Abstract
Strawberry black spot caused by Alternaria is an important foliar disease. In 2016, strawberry leaves with severe black spot lesions were collected from eight districts of Beijing municipality in China. Molecular and morphological criteria were matched to identify the Alternaria species inciting strawberry black spot in Beijing. A total of 102 Alternaria isolates from diseased strawberry leaves were divided into two species: A. tenuissima (61 isolates, 59.8%) and A. alternata (41 isolates, 40.2%). The detached leaves of strawberry showed typical black spot symptoms when inoculated with spore suspensions of isolates of A. tenuissima and A. alternata, with disease incidence ranging from 20.4–100.0% and disease index ranging from 6.0–29.1, respectively. To our knowledge, this is the first report of A. tenuissima and A. alternata causing black spot of strawberry in Beijing, China.
Résumé
La tavelure du fraisier, causée par Alternaria, est une importante maladie foliaire. En 2016, des feuilles de fraisier comportant de graves lésions ont été collectées dans huit districts de la municipalité de Beijing, en Chine. Des critères moléculaires et morphologiques ont été assortis pour identifier les espèces d’Alternaria provoquant la tavelure à Beijing. Cent deux isolats d’Alternaria issus des feuilles infectées ont été divisés en deux espèces: A. tenuissima (61 isolats, 59.8%) et A. alternata (41 isolats, 40.2%). Après avoir été inoculées avec des suspensions de spores des isolats d’A. tenuissima et d’A. alternata, les feuilles détachées de fraisier affichaient les symptômes typiques de la tavelure, et ce, pour une incidence de la maladie variant de 20.4% à 100.0% et un indice de la maladie variant de 6,0 à 29,1, respectivement. À notre connaissance, il s’agit de la première mention d’A. tenuissima et d’A. alternata causant la tavelure du fraisier à Beijing, en Chine.
Introduction
Strawberry (Fragaria sp.) is a popular and economically valuable fruit, which contains significant levels of antioxidants that act against free radicals. China has become the largest producer and consumer of strawberry worldwide with a cultivation area of about 100 000 ha and a total production of ~2 million tons annually (Zhu et al. Citation2016). Strawberry black spot caused by Alternaria alternata was first recorded in Japan in 1977 (Watanabe and Umekawa Citation1977; Watanabe et al. Citation1978). Foliar symptoms are irregular and often circular lesions, with a dark-brown centre surrounded by light-brown rings and yellow halos. The lesions gradually enlarge and become firm with a dark brown-to-black, velvety surface composed of mycelia, conidiophores and conidia (Cho and Moon Citation1980). Alternaria alternata has also been reported to cause strawberry black spot in Italy (Wada et al. Citation1996; Maltoni et al. Citation2000), Korea (Cho and Moon Citation1980) and Pakistan (Mehmood et al. Citation2018). Alternaria tenuissima has been reported to cause strawberry black spot in Iran (Bagherabadi et al. Citation2015), but is more often reported to cause strawberry fruit rot (Howard and Albregts Citation1973; Lee et al. Citation2001; Ko et al. Citation2008). Alternaria alternata and A. tenuissima are the main pathogenic fungi that cause strawberry black spot across the world.
Information about the population structure of Alternaria species causing strawberry black spot is crucial to better understand the distribution and importance of different species and for designing effective management strategies to control this disease. However, the population structure of Alternaria species associated with strawberry black spot in Beijing municipality, which is an important strawberry cultivating region in China, is unclear. Thus, the aim of this study was to identify the Alternaria species composition causing strawberry black spot in Beijing using morphological and molecular methods.
Materials and methods
Sampling and fungal isolation
Strawberry leaves with black spot symptoms were sampled from 31 fields in eight districts of Beijing in 2016. Fungal isolates were recovered from the diseased leaves using the methods of Zheng et al. (Citation2015). Briefly, a small piece of leaf tissue (about 5 × 5 mm2) was taken from the margin of the healthy and diseased tissues. Surface-disinfection of leaf tissues was performed with 70% ethanol for 30 s, followed by soaking in 0.5% sodium hypochlorite for 3 min, and then rinsing with sterile distilled water three times. The leaf pieces were then incubated in Petri dishes containing potato dextrose agar (PDA) amended with streptomycin sulphate (50 mg L−1) for 7 days at 25°C in the dark. Multiple fungal colonies were obtained from each leaf lesion and only those with different morphologies were selected as individual isolates.
Morphological examination and identification
The fungal isolates obtained were grown on potato dextrose agar (PDA) and potato carrot agar (PCA). To observe colony colour, margin and texture, the isolates were incubated on PDA at 25°C in darkness for 7 days (Andersen et al. Citation2001). To stimulate sporulation, the isolates were incubated at 25°C for 7 days on PCA positioned 40 cm below cool white fluorescent bulbs with 8 h/16 h periods of light/dark, with the illumination intensity at the agar surface being ~4000 lux (Andersen et al. Citation2001; Simmons Citation2007). The sporulation pattern was examined with a dissecting microscope and isolates were categorized to a sporulation group according to Simmons (Citation2007).
DNA extraction and polymerase chain reaction (PCR) amplification
Genomic DNA was extracted from 7-day-old mycelia of 102 isolates grown on PDA at 25°C according to the method described by Stenglein and Balatti (Citation2006). In brief, mycelia were frozen in liquid nitrogen and ground into fine powder, which was mixed with 800 μL of cetyltrimethyl-ammonium bromide (CTAB) extraction buffer [final concentration: 2% (w/v) CTAB, 200 mM Tris-HCl pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl, 1% (w/v) polyvinylpyrrolidone, 1% (v/v) β-mercaptoethanol]. Extraction was done in accordance with the method of Lee and Taylor (Citation1990). Finally, the DNA pellet was dissolved in 30 μL Tris-EDTA buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) and stored at −20°C until further use.
PCR amplification of the internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) of all the fungal isolates was assayed using primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′), according to the method of White et al. (Citation1990). Amplification reactions were performed in a Mastercycler Gradient Thermal Cycler (Eppendorf AG, Hamburg, Germany) with an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 40 s, annealing at 58°C for 40 s, and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. Another set of PCR primers, H3-1a (5′-ACT AAG CAG ACC GCC CGC AGG-3′) and H3-1b (5′-GCG GGC GAG CTG GAT GTC CTT-3′), was used to amplify partial coding sequences of the histone 3 gene (Glass and Donaldson Citation1995). PCR amplification was carried out with an initial denaturation at 96°C for 2 min, followed by 30 cycles of denaturation at 96°C for 15 s, annealing at 55°C for 30 s, and extension at 75°C for 35 s, and a final extension at 72°C for 2 min (Kang et al. Citation2002). The 25 μL PCR reaction mixture was comprised of 10.5 μL ddH2O, 12.5 μL Premix Ex Taq (v. 2.0, TaKaRa; containing 0.625 U Taq DNA polymerase, 200 μM dNTP and 2 mM MgCl2), 0.5 μL each of the two primers (10 μM), and 1 μL DNA template (100 μg mL−1). Negative controls containing the same reagents but without DNA template were included in all PCR reaction sets.
DNA sequencing and phylogenic analysis
The PCR amplification products were purified with an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Hangzhou, China) and ligated to the vector pMD19-T (TaKaRa Biotechnology, Dalian, China) according to the manufacturers’ instructions. The ligation reaction mixture was used to transform high-efficiency competent cells of Escherichia coli MC1022 and the resulting transformants were selected on Luria-Bertani broth (LB) (Thammason et al. Citation2015) agar plates containing ampicillin (50 μg mL−1), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal, 100 μg mL−1), and isopropyl-β-d-thiogalactopyranoside (IPTG, 100 μg mL−1). White colonies with the target DNA insertion, verified by PCR, were sent to Beijing Tianyihuiyuan Biotech Co. Ltd (Beijing, China) for sequencing. Sequence similarity searches were performed using the nucleotide BLAST program at the National Centre for Biotechnology Information (NCBI). Sequence alignment was done using Clustal W (v. 1.83) (Thompson et al. Citation1994) with manual adjustment. Phylogenetic analysis for each of the two sequences (rDNA ITS and histone 3 gene) was carried out with MEGA5 version 5.2.2 (http://www.megasoftware. net/) using the Maximum likelihood method (ML).
Pathogenicity tests
Forty-five isolates were selected and tested for their pathogenicity on strawberry leaflets according to a revised procedure described previously (Wada et al. Citation1996; Pryor and Michailides Citation2002; Zheng et al. Citation2015) (). The experiment was conducted using fully expanded compound leaves from 45-day-old plants of strawberry (cv. ‘Benihoppe’).
Table 1. Numbers of Alternaria isolates collected from diseased strawberry leaves collected in different geographic districts of Beijing, China.
The isolates were incubated on PDA for 7 days in the dark at 25°C. Spore suspensions of the isolates were prepared by flooding the cultures with sterile distilled water, gently scraping the colony surface and collecting the suspension. After filtering through four layers of cheesecloth, the spore suspension was adjusted to 106 spores mL−1 using a hemocytometer.
Thirty detached apical leaflets of the fully expanded leaves were inoculated per isolate and then placed in Petri dishes in five plastic boxes (19 × 14 × 5 cm3; length × width × height) with three leaflets per dish and two dishes per plastic box. A drop of 20 μL spore suspension was inoculated on each side of the main vein of the upper surface of each leaflet (two points per leaflet). For the controls, leaflets were inoculated with sterile distilled water. The plastic boxes were covered to maintain high humidity and incubated in a growth chamber at 25°C and 90% RH for 14 days with a daily 12 h fluorescent light photoperiod. Symptoms on the inoculated leaflets were observed and compared with those occurring on naturally infected leaflets in the field. The experiments were conducted twice. Disease severity (DS) was scored on a 4-point rating system (Pryor and Michailides Citation2002), where: 1 = no lesions, 2 = lesions <1 mm in diameter, 3 = lesions 1–5 mm in diameter and 4 = lesions >5 mm in diameter. A disease index (DI) was calculated according to the following formula: DI = [100 × Σ (n × corresponding DS)]/4N, where n is the number of the inoculation points on the leaflets corresponding to each disease rating, and N is the total number of inoculation points. Statistical significance was determined at P < 0.05, using the least significant difference test.
Results
Isolation and identification of Alternaria species associated with strawberry black spot
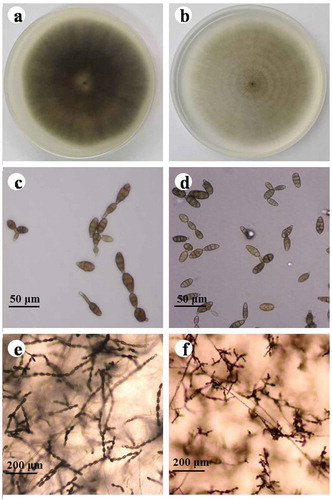
A total of 102 Alternaria isolates were obtained from diseased strawberry leaves (). The colony appearance of 61 (59.8%) isolates was initially greyish-green and eventually became olive brown on PDA. On PCA plates, these isolates formed up to 12 conidia in unbranched conidial chains with one or two lateral branches. Conidiophores were short, and arose singly, measuring 15.2–61.6 μm long and 3.0–6.8 μm thick. The conidia were ovoid to obclavate in shape, ranging from 17.3–49.0 μm in length and 7.2–21.9 μm in width, and had one to five transverse and zero to two longitudinal septa. Based on these features, these isolates were tentatively identified as A. tenuissima (Ness) Wiltshire (,c,e). The colony appearance of 41 (40.2%) isolates incubated on PDA was initially dark grey and then became black-brown with ageing. On PCA plates, these isolates formed 8–12 spores in conidial chains with numerous secondary and occasionally tertiary chains branching from apical and median cells. Conidiophores were single or fasciculate, straight or knee curved with regular septa, measuring 3.7–12.5 μm long and 2.4–6.9 μm thick. Conidia were ovoid, ellipsoid or obpyriform, 15.9–43.5 μm long and 6.9–18.3 μm wide, with 1–5 transverse septa and 0–5 longitudinal septa. These isolates were tentatively identified as A. alternata (Fr.) Keissl (,d,f). Alternaria tenuissima and A. alternata were isolated from Yanqing, Changping, Fangshan, Shunyi and Tongzhou districts. However, in Miyun and Huairou districts, only A. tenuissima was obtained; and in Daxing district, only A. alternata was found ().
Fig. 1 (Colour online) Morphology of Alternaria tenuissima and A. alternata after 7 days of growth on PDA or PCA. a–b, Colonies of isolates representing A. tenuissima and A. alternata on PDA, scale bars: 40 μm; c–d, Conidia of isolates representing A. tenuissima and A. alternata on PCA, scale bars: 40 μm; e–f, sporulation patterns of isolates representing A. tenuissima and A. alternata on PCA. Scale bars: 200 μm.
DNA sequencing and phylogenetic analysis
The PCR amplifications of the rDNA-ITS regions produced a 570-bp fragment for all the 102 Alternaria isolates, which were over 99% identical to each other, and to A. tenuissima (GenBank accession no. KR867207), A. alternata (GenBank accession nos. GU566303, KR867035 and MH862229), and other Alternaria species, such as A. brassicae (GenBank no. FJ869872) and A. longipes (GenBank no. FJ459960) included in the analysis. Analyses of the partial coding sequences of the histone 3 gene indicated the 102 Alternaria isolates could be divided into two species, A. tenuissima and A. alternata. Sixty-one of the Alternaria isolates had a 546-bp fragment that exhibited more than 99% similarity with those of A. tenuissima isolates retrieved from the GenBank database (AF404634, JX495167 and JX495168). The other 41 isolates had a 440-bp fragment with more than 99% similarity to those of A. alternata isolates AF404624 and AF404638.
The sequences of the tested Alternaria isolates were deposited in GenBank (Supplementary Table 1). Phylogenetic analysis of the partial coding sequences of the histone 3 gene divided all 45 Alternaria isolates used in the pathogenicity tests into two distinct clades, A. tenuissima and A. alternata (), but all the isolates were grouped in one clade using the ITS sequence data (). Phylogenetic analysis based on the rDNA-ITS region was not able to differentiate between A. tenussima and A. alternata.
Fig. 2 Phylogenetic tree based on the histone 3 gene of 45 Alternaria isolates used in the pathogenicity tests in this study and seven reference sequences retrieved from GenBank using the Maximum likelihood method (ML). Bootstrap values (1000 replicates) greater than 60% are shown for major lineages within the tree. The marker denotes a measurement of relative phylogenetic distance. Alternaria solani isolate CNU3072 (GenBank no. JX213319) and Alternaria infectoria isolate CR30 (GenBank no. AF404629) were used as outgroups.
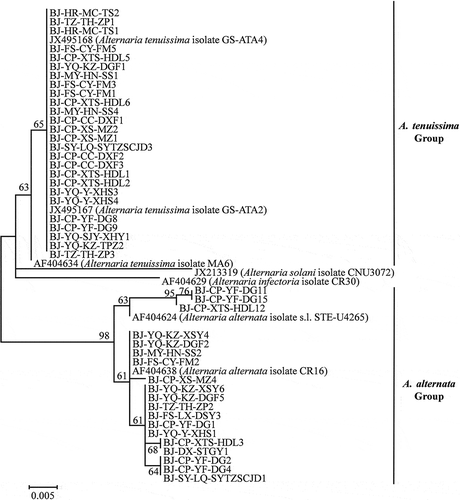
Fig. 3 Phylogenetic tree based on the ITS gene of 45 Alternaria isolates used in the pathogenicity tests in this study and six reference sequences retrieved from GenBank using the Maximum likelihood method (ML). Bootstrap values (1000 replicates) greater than 90% are shown for major lineages within the tree. The marker denotes a measurement of relative phylogenetic distance. Alternaria brassicae isolate AB11 (GenBank no. U05253) and Alternaria infectoria isolate STE-U4271 (GenBank no. AF397248) were used as outgroups.

Pathogenicity tests
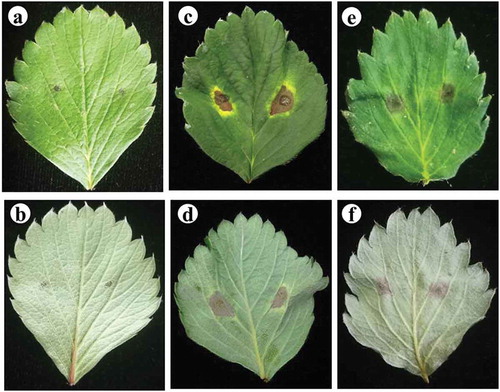
The symptoms of black spot such as a distinct circular spot, dark brown lesions and a typical yellow border developed on the detached leaflets 14 days after inoculation with spore suspensions of the 45 isolates (). Fungi isolated from lesions on inoculated leaflets were similar to the isolates used for inoculation as identified using the morphological and molecular methods mentioned above, fulfilling Koch’s postulates. The disease incidence and disease index of strawberry leaves inoculated with A. tenuissima were 47.2–100.0% (73.6 ± 26.4%) and 12.2–28.1 (20.2 ± 7.9), respectively. The disease incidence and disease index of the diseased leaves caused by A. alternata were 20.4–100.0% (60.2 ± 19.8%) and 6.0–29.1 (17.6 ± 11.6), respectively. There were no significant differences in disease incidence or disease index between isolates from different geographic origins or between the two species.
Fig. 4 (Colour online) Pathogenicity of the isolates of A. tenuissima and A. alternata on the detached leaves of strawberry. a–b, sterile water controls; c–d, A. tenuissima; e–f, A. alternata. The experiment was conducted using detached apical leaflets of the fully expanded leaves from 45-day-old plants of strawberry cv. ‘Benihoppe’. A drop of 20 μL spore suspension of 106 spores mL−1 was inoculated on each side of the main vein of the upper surface of each leaflet (two points per leaflet).
Discussion
Black spot caused by Alternaria is a common disease in the major strawberry-growing countries worldwide. Alternaria alternata and A. tenuissima were described as the major pathogens of black spot in Japan (Watanabe and Umekawa Citation1977; Watanabe et al. Citation1978; Takahashi et al. Citation1990, Citation2008; Misawa et al. Citation2012), Italy (Wassenaar and Van der Scheer Citation1989; Wada et al. Citation1996), Korea (Cho and Moon Citation1980), Iran (Bagherabadi et al. Citation2015) and Pakistan (Mehmood et al. Citation2018). This disease usually reduces both quality and yield on an annual basis. In this study, it was shown that A. tenuissima was the most prevalent (59.8%) species causing black spot of strawberry, followed by A. alternata (40.2%), in Beijing municipality. To our knowledge, this is the first report of A. tenuissima and A. alternata causing black spot on strawberry in Beijing.
Alternaria is a ubiquitous fungal genus, which can infect several crops, including sunflower (Wang et al. Citation2014), potato (Zheng et al. Citation2015; Zhao et al. Citation2018), tomato (Pose et al. Citation2004), watermelon (Zhao et al. Citation2016a), muskmelon (Zhao et al. Citation2016b), apple (Robiglio and Lopez Citation1995), wheat (Patriarca et al. Citation2007) and citrus (Peres et al. Citation2003). Watermelon, muskmelon and tomato are common crops, which are included in rotations with strawberry in the main production areas of China, including Beijing municipality. Watermelon and muskmelon have been reported to be infected by A. tenuissima and A. alternata in Beijing (Zhao et al. Citation2016a, Citation2016b). Therefore, cross-pathogenicity tests of Alternaria species isolated from strawberry and these common rotation crops need to be conducted, to determine if selected crops should be included as part of a crop rotation.
Previous studies have shown that the A. tenuissma isolates grown on PCA plates were characterized by the formation of unbranched conidial chains of up to 12 conidia with one or two lateral branches, while the A. alternata isolates produced conidial chains of 8–12 spores in length with numerous secondary and occasionally tertiary chains branching from apical and median cells (Wang et al. Citation2014; Zheng et al. Citation2015; Zhao et al. Citation2016a, Citation2016b, Citation2018). Based on these morphological traits, we identified two different small-spored species: A. tenuissima and A. alternata. Moreover, sequence analysis of the histone 3 gene effectively discriminated between these two small-spored Alternaria species (Deng et al. Citation2012; Wang et al. Citation2014; Zheng et al. Citation2015; Zhao et al. Citation2016a, Citation2016b, Citation2018; Landschoot et al. Citation2017). Combining morphological characteristics with molecular features, we have successfully distinguished between A. tenuissima and A. alternata on potato, watermelon, muskmelon and sunflower in the past five years (Wang et al. Citation2014; Zheng et al. Citation2015; Zhao et al. Citation2016a, Citation2016b, Citation2018). In the current study, A. tenuissima was also differentiated from A. alternata on strawberry on the basis of the combination of morphological and molecular traits.
Strawberry has been widely sown throughout China. Until now, there has been no report of Alternaria causing strawberry black spot in regions other than Beijing municipality. Further studies on Alternaria causing black spot of strawberry are needed across the strawberry growing regions of China. The sensitivity of A. tenuissima and A. alternata from strawberry to different fungicides currently used in the field to control black spot should also be investigated. Additional information on the Alternaria species infecting strawberry plants in other regions, along with the results of fungicide efficacy trials to control these species, will help to develop effective disease management strategies for strawberry black spot.
Supplementary Table 1
Download MS Word (209 KB)Acknowledgements
This work was supported by the Chinese Universities Scientific Fund (2015NX005). Mention of trade names or commercial products in this report is solely for the purposes of providing specific information and does not imply a recommendation or endorsement.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Andersen B, Kroger E, Roberts RG. 2001. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol Res. 105(3):291–299.
- Bagherabadi S, Zafari D, Soleimani MJ. 2015. First report of leaf spot of strawberry caused by Alternaria tenuissima in Iran. J Plant Pathol Microb. 6(3):258.
- Cho JT, Moon BJ. 1980. The occurrence of strawberry black leaf spot caused by Alternaria alternata (Fr.) Keissler in Korea. Korean J Plant Prot. 19(4):221–227.
- Deng JX, Paul NC, Park MS, Yu SH. 2012. Molecular characterization, morphology, and pathogenicity of Alternaria panax from araliaceous plants in Korea. Mycol Prog. 12(2):383–396.
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb. 61(4):1323–1330.
- Howard CM, Albregts EE. 1973. Strawberry fruit rot caused by Alternaria tenuissima. Phytopathology. 63(7):938–939.
- Kang JC, Crous PW, Mchau GR, Serdani M, Song SM. 2002. Phylogenetic analysis of Alternaria spp. associated with apple core rot and citrus black rot in South Africa. Mycol Res. 106(10):1151–1162.
- Ko Y, Chen CY, Yao KS, Liu CW, Lin CH, Maruthasalam S. 2008. First report of fruit rot of strawberry caused by an Alternaria sp. in Taiwan. Plant Dis. 92(8):1248.
- Landschoot S, Vandecasteele M, De Baets B, Höfte M, Audenaet K, Haesaert G. 2017. Identification of A. arborescens, A. grandis and A. protenta as new members of the European Alternaria population on potato. Fungal Biol. 121(2):172–188.
- Lee H, Kim CJ, Yu SH. 2001. First report of strawberry fruit rot caused by Alternaria tenuissima in Korea. Plant Dis. 85(5):563.
- Lee SB, Taylor JW. 1990. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; p. 282–287.
- Maltoni ML, Magnani S, Baruzzi G. 2000. Screening for Alternaria balck spot resistance in strawberry (in Italian). J Fruit Cultivation Hortic. 62:95–97.
- Mehmood N, Riaz A, Naz F, Hassan I, Jaabeen N, Anwaar S, Rosli H, Gleason ML. 2018. First report of strawberry leaf spot caused by Alternaria alernata in Pakistan. Plant Dis. 102(4):820.
- Misawa T, Nishikawa J, Kayamori M. 2012. Occurrence of black leaf spot in strawberry cultivar “HS-138” and morphological characteristics of causal fungus. Ann Rept Plant Prot North Jpn. 63:92–96.
- Patriarca A, Azcarate M, Terminiello L, Pinto VF. 2007. Mycotoxin production by Alternaria strains isolated from Argentinean wheat. Int J Food Microbiol. 119(3):219–222.
- Peres NAR, Agostini JP, Timmer LW. 2003. Outbreaks of Alternaria brown spot of citrus in Brazil and Argentina. Plant Dis. 87(6):750.
- Pose G, Ludemann V, Segura J, Pinto VF. 2004. Mycotoxin production by Alternaria strains isolated from tomatoes affected by Blackmold in Argentina. Mycotoxin Res. 20(2):80–86.
- Pryor BM, Michailides TJ. 2002. Morphological, pathogenic, and molecular characterization of Alternaria isolates associated with Alternaria late blight of pistachio. Phytopathology. 92(4):406–416.
- Robiglio AL, Lopez SE. 1995. Mycotoxin production by Alternaria alternata strains isolated from red delicious apples in Argentina. Int J Food Microbiol. 24(3):413–417.
- Simmons EG. 2007. Alternaria: an indentification manual. CBS biodiversity series 6. Utrecht (the Netherlands): CBS Fungal Biodiversity Centre.
- Stenglein SA, Balatti PA. 2006. Genetic diversity of Phaeoisariopsis griseola in Argentina as revealed by pathogenic and molecular markers. Physiol Mol Plant P. 68:158–167.
- Takahashi H, Furuya H, Takai T, Matsumoto T. 2008. Characteristics of Alternaria alternata strawberry pathotype isolated in New Zealand and the resistance of the “Akita Berry” strawberry to the fungus (in Japanese). Engei Gakkai Zasshi. 65:785–790.
- Takahashi H, Takai T, Matsumoto T. 1990. Susceptible strawberry cultivars to Alternaria black spot of strawberry (Alternaria alternata strawberry pathotype) in Japan (in Japanese). Engei Gakkai Zasshi. 59:539–544.
- Thammasorn T, Sangsuriya P, Meemetta W, Senapin S, Jitrakorn S, Rattanarojpong T, Saksmerprome V. 2015. Large-scale production and antiviral efficacy of multi-target double-stranded RNA for the prevention of White spot syndrome virus (WSSV) in shrimp. BMC Biotechnol. 15(1):110.
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.
- Wada H, Cavanni P, Bugiani R, Kodama M, Otani H, Kohmoto K. 1996. Occurrence of the strawberry pathotype of Alternaria alternata in Italy. Plant Dis. 104(6):715–716.
- Wang TY, Zhao J, Sun P, Wu XH. 2014. Characterization of Alternaria species associated with leaf blight of sunflower in China. Eur J Plant Pathol. 140(2):301–315.
- Wassenaar LM, Van der Scheer HAT. 1989. Alternaria leaf spot in strawberry. Acta Hortic. 265:575–578.
- Watanabe Y, Umekawa M. 1977. On a new black leaf spot of strawberry caused by Alternaria sp. Ann Phytopathol Soc Jpn. 43:82. (in Japanese).
- Watanabe Y, Umekawa M, Nishimura S. 1978. On the causal pathogen of black leaf spot of strawberry (in Japanese). Ann Phytopathol Soc Jpn. 44:363.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; p. 315–322.
- Zhao J, Bao SW, Ma GP, Wu XH. 2016a. Characterization of Alternaria species associated with muskmelon foliar diseases in Beijing municipality of China. J Gen Plant Pathol. 82(1):29–32.
- Zhao J, Bao SW, Ma GP, Wu XH. 2016b. Characterization of Alternaria species associated with watermelon leaf blight in Beijing municipality of China. J Plant Pathol. 98(1):135–138.
- Zhao J, Ma GP, Liu YY, Wu XH. 2018. Alternaria species infecting potato in southern China. Can J Plant Pathol. 40(2):312–317.
- Zheng HH, Zhao J, Wang TY, Wu XH. 2015. Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathol. 64(2):425–433.
- Zhu XB, Xia MR, Li YL, Li YZ. 2016. Food safety problems and preventive measures in strawberry production (in Chinese). Farm Products Processing. 4:56–58.
