 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Plasmodiophora brassicae causes clubroot on Brassica vegetables and canola around the world. A diverse assemblage of new pathotypes have been identified on canola in the Canadian province of Alberta since 2012, but the pathotypes present in the rest of Canada have not been assessed in detail in recent years. Brassica vegetable and canola fields in Ontario were surveyed for clubroot in 2017–2019. The pathotype of isolates collected was determined using the Williams differential set. Clubroot samples or soil from six other Canadian provinces were also assessed. Clubroot appears to be spreading on canola in Ontario. Pathotype 6 was found only on vegetables and pathotype 2 was predominant on canola; pathotypes 2X and 3X were also identified on canola. The ‘X’ indicates collections that were virulent on the first generation of clubroot-resistant canola cultivars in Canada. Prior to 2012, the pathotypes in eastern Canada were 1, 2, 3, 4, 5, and 6 and the pathotypes in western Canada were 3, 5, and 8 with 6 found on vegetables in British Columbia. Additional virulent pathotypes 2X, 3X, and 5X were identified in eastern Canada and pathotype 9 was found in Manitoba. Two instances of rapid shifts in pathotype at research sites were demonstrated, one in Ontario (pathotype 6 to 2) and one in Quebec (pathotypes 2 and 5 to 2X and 5X).
Résumé
Plasmodiophora brassicae cause la hernie chez les légumes du genre Brassica et le canola partout dans le monde. Un assemblage différent de nouveaux pathotypes a été détecté chez le canola en 2012 en Alberta, au Canada, par ailleurs les pathotypes trouvés dans les autres provinces canadiennes n’ont pas été évalués en détail au cours des dernières années. En Ontario, les champs de légumes du genre Brassica et de canola ont été testés en 2017-2019 pour la hernie. Le pathotype des isolats collectés a été identifié à l’aide de la série différentielle de William. Des échantillons de hernie ou de sol provenant de six autres provinces canadiennes ont également été analysés. La hernie semble se répandre dans le canola en Ontario. Le pathotype 6 a été trouvé seulement chez les légumes et le pathotype 2 prédominait chez le canola; les pathotypes 2X et 3X ont également été détectés chez le canola. Le « X » indique des collections qui étaient virulentes à l’égard de la première génération de cultivars de canola résistants à la hernie au Canada. Avant 2012, les pathotypes de l’est du Canada étaient les pathotypes 1, 2, 3, 4, 5 et 6 et ceux de l’ouest du Canada, les pathotypes 3, 5 et 8, y compris le pathotype 6 trouvé chez les légumes en Colombie-Britannique. Les pathotypes supplémentaires 2X, 3X et 5X ont été identifiés dans l’est du Canada, tandis que le pathotype 9 a été trouvé au Manitoba. Deux cas de changements rapides chez les pathotypes ont été démontrés à des centres de recherche, un en Ontario (pathotypes 6 à 2) et un au Québec (pathotypes 2 et 5 à 2X et 5X).
Introduction
Plasmodiophora brassicae Woronin causes the disease clubroot, which affects canola (Brassica napus L.) and many brassica vegetable crops (including B. oleracea L). Pathogen development in infected roots results in club-shaped growths that are characteristic of the disease. Heavy infestations can cause yield losses of up to 100% (Strelkov et al. Citation2007; Xue et al. Citation2008). Infected roots can contain a mixture of pathotypes of P. brassicae (Cao et al. Citation2009; Sedaghatkish et al. Citation2019; Fu et al. Citation2020).
Several systems have been developed to identify pathotypes of P. brassicae, including the Williams’ differential hosts (Williams Citation1966), the European Clubroot Differential (ECD) series (Buczacki et al. Citation1975), and the differentials of Somé et al. (Citation1996). The most recent is the Canadian Clubroot Differential system, developed to characterize the range of pathogenic diversity observed in western Canada on canola since 2013, when a breakdown in resistance to clubroot was first observed in the region (Strelkov et al. Citation2018).
The original paper by Williams (Citation1966) showed P. brassicae pathotype 6 on cabbage in British Columbia, pathotype 2 on cabbage in Quebec and pathotype 4 on rutabaga also from Quebec. An early survey by Ayers (Citation1972) of pathotypes using the Williams’ differential system, identified pathotypes 1, 2, 3, and 4 in the Maritime provinces, pathotypes 2 and 6 in Quebec, and pathotypes 6 in Ontario and British Columbia (). Pathotype 2 was found on turnip in Ontario in the 1960s (Reyes Citation1969). Clubroot has been present on vegetable Brassicas in southwestern Ontario since 1921 and was found across the province in 1924 (Drayton Citation1926). Clubroot was first reported on canola in Ontario in 2016 and was identified as pathotype 2 (Al-Daoud et al. Citation2018). In Alberta, clubroot was first reported on canola in 2003 (Tewari et al. Citation2005), with pathotype 3 as the predominant pathotype (Strelkov and Hwang Citation2014). Since then, a range of new, virulent pathotypes have been identified in Alberta (Strelkov et al. Citation2016, Citation2018). The first of these pathotypes was called ‘5X’ because the Williams’ differential hosts did not distinguish between the old pathotype 5, that did not overcome resistance, and the new pathotype 5 that did (Strelkov et al. Citation2016). In this paper, an ‘X’ after the pathotype number indicates that the virulence of the pathotype was not revealed by the Williams’ differentials, and that the pathotype is virulent on the first generation of previously resistant canola cultivars as represented by canola cv. ‘45H29’ (Corteva Agriscience). The naming of these new virulent pathotypes has recently been improved with the development of a Canadian Clubroot Differential set (Strelkov et al. Citation2018). However, seed of the differentials was not available, so use of the older Williams’ system was retained for this study.
Fig. 1 The predominant pathotypes of Plasmodiophora brassicae across Canada (Williams Citation1966; Ayers Citation1972; Strelkov and Hwang Citation2014; Strelkov et al. Citation2018; this study). Pathotype nomenclature is according to Williams’ differential system (Williams Citation1966). ”X” pathotypes are able to overcome resistance in 45H29. The pathotypes reported to be present only after 2013 are presented in bold lettering. An Asterix indicates that the pathotype was present in a mixture of two pathotypes: see Ayers (Citation1972)
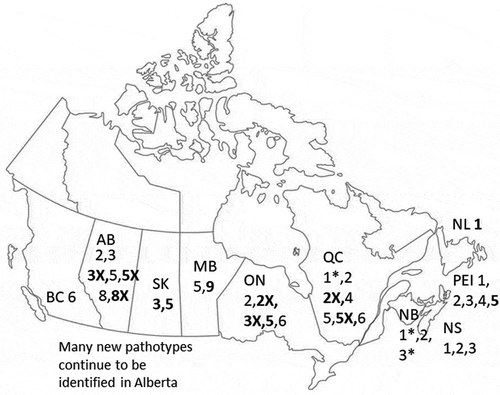
Since 2003, P. brassicae has spread quickly within Alberta and then beyond (Gossen et al. Citation2015). The pathogen was first identified in Saskatchewan in 2010 (Dokken-Bouchard et al. Citation2010) and is now spreading within that province (Ziesmann Citation2019). Pathotype 5 was reported on canola in Manitoba (Cao et al. Citation2009) ().
Clubroot management relies primarily on resistant cultivars. A number of commercial brassica vegetable and canola cultivars carry clubroot resistance (Saude et al. Citation2012; Deora et al. Citation2013; Strelkov et al. Citation2018). However, resistance is pathotype-specific and so may not provide protection against all the pathotypes present in an infested field (Xue et al. Citation2008). Also, the predominant pathotype in a field can change over time, leading to the emergence of new pathotypes that overcome host resistance (Strelkov et al. Citation2018; Sedaghatkish et al. Citation2019).
The objectives of this study were to: (a) survey the distribution of clubroot in brassica vegetable and canola fields in Ontario and determine the predominant pathotype of P. brassicae, (b) determine the pathotype of P. brassicae from other locations across Canada, and (c) document an observed change in pathotypes at the MCRS and Normandin Research Farm.
Materials and methods
Clubroot survey
In the summer and autumn of 2017, staff of the Ontario Ministry of Agriculture and Rural Affairs (OMAFRA) visited fields that had or had recently had canola crops and sampled plants and soil. Clubbed roots of brassica vegetables were submitted by growers directly to OMAFRA. Canola fields were sampled within 30 m of the field entrance, because that is where infestations usually begin in canola fields in Alberta (Gossen et al. Citation2015). Soil probes with 2.5-cm-dia. were used to sample 25 to 30 cores to a depth of 20 cm. All tools and footwear were cleaned with a 2% bleach solution after sampling each field. Cores from single fields were bulked and samples were submitted to the Pest Diagnostics Clinic (University of Guelph, Guelph, ON) for testing for the presence of P. brassicae DNA. DNA extractions were performed using the PowerSoil DNA Extraction Kit (Qiagen) following the procedures recommended by the manufacturer. PCR analyses were performed according to Cao et al. (Citation2007) using the TC1 primer sequences.
All canola fields that were sampled had canola within the past three years; often they were planted to canola the year of sampling or the year before. If canola was being grown, roots were assessed for clubroot symptoms. Most fields had been sown to clubroot-susceptible Bayer InVigor canola cultivars, grown in a 3–5 year rotation. A sample collected in 2016 from a field in Verner, ON, where the first symptoms of clubroot on canola in Ontario were found (Al-Daoud et al. Citation2018), was tested for the ability to overcome clubroot resistance in the canola ‘45H29’, as described below.
Collections across Canada
Clubbed roots of brassica vegetable and canola plants from six provinces across Canada were obtained from researchers and extension staff. Clubs were sampled in 2015 from canola fields near Swan River, Manitoba, in 2016, from a site in northwest Saskatchewan and from Alberton and Emerald, Prince Edward Island. In 2017 soil was collected from Spruce Grove, Alberta. Four samples of clubbed canola were collected from Saskatchewan (CN1, CJC-Hills, LLH2, SD1) in 2018. The fields in Prince Edward Island were experimental sites where a mixture of susceptible (‘45H73’, Corteva Agrisciences) and resistant canola were grown. The samples of vegetable brassicas from Newfoundland and British Columbia were from another study (Sedaghatkish et al. Citation2019). One sample from near Orangeville, Ontario, was submitted by an industry field representative in 2019.
Pathotyping
Pathotype assessments were conducted in a growth room at the University of Guelph in Guelph, ON (25/20°C day/night, 16 hr photoperiod, 50% relative humidity). Prior to assessment, each collection was increased on a susceptible host, Shanghai pak choy (B. rapa var. chinensis) cv. ‘Mei Qing Choi’ (Stokes Seeds, St Catharines, ON), as described below.
Pathotype assessments consisted of three or four replicates with 6–12 plants per replicate. They included the differential hosts of Williams (Citation1966): B. napus var. rapifera ‘Wilhelmsburger’ (rutabaga, ECD 10), B. oleraceae var. capitata L. ‘Badger Shipper’ (cabbage, ECD 11) and ‘Jersey Queen’ (cabbage, ECD 13), and B. napus var. napobrassicae Mill. ‘Laurentian’ (rutabaga). Seed of ECD 10, 11, and 13 were obtained from the Genetic Resources Unit, Warwick Crop Centre, the University of Warwick, Wellesbourne, UK, and ‘Laurentian’ was purchased from Wisconsin Fast Plants, University of Wisconsin, Madison, WI. Other hosts were susceptible checks, canola ACS-N39 (Agriculture and Agri-Food Canada) or Shanghai pak choy and the clubroot-resistant canola ‘45H29’ (Corteva AgriScience). Any pathotypes able to overcome resistance in ‘45H29’ were designated as an X pathotype to differentiate them from collections that did not overcome this resistance (Strelkov et al. Citation2016). Seed of the differential cultivars of the CCD set (Strelkov et al. Citation2018) was not available when the collections were assessed.
Plants were grown in 19-cm-tall plastic pots (conetainers, Stuewe and Sons Inc., OR), except for collections from NL and BC, which were grown in 24 cell seed nursery trays. LA4 Sunshine Mix (Sun Gro Horticulture, MA, USA) was the growth medium. Plants were watered daily with water adjusted to pH 6.0 with food-grade white vinegar, and fertilized weekly with 0.1% nitrogen, phosphate, potassium (20–20-20; Plant Products, ON, Canada) supplemented with magnesium sulphate (K + S; Kali GmbH, Germany). One-week-old plants were inoculated with 5 mL of 107 resting spores mL−1 applied to the base of the seedling. Each plant was rated for clubroot symptoms on a 0–3 scale at 5–6 weeks after inoculation and separated into classes. The classes were: 0 = no clubroot, 1 = < 1/3 of root with clubroot symptoms, 2 = 1/3-2/3 of root with clubroot, and 3 = > 2/3 of root with clubroot symptoms. A disease severity index (DSI) was calculated (Crête et al. Citation1963) as follows:
A host was considered resistant if DSI + 95% confidence interval was less than 50; otherwise it was susceptible (Strelkov et al. Citation2006, Citation2016). The 95% confidence interval was calculated using Microsoft Excel 2010.
Changes in pathogenicity
Clubroot research has been conducted for many years at the Muck Crops Research Station (MCRS) of the University of Guelph, located in the Holland Marsh, Ontario (44º 02ʹ 27.7'' N, 79º 35ʹ 43.9'' W). The predominant pathotype was 6 (Deora et al. Citation2013).
Clubroot has been endemic in the Holland Marsh for decades. It was first observed in this area in 1953 in a field of cauliflower. In1954, Hurricane Hazel caused flooding in the Marsh, which resulted in spread of P. brassicae and extensive damage to brassica vegetable crops in the Holland Marsh in following years (Canadian Plant Disease Survey Citation1956).
Similarly, clubroot has been present at the Agriculture and Agri-Food Canada Research Farm at Normandin, QC (48º 50ʹ 43.17'' N, 72º 32ʹ 20.8176'' W), since 1997 (Pageau et al. Citation2006), and the predominant pathotypes in that area were pathotypes 2 and 5 (Williams Citation1966; Ayers Citation1972; Peng et al. Citation2015). Clubbed roots of the susceptible canola line ACS-N39 were collected from field trials in range 4 of the MCRS in 2011, 2014, and 2016, and soil was collected from range 6 (~ 35 m distant) prior to planting in the spring of 2017. Clubbed roots of normally resistant canola ‘45H29’ were collected in 2016 and 2017 from the Normandin Research Farm.
Results and Discussion
Clubroot survey
Soil samples from 13 of 33 canola fields tested positive for the presence of DNA of P. brassicae, and eight additional fields had clubroot symptoms on canola (). Only five of the eight samples with clubs could be identified to pathotype; results were inconclusive for the remaining three samples. Four of the five samples assessed were pathotype 2 and one was pathotype 5 (Supp Table 1). The samples were tested for virulence on ‘45H29’, including samples from the field where clubroot was first reported on canola in Verner, Ontario (Al-Daoud et al. Citation2018). Samples from a field in New Liskeard and one near Earlton produced abundant clubroot symptoms on ‘45H29’ and were designated pathotype 2X (Supp Table 1). Both of these fields were in clubroot-susceptible canola when sampled, but the grower at New Liskeard had grown ‘45H29’ in nearby fields for many years. A sample from near Orangeville, Ontario, submitted in 2019 was pathotype 3X, which was the first time that pathotype 3 has been identified in Ontario.
Fig. 2 Map of canola and brassica vegetable fields in Ontario surveyed for clubroot symptoms and presence of the pathogen in soil in 2017. Fields where the pathotype (P) of P. brassicae was identified are indicated. The field where clubroot was first reported on canola in Ontario in 2016 is circled
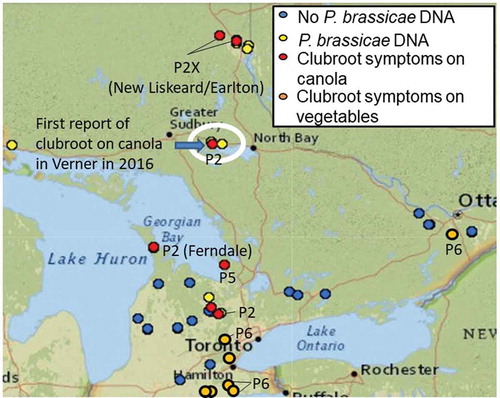
Seven brassica vegetable fields in the survey were in southern Ontario, and one was in the east, near Ottawa. Clubroot symptoms were observed in all samples, but only four could be pathotyped; all were pathotype 6 (Supp Table 1; ). Pathogen collections from vegetables were consistently less aggressive (produced lower severity) on the susceptible canola check, ACS-N39, than collections from canola (30–65 vs. 80–100 DSI) (Supp Table 1).
Clubroot had been present in vegetables fields in Ontario since at least the early 1920s (Drayton Citation1926) and was identified as pathotype 6 in the 1960s (Reyes Citation1969; Ayers Citation1972; Reyes et al. Citation1974) and there has been no change in the cruciferous vegetables such as cabbage (B. oleracea). Pathotype 2 was found on turnip and rutabaga in the surveys of Reyes (Citation1969, Citation1974) and this was the pathotype found consistently on canola since 2016, until 3X was identified in 2019.
The presence of pathotype 2X (virulent on ‘45H29’) in two Ontario canola fields was unexpected because susceptible canola cultivars were being grown in the fields when sampled. However, one grower had planted ‘45H29’ in nearby fields for many years. It is possible that pathotype 2X increased in nearby fields and was transported to the sampled field on equipment as described in Gossen et al. (Citation2015).
Collections across Canada
Collections from experimental plots in Prince Edward Island, where a mixture of susceptible and resistant canola had been grown, were identified as pathotypes 2 and 5 (Supp Table 2; ). A collection from a canola field in MB was pathotype 9, and samples from Saskatchewan were pathotypes 5 and 3 (Table 2; ). Collections from brassica vegetable fields in Newfoundland were pathotype 1 and collections from British Columbia were pathotype 6 (Supp Table 2; ). A collection that was grown out from soil from an experimental field in Alberta was identified as pathotype 3X because it was pathotype 3 and virulent on ‘45H29’.
Results from pathotyping of samples in Ontario support previous observations that pathotype 6 is more aggressive on, or better adapted to, brassica vegetables (especially B. oleracea vegetables), than pathotypes common on canola in Canada (and vice versa) (Deora et al. Citation2013). Pathotype 6 has remained consistent on these vegetables in Ontario and British Columbia, even though a range of pathotypes has been common on vegetables in the Atlantic provinces (pathotypes 1, 2, 3 and 4, ).
Changes in pathotype
Collections of clubbed roots of a susceptible canola line collected from range 4 at the MCRS in 2011 were identified as pathotype 6. Collections from the same range in 2014 and 2016 were pathotype 2. A sample from range 6 at the MCRS in 2017 was also pathotype 2 (Supp Table 3). None of these collections were able to overcome resistance, but severity on ‘45H29’ increased from 2011 (0 DSI) to 2017 (39 DSI, Supp Table 3).
Prior to 2009, studies at the MCRS were focused on vegetable brassicas, and the pathotypes was stable for many years. However, when studies shifted to canola at this site, the predominant pathotype shifted rapidly to pathotype 2, which is more aggressive on canola than pathotype 6 (Deora et al. Citation2013). It is interesting to note that very few clubroot-resistant lines have been produced in the field at the MCRS, and at the time of writing, neither pathotypes 6 or 2 were virulent on ‘45H29’.
Similarly, collections from the Normandin Research Farm were identified as 5X in 2016 and 2X in 2017. These samples were harvested from ‘45H29’, which had previously been resistant at the site (D. Pageau, personal communication). The virulence of the collections on ‘45H29’ was confirmed (Supp Table 3). Canola ‘45H29’ began to exhibit susceptibility in 2014 in the long-term rotation trial at this site (Gossen et al. Citation2019). Prior to 2014, there was a mixture of pathotypes 2 and 5 in this field (Cao et al. Citation2009; Peng et al. Citation2015) and the virulent ‘X’ pathotype 5X was predominant in 2016, and pathotype 2X was predominant in 2017. Trials at this site included clubroot-resistant canola cultivars over several years. It appears likely that selection for virulence on ‘45H29’ resulted in a pathotype shift in this trial site.
Jones et al. (Citation1982) showed that clubs from the same field can contain different pathotypes of P. brassicae. This observation was recently confirmed based on molecular assessment (Fu et al. Citation2020). A recent whole-genome sequencing study presented supporting evidence for balancing selection in P. brassicae (Sedaghatkish et al. (Citation2019), which provides an explanation of how many different genotypes and pathotypes could be present in a single club.
This study and other recent studies (Gossen et al. Citation2015; Strelkov et al. Citation2018) have documented important changes in the diversity and distribution of P. brassicae in several regions of Canada since pathotypes were assessed in the 1970’s by Ayers (Citation1972). Clubroot on canola is spreading in Ontario, new pathotypes were added for several provinces, and pathotype shifts at two experimental sites were documented.
For Prince Edward Island, pathotype 5 was added to the pathotypes 1, 2, 3 and 4 that were identified previously (Ayers Citation1972). In Quebec, pathotypes 2X and 5X were added to pathotypes 1, 2 and 5. Pathotype 2 had only recently been identified on canola in northern Ontario, but the current survey added pathotypes 2X, 3X and 5. Pathotype 5 is a new report in Saskatchewan, and pathotype 9 was identified in Manitoba. Pathotype 3 (the same one that is predominant in Alberta) had previously been reported in Saskatchewan (Strelkov and Hwang Citation2014), consistent with this study. Pathotype 6 was confirmed in British Columbia, which is unchanged from previous reports.
The rapid shift in pathotype documented by this study is consistent with the rapid development of new virulent pathotypes in Alberta (Strelkov et al. Citation2018), and both demonstrate how quickly the pathogen can respond to selection pressure. This emphasizes the importance of measures to reduce the populations of resting spores in soil to reduce selection pressure. The rapid changes in pathotype also present huge challenges to plant breeders to respond with new sources of resistance. The different and varied pathotypes in the Atlantic provinces suggest that there were several introductions of the pathogen to this region.
Table_S3_2020.docx
Download MS Word (17.3 KB)Table_S2__2020.docx
Download MS Word (21.3 KB)Table_S1__2020.docx
Download MS Word (18 KB)Acknowledgements
We thank J. Horsman and the staff at the MCRS for technical assistance, Drs. J. Elmhirst, S. De Boer, L. L. Jewel and D. Pageau, and C. Jurke, I. Epp, H. Derksen, and others who provided collections of P. brassicae from across Canada.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Al-Daoud F, Moran M, Gossen BD, McDonald MR. 2018. First repost of clubroot (Plasmodiophora brassicae) on canola in Ontario. Can J Plant Pathol. 40:96–99. doi:10.1080/07060661.2017.1393696.
- Ayers GW. 1972. Races of Plasmodiophora brassicae infecting crucifer crops in Canada. Can Plant Dis Surv. 52:77–81.
- Buczacki ST, Toxopeus H, Mattusch P, Johnston TD, Dixon GR, Hobolth LA. 1975. Study of physiologic specialisation in Plasmodiophora brassicae: proposals for attempted rationalization through an international approach. Trans Brit Mycol Soc. 65:295–303. doi:10.1016/S0007-1536(75)80013-1.
- Canadian Plant Diseases Survey. 1956. Diseases of vegetables and field crops. Canadian Plant Disease Survey. 36:48–97.
- Cao T, Manolii VP, Hwang S-H, Howard RJ, Strelkov SE. 2009. Virulence and spread of Plasmodiophora brassicae (clubroot) in Alberta, Canada. Can J Plant Pathol. 31:321–329. doi:10.1080/07060660909507606.
- Cao T, Tewari J, Strelkov SE. 2007. Molecular detection of Plasmodiophora brassicae Woronin, causal agent of clubroot of crucifers, in plant and soil. Plant Dis. 91:80–87. doi:10.1094/PD-91-0080.
- Crête R, Laliberté J, Jasmin JJ. 1963. Lutte chimique contre la hernie, Plasmodiophora brassicae Wor., des cruciferes en sols mineral et organique. Can J Plant Sci. 43:349–354. doi:10.4141/cjps63-065.
- Deora A, Gossen BD, McDonald MR. 2013. Cytology of infection, development and expression of resistance in canola. Ann Appl Biol. 163:56–71. doi:10.1111/aab.12033.
- Dokken-Bouchard FL, Bouchard AJ, Ippolito J, Peng G, Strelkov SE, Kirkham CL, Kutcher HR. 2010. Detection of Plasmodiophora brassicae in Saskatchewan, 2008. Can Plant Dis Surv. 90:126.
- Drayton FL. 1926. A summary of the prevalence of plant diseases in the dominion of Canada 1920–1924. Vol. 71. Ottawa: F.A. Acland.
- Fu H, Yang Y, Mishra V, Zhou Q, Zuzak K, Feindel D, Harding M, Feng J. 2020. Most Plasmodiophora brassicae populations in single canola root galls from Alberta fields are mixtures of multiple strains. Plant Dis. 104:116–120. doi:10.1094/PDIS-06-19-1235-RE.
- Gossen BD, Al-Daoud F, Dumonceaux T, Dalton JA, Peng G, Pageau D, McDonald MR. 2019. Comparison of techniques for estimation of resting spores of Plasmodiophora brassicae in soil. Plant Pathol. 68:954–961. doi:10.1111/ppa.13007.
- Gossen BD, Strelkov SE, Manolii VP, Cao T, Hwang S-F, Peng G, McDonald MR. 2015. Spread of clubroot on canola in Canada, 2003–2014. Old pathogen, new home. Can J Plant Pathol. 37:403–413. doi:10.1080/07060661.2015.1105871.
- Jones DR, Ingram DS, Dixon GR. 1982. Characterization of isolates derived from single resting spores of Plasmodiophora brassicae and studies of their interaction. Plant Pathol. 31:239–246. doi:10.1111/j.1365-3059.1982.tb01274.x.
- Pageau D, Lajeunesse J, Lafond J. 2006. Impact de l’hernie des crucifères (Plasmodiophora brassicae) sur la productivité et la qualité du canola. Can J Plant Pathol. 28:137–143. doi:10.1080/07060660609507280.
- Peng G, Pageau D, Strelkov SE, Gossen BD, Hwang S-F, Lahlali R. 2015. A > 2-year crop rotation reduces resting spores of Plasmodiophora brassicae in soil and the impact of clubroot on canola. Eur J Agron. 70:78–84. doi:10.1016/j.eja.2015.07.007.
- Reyes AA. 1969. Detection of Plasmodiophora brassicae races 2 and 6 in Ontario. Plant Dis Rptr. 53:223–224.
- Reyes AA, Davidson TR, Marks CF. 1974. Races, pathogenicity and chemical control of Plasmodiophora brassicae in Ontario. Phytopathology. 64:173–177.
- Saude C, McKeown A, Gossen BD, McDonald MR. 2012. Effect of host resistance and fungicide application on clubroot pathotype 6 in green cabbage and napa cabbage. Hort Technol. 22:311–319. doi:10.21273/HORTTECH.22.3.311.
- Sedaghatkish A, Gossen BD, Yu F, Torkamaneh D, McDonald MR. 2019. Whole-genome DNA similarity and population structure of Plasmodiophora brassicae strains from Canada. BMC Genom. 20:744. doi:10.1186/s12864-019-6118-y.
- Somé A, Manzanares MJ, Laurens F, Baron F, Thomas G, Rouxel F. 1996. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 36:27–36.
- Strelkov SE, Hwang SF. 2014. Clubroot in the Canadian canola crop: 10 years into the outbreak. Can J Plant Pathol. 40(36):27–36. doi:10.1080/07060661.2013.863807.
- Strelkov SE, Hwang S-F, Manolii VP, Cao T, Feindel D. 2016. Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. Eur J Plant Pathol. 145:517–529. doi:10.1007/s10658-016-0888-8.
- Strelkov SE, Hwang SF, Manolii VP, Cao T, Fredua-Agyeman R, Harding MW, Peng G, Gossen BD, McDonald MR, Feindel D. 2018. Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can J Plant Pathol. 40:284–298. doi:10.1080/07060661.2018.1459851.
- Strelkov SE, Manolii VP, Cao T, Xue S, Hwang S-F. 2007. Pathotype classification of Plasmodiophora brassicae and its occurrence in Brassica napus in Alberta, Canada. J Phytopathol. 155:706–712. doi:10.1111/j.1439-0434.2007.01303.x.
- Strelkov SE, Tewari JP, Smith-Degenhardt E. 2006. Characterization of Plasmodiophora brassicae populations from Alberta, Canada. Can J Plant Pathol. 28:467–474. doi:10.1080/07060660609507321.
- Tewari JP, Strelkov SE, Orchard D, Hartman M, Lange RM, Turkington TK. 2005. Identification of clubroot of crucifers on canola (Brassica napus) in Alberta. Can J Plant Pathol. 27:143–144. doi:10.1080/07060660509507206.
- Williams PH. 1966. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology. 56:624–626.
- Xue S, Cao T, Howard RJ, Hwang S-F, Strelkov SE. 2008. Isolation and variation in virulence of single-spore isolates of Plasmodiophora brassicae from Canada. Plant Dis. 92:456–462. doi:10.1094/PDIS-92-3-0456.
- Ziesmann B 2019. Clubroot survey plan- 2019. Vol. 4. Crop Production News. https://www.saskatchewan.ca/business/agriculture-natural-resources-and-industry/agribusiness-farmers-and-ranchers/sask-ag-now/crops/crop-production-news/cpn-2019-issue-4/clubroot-survey.
