic tool (Foster et al. Citation2002). An alternative PCR-based detection method developed by Karolewski et al. (Citation2006) that amplified a region in the internal transcribed spacer (ITS) detected P. brassicae in asymptomatic tissue without the requirement for nested PCR. Because the novel outbreak of light leaf spot in the Pacific Northwest in the USA in Brassica crops occurred in fields where blackleg and white leaf spot were occurring on the same plants, a single-step PCR screen was needed for detection and diagnosis of light leaf spot. Moreover, the value of a single-step PCR could be further increased using a real-time PCR assay that could be adapted in the future as a multiplex PCR screen for the other economically important Brassica diseases co-occurring with P. brassicae. Our objectives were to 1) conduct a disease survey in commercial Brassica seed fields to understand the geographic occurrence of light leaf spot, blackleg, and white leaf spot on Brassica crops in western Oregon; and 2) develop new primers for a real-time PCR assay for P. brassicae to use as a diagnostic tool for detecting outbreaks in the Pacific Northwest in the USA.
Materials and methods
Disease surveys in commercial plantings
During the 2013–2014 and 2014–2015 cropping cycles, winter oilseed rape, daikon radish, turnip, and forage rape (Supplementary Table 1) seed fields were surveyed for disease. Disease surveys consisted of examination of plants along a 3-m length of row in each of 10 transects arranged in a V-shaped pattern along one side of each field. Disease incidence and severity were estimated in the field; symptomatic plant samples were collected for disease confirmation in the laboratory. Leaf severity ratings were estimated using the leaf severity models developed by Conn et al. (Citation1990). Disease surveys were conducted in additional Brassica fields, both seed and vegetable crops, in order to determine the extent of the light leaf spot outbreak in western Oregon. These additional fields were typically much smaller in size, ranging from 0.1 to 6 ha in area. Rows of plants were visually assessed along the length on two sides of each of these additional fields; the individual field survey was halted after 5 to 10 plants with suspected light leaf spot were collected for disease confirmation.
Plant samples were incubated in moist chambers at 5ºC in the dark for 2 to 6 weeks in order to induce sporulation. Light leaf spot was identified by the presence of gelatinous masses of hyaline, smooth, thin-walled, aseptate conidia (10 to 16 μm × 3 to 4 μm in size) that formed along lesion margins. Light leaf spot found during 2014 was confirmed in each field by fungal isolation from plant samples and PCR testing of isolated fungi with β-tubulin gene-specific primers (, Glass and Donaldson Citation1995). Blackleg was identified by pycnidia and pinkish to purplish cirrhus containing unicellular, hyaline pycnidiospores (1.2 to 2 μm × 3 to 5 μm in size). White leaf spot was identified via characteristic hyaline, cylindrical, one to five septate conidia (2 to 4 μm × 30 to 90 μm in size) produced in small clusters on the leaf surface.
Table 1. Primers used for detection and confirmation of fungal species.
Development and specificity of the PCR assays
Fungal isolates were collected from infected plant samples from surveys conducted during 2014 and 2015. Pyrenopeziza brassicae isolates were obtained from turnip and weed hosts () and used for PCR development and plant inoculations. A sterile scalpel tip was used to aseptically remove a small portion of the spore mass from sporulating host tissue. Spores were placed in a sterile tube containing 3 mL of sterile reverse-osmosis water. Tubes were vortexed for 10 s, and 100 µL was aseptically spread across a half-strength potato dextrose agar (PDA) plate. After 4 to 7 days of incubation at 25ºC, single, germinating spores were transferred to individual PDA plates to obtain pure cultures of P. brassicae; cultures were grown on malt extract agar (CriterionTM, Hardy Diagnostics, Santa Maria, CA) at 18ºC with no light for 8 weeks and then subsequently stored at 5ºC.
Table 2. Isolates of Pyrenopeziza brassicae, Leptosphaeria maculans, and Mycosphaerella capsellae from Oregon used in PCR development.
Total genomic DNA was obtained from single-spore cultures of isolated fungi. The mycelial mat was harvested from the surface of a culture plate, and 100 mg of mycelial tissue was transferred to Lysing Matrix A (MP Biomedicals) for homogenization in a FastPrep 24 (MP Biomedicals) set to 6.0 m/s for 1 min. Total genomic DNA was isolated from homogenized fungal cells using the DNeasy Plant Minikit (Qiagen) according to manufacturer’s instructions. The identity of fungal isolates of L. maculans and M. capsellae were confirmed via sequencing of the internal transcribed spacer (ITS) region amplified by primers ITS1 and ITS4 (; White et al. Citation1990). However, in P. brassicae this region shares 100% identity with the same region in species of the genus Cadophora, a fungal pathogen of apple and pear; therefore, we also sequenced a region of the β-tubulin gene Bml amplified by primers Bt1a and Bt1b (, Glass and Donaldson Citation1995). PCR amplifications of the ITS region were conducted using the thermocycler protocol described by White et al. (Citation1990), and PCR amplification of the β-tubulin gene Bml was conducted using the thermocycler protocol described by Glass and Donaldson (Citation1995). PCR products were purified using the Qiagen PCR Purification kit and were sequenced at the Center for Genome Research and Biocomputing (Oregon State University, Corvallis). The sequences obtained from the ITS amplicons were specific to the corresponding sequences in L. maculans and M. capsellae. Likewise, the sequence obtained from the Bml amplicon was specific to P. brassicae, and therefore was suitable to confirm the identity of P. brassicae isolates. However, the Bml sequence was not suitable for detecting the fungus in infected plant tissues, as primers Bt1a and Bt1b amplified an ortholog of Bml from the genomic DNA of the host plant.
The gene Pbc1 encodes a unique cutinase that functions as an important virulence factor for P. brassicae (GenBank: AJ009953.1; Davies et al. Citation2000; Li et al. Citation2003). Unlike the ITS region or β-tubulin, which are ubiquitous among fungal plant pathogens, Pbc1 lacks homologs among the majority of non-target organisms, and shares limited homology with other fungal cutinases, making it an ideal target for a single-step diagnostic PCR. We designed P. brassicae-specific primers, OrSU677 and OrSU678, and dual-labelled probe OrSU681 to target Pbc1 in a real-time PCR assay for the detection of P. brassicae in plant tissue (). For the isolation of total genomic DNA from symptomatic plant samples, approximately 100 mg of tissue was homogenized using Lysing Matrix A in the FastPrep 24 set to 6.0 m/s for 1 min. Plant tissue samples included a portion of a lesion as well as surrounding healthy-looking tissue. Total genomic DNA was isolated from the homogenized plant tissue using the DNeasy Plant Minikit. Because many of the Brassica hosts for P. brassicae produce inhibitory metabolites, conventional PCR assays using plant samples were optimized with the inhibitor-resistant 2X Accustart II PCR ToughMix (Quanta Biosciences). In accordance with the manufacturer’s recommendations, PCR reactions were carried out in a 25-µL volume consisting of 12.5 µL 2X Accustart II PCR ToughMix, 200 nM each of the appropriate primers, and 2.0 µL of template DNA (100 ~ 200 ng). Thermocycler protocol for the detection of P. brassicae via conventional PCR consisted of an initial 3-min denaturation step at 95ºC, followed by 35 cycles of 20-s denaturation at 95ºC, 30-s annealing at 54ºC, and 20-s extension at 72ºC, with a final 2-min extension at 72ºC. The real-time PCR assay designed for this study employed the inhibitor-resistant 2X PerfeCTa qPCR ToughMix (Quanta Biosciences).
We tested the efficacy of the PCR assay using the recommended 25-µL reactions consisting of 12.5 µL of the ToughMix master mix, 300 nM each of primers OrSU677 and OrSU678, 200 nM of the real-time probe OrSU681, and 1.0 µL of template DNA. The specificity of the primers and probe to P. brassicae were evaluated using total genomic DNA isolated from single-spore fungal cultures of 13 different fungal species; for these experiments, the final concentration of template DNA used in each reaction was 1 ng. The 13 fungal species examined were P. brassicae, L. maculans, M. capsellae, Penicillium radicum, Diplodia mutila, Phaeoacremonium viticola, Pestalotiopsis cocculi, Phomopsis viticola, Pythium undulatum, Diaporthe eres, Dothiorella sarmentorum, Cytospora cinta, and Alternaria sp. The optimized thermocycler protocol for real-time PCR detection of P. brassicae consisted of an initial 2-min denaturation step at 95ºC followed by 40 cycles of 15-s denaturation at 95°C, 30-s annealing at 58°C, and 15-s extension at 72°C, with a final 2-min extension at 72°C. Changes in fluorescence were measured during the annealing step of each cycle.
The existing primer sets Pb1/Pb2 (Foster et al. Citation1999) and PbITSF/PbITSR (Karolewski et al. Citation2006) () were used in conventional screens of total genomic DNA isolated from pure fungal cultures or from symptomatic plant tissues. The reaction mixes and thermocycler protocols described in Foster et al. (Citation1999) and Karolewski et al. (Citation2006) were optimized for use with the Accustart II PCR ToughMix used in this study. For both assays, PCR reactions were carried out in a 25-µL volume consisting of 12.5 µL 2X Accustart II PCR ToughMix (Quanta Biosciences), 200 nM each of the appropriate primers, and 1.0 µL of template DNA. For the experiments in which total DNA isolated from 13 different fungal species was used as a template, the final concentration of template DNA used was 100 ng per reaction. The optimized thermocycler protocol for both primer sets consisted of an initial 2-minute denaturation step at 95ºC, followed by 40 cycles of 60-s denaturation at 95ºC, 60-s annealing at 58ºC, and 60-s extension at 72ºC, with a final 3-min extension at 72ºC.
Sensitivity of PCR primers in detection of P. brassicae DNA
Tenfold serial dilutions of total genomic DNA prepared from single-spore P. brassicae cultures were used as a template for PCR assays. Four Oregon isolates of P. brassicae were used: two isolates identified as mating type 1 (MAT-1) and two isolates identified as mating type 2 (MAT-2) (Foster et al. Citation2002). Final concentrations of DNA ranged from 50 ng to 5.0 pg of total genomic DNA per 25-µL reaction. The serially diluted genomic DNA was used as a template for real-time PCR analysis with primer/probe set OrSU677/OrSU678/OrSU681, and for separate conventional PCR assays using primer sets Pb1/Pb2 (Foster et al. Citation2002) and PbITSF/PbITSR (Karolewski et al. Citation2006). Three replicates of each PCR assay were conducted for each of the four fungal isolates.
Detection of pathogen in planta using PCR
Seedlings of ‘Baldur’ oilseed rape, ‘Barkant’ turnip, ‘Pak choi’ Chinese cabbage, ‘Golden Acre’ green cabbage, ‘Green sprouting Calabrese’ broccoli, and ‘Southern giant curled’ mustard greens (Supplementary Table 1) were initiated by sowing seeds in moistened Metro-Mix 840 (Sun Gro Horticulture, Agawam, MA) placed in 180-celled (70 mL volume cells) Superblock styrofoam trays (Beaver Plastics, Portland, OR, USA). Seedling trays were placed in an incubator set at 15ºC with a 12-h photoperiod. Cotyledons of approximately 7-day-old seedlings were inoculated with 30-µL drops of spore suspensions of P. brassicae (106 conidia/mL). Symptomatic tissues of inoculated plants were identified based on chlorosis and/or necrosis of the cotyledon and/or first true leaf tissue 10 to 15 days post-inoculation. Symptomatic cotyledons were detached, placed in a moist chamber, and incubated at 5ºC for up to four weeks to induce sporulation. Tissue samples were then placed in sterile microfuge tubes and stored at −20ºC prior to DNA extraction. Total genomic DNA was isolated from symptomatic tissues of oilseed rape, turnip, green cabbage, broccoli, mustard greens, and pak choi. Tenfold serial dilutions were prepared as a template for PCR assays with final concentrations of DNA ranging from 10.0 ng to 10.0 pg of total genomic DNA per 25-µL reaction. Real-time PCR analysis was done using the OrSU677/678/681 primer/probe set, and conventional PCR assays were conducted using the Pb1/Pb2 and PbITSF/PbITSR primer sets.
Results
Disease surveys in commercial plantings
By early June 2014, light leaf spot was detected in 17 crop field sites surveyed in Benton, Linn, Marion, Polk, and Yamhill counties located in the Willamette Valley of western Oregon (). Sites found with light leaf spot were primarily fall-planted commercial seed fields of winter oilseed rape and turnip (), but light leaf spot was also observed on red radish and mizuna grown for seed as well as in Brassica forage and cover crop fields (Supplementary Table 2).
Table 3. Disease occurrence as % plants affected (DI: disease incidence) and range of percentage of leaf area affected on diseased leaves (DS: disease severity) in survey of Brassica seed fields during April-May 2014 and daikon radish during June–July 2014.
Light leaf spot resulted in olive-brown discolouration on the underside of leaves along portions of leaf veins, especially in turnip, prior to development of necrotic leaf spots (). Affected portions of leaves developed into irregular brown lesions, sometimes with cracked centres. Asexual sporulation developed on the margins of lesions but lacked the white concentric rings described for light leaf spot sporulation in Europe and elsewhere (McCartney and Doughty Citation2007). Leaf lesions coalesced and often resulted in premature defoliation. Stem lesions often developed on affected plants and were superficial, elongated brownish streaks with greyish-black margins (). Light leaf spot-affected plants were sometimes severely stunted.
Fig. 1 (Colour online) Light leaf spot in turnip (Brassica rapa subsp. rapa) seed fields resulted in olive-brown discoloration of turnip leaf veins, prior to lesion development as well as of veins surrounding leaf lesions (a). Blackleg (BL) and light leaf spot (LLS) can both occur on the same leaf, as seen on a turnip leaf above (b). Light leaf spot on the lower stem of turnip (c). Blackleg on winter oilseed rape (Brassica napus subsp. oleifera) in western Oregon can result in stunting (≤ 1-meter in height) and canker development along the entire length of the main stem as seen in photograph (d) taken on 21 April 2015, while other plants are in bloom with a more limited canker development at the base of the plant (e).
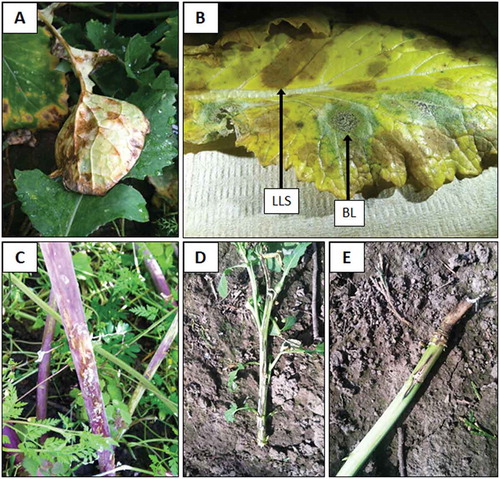
Light leaf spot also was detected during 2014 on volunteer crop plants and weeds, including birdsrape mustard, black mustard, and wild radish (Supplementary Table 2). During the subsequent cropping cycle, light leaf spot symptoms developed during January and February 2015 in oilseed rape and turnip seed fields planted during the previous fall season (). Light leaf spot also was detected on daikon radish seed crops during 2015 (). Additional Brassicaceae species were found with light leaf spot in western Oregon during 2015 and included: cabbage, Chinese cabbage (napa), and forage rape seed fields; bok choi and kale fresh-market vegetable crops; and wild mustard (Supplementary Tables 1 & 2).
Blackleg and white leaf spot co-occurred with light leaf spot on plants in winter oilseed rape, turnip and forage rape seed fields during 2014 and 2015. Some winter oilseed rape fields had blackleg and white leaf spot incidence levels of at least 50% by May 2014; while all the turnip and forage rape seed fields were found to have blackleg, not all of these fields had light or white leaf spot (). Blackleg incidence was severe during the second year survey in winter oilseed rape, turnip and forage rape seed fields, and most turnip seed fields had severe incidences of white leaf spot (). Initial leaf spots caused by L. maculans appeared by mid-October 2014. Oilseed rape appeared very susceptible to L. maculans during 2014 and 2015 in western Oregon, stem cankers greater than 30 cm in length developed on lower stems and were often accompanied by dramatic plant stunting () or a more limited canker developed at the base of the plant (). Turnip storage roots were severely decayed by March 2015, and two fields had 70–80% incidence of plants with severe storage root decay due to L. maculans. It was very common to find individual plants with both blackleg and light leaf spot (). Other diseases previously observed in surveys of Brassica vegetable seed crops were also encountered and included alternaria black spot (Alternaria brassicae (Berk.) Sacc., A. brassicicola (Schwein.) Wiltshire), downy mildew (Hyaloperonospora parasitica (Pers.) Constant.), botrytis grey mould (Botrytis cinerea Pers.), powdery mildew (Erysiphe polygoni DC.), sclerotinia stem rot (Sclerotinia sclerotiorum (Lib.) de Bary), and white rust (Albugo candida (Pers.) Roussel) (data not shown; Farr and Rossman Citation2019).
Development and specificity of the PCR assays
The real-time PCR assay used in this study was designed to target the virulence factor, Pbc1, which encodes a cutinase that has few homologs in other fungal plant pathogens and very limited homology to other cutinases for which sequence data is available (Davies et al. Citation2000). To evaluate the specificity of the real-time PCR assay to existing conventional PCR assays, we performed a side-by-side comparison of three methods: the real-time PCR targeting Pbc1; the conventional PCR assay targeting the conserved ITS (; Karolewski et al. Citation2006); and the conventional PCR assay that targets a genomic region downstream of the mating type genes PHB1 and PHB2 (; Foster et al. Citation1999). For the real-time PCR method, we used primers OrSU677 and OrSU678 with dual-labelled fluorescent probe OrSU681 to interrogate total genomic DNA isolated from single-spore cultures of P. brassicae, L. maculans, M. capsellae, Penicillium radicum, Diplodia mutila, Phaeoacremonium viticola, Pestalotiopsis cocculi, Phomopsis viticola, Pythium undulatum, Diaporthe eres, Dothiorella sarmentorum, Cytospora cinta, and Alternaria sp. For the conventional PCR methods, the same DNA was examined using primer set Pb1/Pb2 (Foster et al. Citation1999) or PbITSF/PbITSR (Karolewski et al. Citation2006). In these experiments, the performance of the real-time PCR assay was comparable to that of the conventional PCR assays (data not shown). Pyrenopeziza brassicae was detected by all three PCR methods; moreover, no other fungal species elicited a positive result from any of the three methods. Visualization of the real-time PCR products via agarose gel electrophoresis confirmed that the amplicon detected in the P. brassicae reaction corresponded to the expected size of 216 base pairs, and no off-target amplification was observed in any of the other 12 fungal species examined.
Sensitivity of real-time PCR primers in detection of Pyrenopeziza brassicae DNA
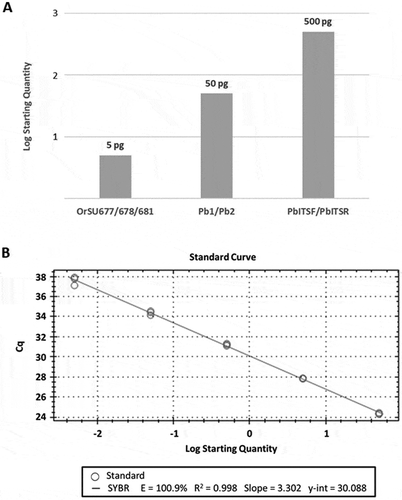
We evaluated the sensitivity of the real-time PCR assay using total genomic DNA isolated from pure cultures of P. brassicae. Two isolates of each of the P. brassicae mating types, MAT-1 and MAT-2, were used for this experiment. Since conventional PCR is routinely used to screen for P. brassicae in Europe, we conducted a side-by-side experiment comparing our real-time PCR assay to the conventional PCR assays developed by Foster et al. (Citation1999) and Karolewski et al. (Citation2006). All three of the PCR screens employed in this study were robust in detecting as little as 500 pg of P. brassicae DNA; in addition, both the real-time PCR assay designed for this study and the conventional PCR assay designed by Foster et al. (Citation1999) detected P. brassicae in PCR reactions using as little as 50 pg of DNA (). However, only the real-time assay consistently detected P. brassicae in 5 pg of genomic DNA isolated from pure culture of P. brassicae ().
Fig. 2 Sensitivity of Pyrenopeziza brassicae-specific primers in detecting Oregon isolates. (a) Pathogen-specific primers () were used to screen 10-fold serial dilutions of total genomic DNA isolated from pure cultures of P. brassicae via conventional PCR (Pb1/Pb2 and PbITSF/PbITSR) or real-time PCR (OrSU677/OrSU678). The values shown indicate the minimum amount of P. brassicae genomic DNA required for detection of the pathogen by each method. This experiment was conducted using genomic DNA isolated from multiple isolates of both mating types of P. brassicae (MAT-1 and MAT-2) with identical results for both mating types over three replicates. (b) A series of 10-fold dilutions wereused to generate a standard curve via real-time PCR. Primers OrSU677/OrSU678 were used to screen total genomic DNA isolated from pure cultures of P. brassicae. Quantities shown range from 5 pg to 50 ng of total genomic DNA.
Detection of pathogen in planta using PCR
To evaluate the utility of our real-time PCR assay as a diagnostic tool, it was necessary to determine its sensitivity in planta. Moreover, it was also desirable to determine the efficacy of the real-time assay as compared to conventional PCR assays already reported. For this purpose, we isolated total genomic DNA from symptomatic plant tissue as a template for both real-time and conventional PCR. The symptomatic tissue samples used in these experiments were collected from green cabbage, oilseed rape, turnip, broccoli, mustard greens, and pak choi. Tenfold dilutions of the template DNA were used, such that the final concentrations of DNA used in the assay ranged from 10 ng to 10 pg of total genomic DNA per reaction.
All three of the PCR assays used in this experiment detected P. brassicae in green cabbage, oilseed rape, turnip, broccoli, and pak choi; however, the real-time PCR assay and the conventional PCR assay using primers Pb1/Pb2 also detected P. brassicae in the mustard greens samples (). Furthermore, only the real-time PCR assay detected the pathogen in all 20 of the samples tested. By comparison, PCR with Pb1/Pb2 yielded positive results for 15 of the 20 samples tested, while the primer set PbITSF/PbITSR detected P. brassicae in 12 samples (). Of the 15 samples that tested positive for P. brassicae by conventional PCR with Pb1/Pb2, eight of the samples were detected by real-time PCR with 10-fold less template DNA. Similarly, of the 12 samples that screened positive for P. brassicae DNA with PbITSF/PbITSR, seven samples were detected by real-time PCR with 10-fold less DNA. In all cases, the real-time assay was at least as sensitive as conventional PCR assays, and in 12 of 20 samples tested, the sensitivity of the real-time PCR assay exceeded that of the conventional PCR assays used in this experiment.
Table 4. Detection thresholds using total genomic DNA from symptomatic plant tissue in a real-time PCR assay compared to conventional PCR primers for identification of Pyrenopeziza brassicae.
Discussion
The presence of light leaf spot in western Oregon had not been reported in North America prior to the outbreaks discovered in western Oregon during 2014. Light leaf spot was detected in five counties in western Oregon during 2014 and in six counties during 2015. Survey of commercial fields showed that light leaf spot commonly occurred alongside blackleg and white leaf spot and some fields had a high percentage of diseased plants. Blackleg, light leaf spot, and white leaf spot were not observed in any of the disease surveys of vegetable seed crops conducted in the Willamette Valley during 2010 through 2013, but blackleg was observed at low levels in western Oregon in one oilseed rape field during 2008 (C. Ocamb, unpublished). The Oregon Department of Agriculture detected blackleg at low levels in specialty vegetable seed fields that underwent voluntary disease certifications between 2009 and 2013 (N. Osterbauer, pers. comm.).
The winter oilseed rape cultivars, ‘Baldur’ and ‘Sitro’, were highly susceptible to P. brassicae as well as to strains of Leptosphaeria present in western Oregon. Seed crops of the turnip cultivars, ‘Barkant’ and ‘Purple Top White Globe’, were also highly susceptible to the fungi that incite blackleg, light leaf spot, and white leaf spot in western Oregon. It appears that numerous other Brassica and radish crops, as well as weeds present in western Oregon, are susceptible to P. brassicae (Claassen Citation2016).
Windblown ascospores of the fungi that cause blackleg and white leaf spot are produced by infected crop residues on the soil surface (Inman and Fitt Citation2007; Rimmer and van der Berg Citation2007). Successful recombination between mating types of P. brassicae results in ascospore development in apothecia on infected plant residues and can be wind-dispersed over relatively long distances after a wetting period, while conidia can develop in leaf and stem lesions but spread over relatively short distances by rain or splashing water, potentially causing pod infections in seed crops during the following spring (McCartney and Doughty Citation2007). Light leaf spot was initially observed across five counties in western Oregon in 2014, infecting an array of Brassica seed crops, including some planted with seed sourced from outside of the Pacific Northwest of the USA. Although the teleomorph of the light leaf spot fungus has yet to be observed in western Oregon, it is possible that some level of ascospore development occurs on a portion of infected plant residues that have resulted from widespread disease outbreaks in western Oregon. Because it has been shown that seed can remain infected with P. brassicae for at least one year (Carmody Citation2017), there is also an inherent risk of light leaf spot moving into other regions of North America through the transport of infected seeds.
The co-occurrence of light leaf spot with blackleg and white leaf spot in the Brassica seed fields of Oregon requires a PCR method sensitive and specific enough to detect P. brassicae in a mixed-infection background. The PCR primers developed for this study, OrSU677 and OrSU678, are specific to P. brassicae, as was demonstrated by screening genomic DNA isolated from pure cultures of 12 other fungal pathogens. When used with probe OrSU681 in a real-time PCR assay, detection of P. brassicae was sensitive and robust, even in a background of total genomic DNA from a wide range of Brassica hosts. The detection limit of the real-time PCR assay varied with the severity of infection in the tissue sampled; however, even in those samples with low severity of infection, detection of P. brassicae was accomplished with only 10 ng of genomic DNA from the host tissue. Furthermore, the real-time assay developed for this study provided increased sensitivity for pathogen strains occurring in western Oregon when compared to the conventional PCR screens previously developed for the detection of P. brassicae (Foster et al. Citation1999; Karolewski et al. Citation2006).
The development of a sensitive real-time PCR assay is an important tool for high-throughput diagnosis of light leaf spot for commercial Brassica growers in the Pacific Northwest of the USA. The real-time PCR assay, developed for detection of light leaf spot in Oregon, provides a means for rapid and sensitive diagnosis of disease caused by P. brassicae through testing of fungal isolates as well as in planta. The utility of this assay has been further demonstrated in another large-scale survey evaluating the incidence of light leaf spot in Brassica weeds throughout western Oregon (Claassen Citation2016). Another obvious application of this method is quantitative PCR (qPCR); indeed, the protocol described here is being optimized to develop a multiplex qPCR assay to quantify levels of P. brassicae and other fungal pathogens of brassicas. The development of these molecular tools, used in conjunction with extensive field surveys like the ones in this study, will contribute to our understanding of the emergence of light leaf spot in North America.
Supplemental Table 1 and 2
Download MS Word (32 KB)Acknowledgements
Financial support was provided by Oregon State University, as well as the state of Oregon and the Specialty Seed Growers of Western Oregon. We also thank Melodie Putnam of the OSU Plant Clinic for providing some of the fungal DNA samples used in this study and a review of this manuscript. We thank Drs. Jon West and Kevin King of Rothamstead Research for their assistance with determining P. brassicae mating types. Thanks to Dr. David H. Gent and anonymous reviewers for their suggestions and comments that improved this paper. We thank the growers, crop consultants, and seed companies who provided access to Brassica plantings.
Supplementary material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2019.1620337
References
- Carmody S, Ocamb C, Du Toit L. 2016. Potential seed transmission of Pyrenopeziza brassicae (light leaf spot) and Mycosphaerella capsellae (white leaf spot) in brassicas in the Pacific Northwest USA. American Phytopathological Society, Pacific Division, 2016 Annual Meeting; La Conner, WA. [accessed 2019 May 31]. https://www.apsnet.org/members/community/divisions/pac/meetings/Pages/default.aspx.
- Carmody SM. 2017. Light leaf spot and white leaf spot of brassicaceae in Washington State [ MS Thesis]. Washington State University.
- Claassen BJ. 2016. Investigations of black leg and light leaf spot on brassicaceae hosts in Oregon [ M.S. Thesis]. Oregon State University.
- Conn KL, Tewari JP, Awasthi RP. 1990. A disease assessment key for Alternaria blackspot in rapeseed and mustard. Can Plant Dis Surv. 70:19–22.
- CropMonitor. 2016. Winter oilseed rape: yield loss estimates 2005-2014. [accessed 2019 Apr 5]. www.cropmonitor.co.uk.
- Davies KA, De Lorono I, Foster SJ, Li D, Johnstone K, Ashby AM. 2000. Evidence for a role of cutinase in pathogenicity of Pyrenopeziza brassicae. Physiol Mol Plant Pathol. 57:63–75.
- de Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW. 2013. Redisposition of phoma-like anamorphs in Pleosporales. Stud Mycol. 75:1–36.
- Farr DF, Rossman AY. 2019. Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA. [accessed 2019 Apr 5]. http://nt.ars-grin.gov/fungaldatabases.
- Fitt BDL, Rimmer SR, Barbetti MJ, Brun H. 2006. World-wide importance of Phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur J Plant Pathol. 114:3–15.
- Foster S, Ashby A, Fitt B. 2002. Improved PCR-based assays for pre-symptomatic diagnosis of light leaf spot and determination of mating type of Pyrenopeziza brassicae on winter oilseed rape. Eur J Plant Pathol. 108:379–383.
- Foster SJ, Singh G, Fitt BDL, Ashby AM. 1999. Development of PCR based diagnostic techniques for the two mating types of Pyrenopeziza brassicae (light leaf spot) on winter oilseed rape (Brassica napus ssp. oleifera). Physiol Mol Plant Pathol. 55:111–119.
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl Environ Microbiol. 61:1323–1330.
- Gunasinghe N, You MP, Li XX, Banga SS, Banga SK, Barbetti MJ. 2016. New host resistances to Pseudocercosporella capsellae and implications for white leaf spot management in Brassicaceae crops. Crop Prot. 86:69–76.
- Inman AJ, Fitt BDL. 2007. White leaf spot. In: Rimmer SR, Shattuck VI, Buchwaldt L, editors. Compendium of Brassica diseases. St. Paul (MN): APS Press; p. 50–54.
- Karolewski Z, Fitt BDL, Latunde-Dada AO, Foster SJ, Todd AD, Downes K, Evans N. 2006. Visual and PCR assessment of light leaf spot (Pyrenopeziza brassicae) on winter oilseed rape (Brassica napus) cultivars. Plant Pathol. 55:387–400.
- Koike ST, Gladders P, Paulus AO. 2007. Vegetable diseases: a color handbook. St. Paul (MN): APS Press.
- Li D, Ashby AM, Johnstone K. 2003. Molecular evidence that the extracellular cutinase Pbc1 is required for pathogenicity of Pyrenopeziza brassicae on oilseed rape. Mol Plant-Microbe Interact. 16:545–552.
- McCartney HA, Doughty KJ. 2007. Light leaf spot. In: Rimmer SR, Shattuck VI, Buchwaldt L, editors. Compendium of Brassica diseases. St. Paul (MN): APS Press; p. 31–35.
- Ocamb CM, Mallory-Smith C, Thomas WJ, Serdani M, Putnam ML. 2015. New and re-emerging fungal pathogens affecting Brassicaceae plants in western Oregon: black leg, light leaf spot, and white leaf spot. Phytopathology. 105:542–P.
- Rimmer SR, van der Berg CGJ. 2007. Black leg (Phoma stem canker). In: Rimmer SR, Shattuck VI, Buchwaldt L, editors. Compendium of Brassica diseases. St. Paul (MN): APS Press; p. 19–22.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A guide to methods and applications. New York (NY): Academic Press; p. 315–322.
