Abstract
Symptoms of partially bleached and completely white heads were observed in a triticale (× Triticosecale) cultivar ‘BARI Triticale 1ʹ in the research plots at the Regional Agricultural Research Station, Jashore, Bangladesh, during April 2019. Foliar symptoms with elliptical or eye-shaped lesions were also found on plant leaves. The main intention of this study was to isolate and identify the causal organism of the disease. Morphological characteristics of the pathogen showed the presence of pyriform 2-septate hyaline conidia and coloured conidiophores, indicating the presence of the fungus Magnaporthe oryzae. Molecular analyses by ITS sequence analysis and amplification of the product with MoT3 primers confirmed the identity of the fungus as M. oryzae pathotype Triticum. The pathogenicity of the representative isolates was confirmed by inoculation of triticale heads and leaves under laboratory conditions, resulting in similar symptoms as observed in the field. To our knowledge, this is the first report of triticale blast caused by M. oryzae pathotype Triticum in Bangladesh.
Résumé
En avril 2019, des symptômes d’épis partiellement et complètement blanchis ont été observés chez un cultivar de triticale (× Triticosecale), ‘BARI Triticale 1ʹ, dans les parcelles de recherche de la Station régionale de recherche agricole de Jashore, au Bangladesh. Des symptômes se manifestant par des lésions elliptiques ou en forme d’œil ont également été trouvés sur les feuilles. Le principal objectif de cette étude était d’isoler et d’identifier l’organisme responsable de la maladie. Les caractéristiques morphologiques de l’agent pathogène ont révélé des conidies hyalines piriformes à deux parois et des conidiophores colorés, indiquant la présence du champignon Magnaporthe oryzae. Les analyses moléculaires basées sur l’analyse de la séquence de l’ITS et l’amplification du produit avec des amorces MoT3 ont confirmé l’identité du champignon en tant que pathotype Triticum de M. oryzae. La pathogénicité des isolats représentatifs a été confirmée par inoculation d’épis et de feuilles de triticale en laboratoire, ce qui a provoqué l’apparition de symptômes analogues à ceux trouvés au champ. À notre connaissance, il s’agit de la première mention d’échaudage du triticale causé par le pathotype Triticum de M. oryzae au Bangladesh.
Introduction
Triticale (× Triticosecale Wittmack) is the first synthetic cereal grain crop species that resulted from the product of a hybridization between wheat (Triticum spp.) and rye (Secale cereale) (Arendt and Zannini Citation2013). This cross was designed to merge the high yield and good bread-baking quality of wheat with the drought tolerance and disease resistance of rye. Triticale is generally used as animal feed, both as green feed and as grain. In addition, triticale is used in preparing baked goods, offering a better amino acid balance resulting in greater biological value and high-fibre content (Wrigley and Bushuk Citation2017). According to WorldAtlas (Faith Citation2014) the largest triticale producing country is Poland with annual production of 5.2 million metric tons followed by Germany with annual production of 3.0 million metric tons. Triticale production differs from country-to-country based on climatic conditions, soil fertility, and type of triticale grown. In Bangladesh, triticale is mostly grown as a high-quality fodder and feed in small-scale dairy farm operations. It is cultivated in very limited areas located in the north-western and central parts of the country.
From its beginning, triticale was considered resistant to many diseases. Although this view lessened with the expansion of cultivated areas, triticale still is less affected by disease compared to wheat and rye (Arseniuk Citation1996). Its pathogens include different groups of fungi including Bipolaris, Fusarium, Magnaporthe, Microdochium, Puccinia, etc., causing leaf, stem, and root diseases with varying levels of severity. Among the diseases of triticale, blast caused by an ascomycetous fungus Magnaporthe oryzae (anamorph Pyricularia oryzae), previously known as Magnaporthe grisea (anamorph Pyricularia grisea), is most important in the southern cone (especially in Parana state of Brazil) of South America (Urashima et al. Citation2004). The first incidence of triticale blast was noticed in 1995 in the state of Parana, Brazil, with a yield loss of 60% (Urashima et al. Citation2004). This pathogenic fungus can attack a wide range of grasses including rice, wheat, barley, finger millet, pearl millet, etc. (Ou Citation1985; Urashima et al. Citation1993; Pennisi Citation2010; Kharkar et al. Citation2016; Zhang et al. Citation2016). The fungus Magnaporthe oryzae is composed of a group of morphologically similar, but genetically distinct host-specific sub-species that are specific for infecting wheat (Triticum sub-species), rice (Oryza sub-species), perennial and annual ryegrass (Lolium sub-species), foxtail millet (Setaria sub-species), and many other Poaceae hosts. The triticale blast pathogen is reported to have originated from the wheat blast pathogen, with a positive cross infection detected between wheat and triticale isolates (Urashima et al. Citation2004). An outbreak of wheat blast caused by the fungus Magnaporthe oryzae pathotype Triticum was reported in Bangladesh for the first time in February 2016 (Islam et al. Citation2016; Malaker et al. Citation2016). In wheat, this pathogen can affect all aerial parts of the plant including leaves, stem, spike, awn, glumes, and seed, and can spread rapidly in wheat growing areas by wind and splashing water (Cruz and Valent Citation2017).
In April 2019, triticale plants (variety BARI Triticale 1), growing in research plots of the Regional Agricultural Research Station (RARS), Jashore (23°18′ 63.9″ N, 89°18′ 80.3″ E) suffered a previously unobserved disease that resulted in white head symptoms similar to wheat blast, on more than 20% of all plants in an 81 sq. m. area of the field. The objective of this study was to isolate and identify the causal agent of this disease based on the morphological characteristics, molecular analyses, and the completion of Koch’s postulates.
Materials and methods
Collection of blast affected samples
Five to six infected spikes along with infected leaves were randomly collected from the research plots at RARS, Jashore, at noon. They were then put inside brown paper bags and air dried overnight under laboratory conditions at 25°C ± 2°C and 12/12 hr light and darkness cycle. The samples were then brought to the plant pathology laboratory of Bangladesh Wheat and Maize Research Institute (BWMRI), Dinajpur, for further analysis.
Fungal isolation
Infected spike/leaf samples were cut into small pieces (approximately 2 cm to 3 cm), surface sterilized in 2% Sodium hypochloride (NaOCl) solution for about 30–40 sec followed by washing with distilled water for 2 min to 3 min, and air dried. Subsequently, the samples were incubated on 3-layered moist Whatman® blotter paper at room temperature (25°C to 28°C) under a 12-hr continuous light/darkness cycle for about 24 hr, which led to the vegetative and reproductive growth of the fungus. Fungal spores from infected samples were transferred onto potato dextrose agar (PDA) in Petri dishes using a sterile glass rod and were incubated for 5–12 days at 25°C with 40% relative humidity on a clean bench ensuring aseptic conditions. Mono-conidial pure culture of the fungus was obtained from culturing on PDA.
Pathogenicity tests (Koch’s postulates)
One fungal isolate (TrBWMRI-19M6) was used for testing pathogenicity. Healthy seeds of the triticale variety ‘BARI Triticale 1’ were planted in plastic pots (4 cm × 3 cm × 2.5 cm and 15 cm × 13 cm × 7 cm) containing a mixture of field soil (sandy loam with pH 5.8 and OM 2.1%) and well-composed cow dung at 1:1 ratio and grown in the laboratory (26°C ± 2°C). The fungus was cultured on an oatmeal agar (OMA) medium, incubated in the laboratory, and the spores harvested after 8–12 days. The spore suspension was prepared by following a standard protocol: a small amount of water was added into the Petri dishes and the fungal spores were then scraped from the surface of OMA into a beaker using a camel hairbrush, which was then filtered through cheesecloth. Finally, 2–3 drops of 0.01% Tween 20 solution were added and the suspension was stirred gently on an electric stir plate for 5–7 min. Spore concentration was measured with a haemocytometer and adjusted to 20,000 spores/mL with distilled water (Cruz et al. Citation2016). The plants were spray inoculated with the conidial suspension until run-off. Spore suspension was inoculated onto seedlings at the two to three leaf stage (14–15 days old) and fully emerged spikes using a hand sprayer. A control treatment of distilled water spray was also included as a negative control. After inoculation, all treatments were incubated in a confined polyhood for 36 hr in continuous darkness with high relative humidity (80%–90%). All the incubated pots were then taken out of the polyhood chamber and incubated on a laboratory bench to allow symptom development.
Fungal identification and molecular confirmation
Primarily morphological characters (vegetative and reproductive growth) were used to identify the fungal isolate obtained in this study. The morphological features such as the colour of mycelia, as well as the shape and growth of conidia and conidiophores were observed under a stereo and compound microscope (Olympus SZ61, Japan). Molecular methods were used to confirm the identity of the fungal isolates. Molecular confirmations of the isolates were done using ITS sequence analysis and PCR using the previously published MoT3 primer set (Pieck et al. Citation2017). The two isolates (TrBWMRI-19M6 and TrBWMRI-19M7) were grown on PDA at 25°C in continuous fluorescent light for 1 week to allow mycelial growth. Mycelia were then harvested, and the total genomic DNA was extracted from the isolates separately using the Wizard Genomic DNA Purification Kit (Promega Corp., USA) following the manufacturer’s protocol. First genetic confirmation was performed by using internal transcribed spacer ITS5 (GGAAGTAAAAGTCGTAACAAGG) and ITS4 (TCCTCCGCTTATTGATATGC) primers (White et al. Citation1990). Forward and reverse nucleotide sequences were determined from First BASE Laboratories (Malaysia), and were edited and aligned using BioEdit 7.2. For confirmation by the MoT3 primer, genomic DNA of the two isolates (TrBWMRI-19M6 and TrBWMRI-19M7) were amplified by PCR assay using the MoT3 primers: MoT3F (GTCGTCATCAACGTGACCAG) and MoT3R (ACTTGACCCAAGCCTCGAAT) (Pieck et al. Citation2017). The PCR mixture was composed of 12.5 μL of GoTaq® G2 Green Master Mix (2X), 9 μL of nuclease-free water, 1.25 μL each of the forward and reverse primer, and 1 μL of template DNA. The GoTaq® G2 Green Master Mix contains GoTaq® G2 DNA Polymerase, 2X Green GoTaq® G2 Reaction Buffer (pH 8.5), which is 400 μM dATP, 400 μM dGTP, 400 μM dCTP, 400 μM dTTP and 3 mM MgCl2 (Promega Corp., USA). PCR was conducted with a PCR Thermal Cycler (Veriti, Applied Biosystems, USA) following the conditions: initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 62°C for 2 min, extension at 72°C for 1 min 30 sec, and final elongation at 72°C for 10 min. Five micro-litres of each amplification mixture were tested by agarose gel (1% w/v) electrophoresis in 0.5X Tris-borate-EDTA (TBE) buffer. The presence or absence of amplified DNA was recorded for confirmation of MoT. To test the specificity of MoT3 primers, also included was the genomic DNA of isolates of M. oryzae obtained from Triticum aestivum (wheat), Hordeum vulgare (barley), Setaria italica (foxtail millet), Oryza sativa (rice), and Eleusine indica (goose grass) wherein the wheat was used as positive control and sterilized water as negative for the PCR assay.
The sequences used for the phylogenetic analysis were those obtained during this study, along with sequences from nine additional isolates of Magnaporthe oryzae, Magnaporthe grisea, Pyricularia oryzae, Pseudopyricularia cyperi, Magnaporthiopsis panicorum, Magnaporthiopsis agrostidis and Pyricularia grisea retrieved from the GenBank database. A phylogenetic tree was constructed by the neighbour joining likelihood method applying the rapid bootstrapping algorithm for 1000 replications using the MEGA-X model (Kumar et al. Citation2018).
Results and discussion
Field symptoms appearance
The infected plants were observed to have distinguishable partially bleached heads with dark grey to black coloured infection points on the rachis (). Some of the plants observed had completely bleached spikes. Spikes infected at the flowering stage yielded no grains, or grains that were withered, distorted, and had very low test-weight. Moreover, characteristic blast symptoms, elliptical or eye shaped lesions with grey centres and dark brown margins, were observed on the seedling leaves as well as the lower and flag leaves of adult plants ().
Fungal characteristics
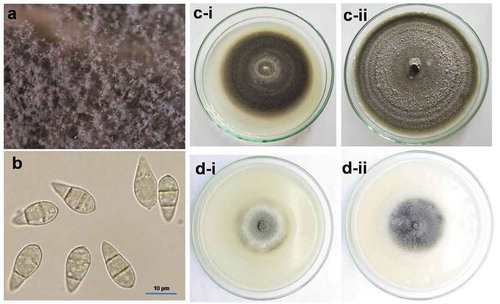
Profuse vegetative and reproductive growth of the fungus was observed on the incubated rachis under a stereomicroscope (). Based on morphological observations, conidia were hyaline but with time became ash coloured, 2-septate, sometimes single celled, and the shape generally pyriform (). Conidiophores were observed as simple, pigmented, erect, cylindrical, and with spores arising from the apex. Isolates TrBWMRI-19M6 and TrBWMRI-19M7 produced grey to dark grey colonies on PDA, covering the plate within 5–7 days ( and c-ii). The colonies also produced concentric rings and puffy mycelia bodies within 10–15 days of incubation. Profuse production of spores was found on OMA within 7–10 days of incubation ( and d-ii). Optimal mycelial growth and sporulation of the fungal isolates were obtained at 25°C–28°C and 40%–45% relative humidity. Conidiophore and conidia (pyriform 2-septate hyaline conidia) morphology of the fungus were consistent with Magnaporthe oryzae (Subramanian Citation1968).
Fig. 2 Vegetative and reproductive growth of Magnaporthe oryzae (pathotype Triticum) under laboratory conditions; (a) ash coloured conidial growth of the fungus on the incubated rachis; (b) 2-septed hyaline pear-shaped conidia under compound microscope (magnification 400X); (c) single-spore colony of both isolates – i. TrBWMRI-19M6, ii. TrBWMRI-19M7 – on potato dextrose agar medium; and (d) sporulation of both isolates – i. TrBWMRI-19M6, ii. TrBWMRI-19M7 – on oatmeal agar medium
Molecular analysis
All the isolates inducing blast in wheat, triticale, barley, rice, foxtail millet and goose grass were amplified by ITS primer sets (ITS5 and ITS4) confirming all isolates were Magnaporthe oryzae (). The ITS sequence results of Bangladesh isolates TrBWMRI-19M6 and TrBWMRI-19M7 (GenBank accession nos. LC520096 and LC520097) showed > 99.75% similarity with the sequence from Magnaporthe oryzae strain BR0032 (KM484887), which was obtained from a Brazilian wheat sample. Likewise, MoT3 primer results showed that triticale blast isolates (TrBWMRI-19M6 and TrBWMRI-19M7) produced a clear band during the PCR assay, highly identical with wheat and barley blast isolates while there was no PCR amplification from rice blast, goose grass blast and foxtail millet blast isolate samples (). Based on the aligned sequences of ITS, molecular phylogenetic analysis by the neighbour joining likelihood method confirmed that isolates TrBWMRI-19M6 and TrBWMRI-19M7 were Magnaporthe oryzae () (Subramanian Citation1968). Both isolates were stored for future study on sterilized blotter paper at −81°C at the laboratory of Plant Pathology Division of Bangladesh Wheat and Maize Research Institute, Dinajpur, Bangladesh. These isolates are available to any researcher upon request.
Fig. 3 PCR analysis of triticale blast isolates and some other blast isolates from different hosts with an ITS primer set (ITS5 and ITS4)
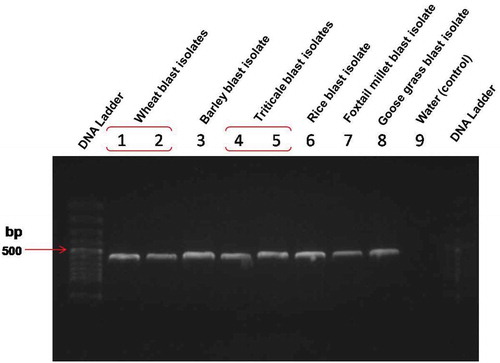
Fig. 4 PCR analysis of triticale blast isolates including some other hosts (wheat, barley, rice, foxtail millet, and goose grass) with MoT3 primers for confirming the Magnaporthe oryzae pathotype Triticum in a molecular detection assay
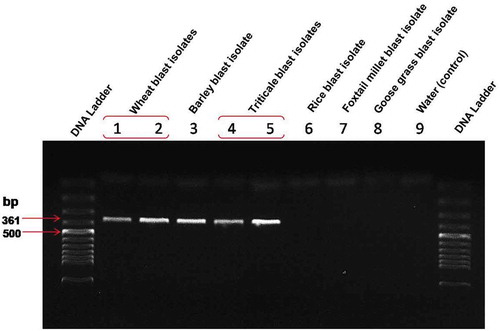
Fig. 5 Phylogenetic tree of Magnaporthe oryzae from triticale plants in Bangladesh and related species based on a neighbour joining likelihood analysis of a combined internal transcribed spacer (ITS) dataset. The ITS sequence data for the other nine isolates were retrieved from the NCBI GenBank database. Numbers beside each branch represent bootstrap values obtained after a bootstrap test with 1,000 replications. The bar indicates the number of nucleotide substitutions. The two triticale blast isolates are indicted by the red diamond shape. This analysis was performed with MEGA X software

Pathogenicity tests
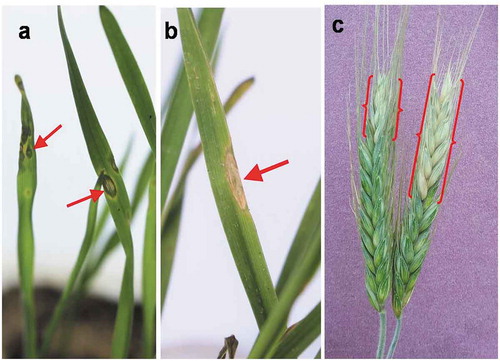
Pathogenicity tests were performed with isolate TrBWMRI-19M6 to fulfil Koch’s postulates. Two to three days after inoculation, symptoms on seedling leaves were observed as water-soaked lesions (), while no spots were found on plants inoculated with distilled water. At 7 days post inoculation, the spots became larger in size, with grey centres surrounded by straw coloured margins. Inoculated heads at 14–15 days after inoculation were found to be partially bleached (). The fungal pathogen (Magnaporthe oryzae) was successfully re-isolated from the symptomatic organs of the inoculated plants, compared with the original, and found as identical, fulfiling Koch’s postulates (Cohen Citation1994).
Fig. 6 Pathogenicity confirmation of the M. oryzae pathotype Triticum fungus (isolate TrBWMRI-19M6) by fulfiling the Koch’s postulates; (a) round to oval-shaped water-soaked lesions developed on the seedling leaves 2–3 days after inoculation with Magnaporthe oryzae (TrBWMRI-19M6); (b) grey centred elongated lesion on seedling leaf at 7 days post-inoculation; and (c) partially bleached head blast symptoms developed 10–12 days after inoculation
Based on morphological characteristics, molecular analyses by ITS sequences, MoT3 markers and pathogenicity tests, the causal agent of triticale head blast was identified as Magnaporthe oryzae pathotype Triticum. To the best of our knowledge, this is the first report of triticale blast caused by Magnaporthe oryzae pathotype Triticum in Bangladesh.
Acknowledgements
The authors highly acknowledge the laboratory staff for technical support.
Additional information
Funding
References
- Arendt EK, Zannini E, editors. 2013. Triticale. In: Cereal grains for the food and beverage industries. Cambridge: Woodhead Publishing; p. 201–229.
- Arseniuk E. 1996. Triticale diseases — a review. In: Guedes-Pinto H, Davey N, Carnide VP, editors. Triticale: today and tomorrow. Dortrecht (Netherlands): Kluwer Academic Press; p. 499–525.
- Cohen J. 1994. Fulfilling Koch’s postulates. Science. 266:1647.
- Cruz CD, Bockus WW, Stack JP, Valent B, Maciel JN, Peterson GL. 2016. A standardized inoculation protocol to test wheat cultivars for reaction to head blast caused by Magnaporthe oryzae (Triticum pathotype). Plant Health Prog. 17:186–187. doi:10.1094/PHP-BR-16-0041.
- Cruz CD, Valent B. 2017. Wheat blast disease: danger on the move. Trop Plant Pathol. 42:210–222. doi:10.1007/s40858-017-0159-z.
- Faith C 2014. Top triticale producing countries in the world. WorldAtlas. [accessed 2020 Jun 29]. https://www.worldatlas.com/articles/top-triticale-producing-countries-in-the-world.html
- Islam MT, Croll D, Gladieux P, Soanes DM, Persoons A, Bhattacharjee P, Hossain MS, Gupta DR, Rahman MM, Mahboob MG. 2016. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 14(1):84. doi:10.1186/s12915-016-0309-7.
- Kharkar H, Kale K, Thakare P. 2016. Molecular characterization of Pyricularia grisea isolates using AFLP DNA fingerprinting. J Pure Appl Microbiol. 10(2):1181+.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Malaker PK, Barma NCD, Tiwari TP, Collis WJ, Duveiller E, Singh PK, Joshi AK, Singh RP, Braun HJ, Peterson GL, et al. 2016. First report of wheat blast caused by Magnaporthe oryzae pathotype Triticum in Bangladesh. Plant Dis. 100(11):2330. doi:10.1094/PDIS-05-16-0666-PDN.
- Ou SH. 1985. Blast. Rice diseases. 2nd ed. Wallingford (UK): CABI; p. 109–201.
- Pennisi E. 2010. Armed and dangerous. Science. 327:804–805.
- Pieck ML, Ruck A, Farman ML, Peterson GL, Stack JP, Valent B, Pedley KF. 2017. Genomics-based marker discovery and diagnostic assay development for wheat blast. Plant Dis. 101:103–109. doi:10.1094/PDIS-04-16-0500-RE.
- Subramanian CV. 1968. Pyricularia oryzae. CMI Desc Patho Fungi Bacteria. 169:1–2.
- Urashima AS, Igarashi S, Kato H. 1993. Host range, mating type and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis. 77:1211–1216. doi:10.1094/PD-77-1211.
- Urashima AS, Lavorent NA, Goulart ACP, Mehta YR. 2004. Resistance spectra of wheat cultivars and virulence diversity of Magnaporthe grisea isolates in Brazil. Fitopatol Bras. 29:511–518. doi:10.1590/S0100-41582004000500007.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. A guide to methods and applications. San Diego: Academic Press; p. 315–322.
- Wrigley C, Bushuk W. 2017. Triticale: grain-quality characteristics and management of quality requirements. In: Wrigley C, Batey I, Miskelly D, editors. Cereal grains. 2nd ed. Cambridge: Woodhead Publishing; p. 179–194.
- Zhang H, Zheng X, Zhang Z. 2016. The Magnaporthe grisea species complex and plant pathogenesis. Mol Plant Pathol. 17(6):796–804. doi:10.1111/mpp.12342.

