Abstract
Horse purslane (Trianthema portulacastrum L.) is a common weed in Mexico, where it competes with different agricultural crops and grows along watercourses, railways and roadsides, abandoned fields, and urban communities. In Sinaloa, this weed grows abundantly during the rainy season (July–September) and competes with summer crops such as corn and sorghum during the fall-winter season, and with common bean and corn mainly in alluvial soils. In recent years, horse purslane has suffered a 100% incidence of stem blight and leaf spot, with a diseased foliar area ranging from 35% to 80%. Ten isolates of an unknown fungus were obtained from symptomatic tissue and identified as Gibbago trianthemae Simmons, based on morphological features of the anamorph, with this identification confirmed in nine isolates by ITS rDNA sequence analysis. The identity of the fungus coincides with previous reports from other parts of the world, where it causes symptoms on horse purslane similar to those described here. In particular, due to its host specificity and aggressiveness towards horse purslane, G. trianthemae has been considered as a potential mycoherbicide. The pathogenicity of six fungal isolates was demonstrated according to Koch’s postulates. Our study provides new research directions for determining host specificity, obtaining aggressive strains, establishing the most appropriate inoculum levels, and demonstrating the influence of climatic conditions on the severity of the disease, which will help to assess the utility of the pathogen as a potential mycoherbicide.
Résumé
Le pourpier courant (Trianthema portulacastrum L.) est une adventice courante au Mexique où elle rivalize avec différentes cultures agricoles et pousse le long des cours d’eau, des voies ferrées et des routes ainsi que dans les champs en friche et les villes. Dans l’État du Sinaloa, cette adventice croît partout durant la saison des pluies (juillet à septembre) et rivalize avec les cultures d’été comme le maïs et le sorgho durant l’automne et l’hiver ainsi qu’avec le haricot commun et le maïs, principalement dans les sols alluviaux. Récemment, le pourpier courant a souffert d’une incidence élevée (100%) de pourriture noire et de tache foliaire, avec un taux de surface foliaire infectée de 35 à 80%. Dix isolats d’un champignon inconnu ont été obtenus de tissus symptomatiques et ont été identifiés en tant que Gibbago trianthemae Simmons, en se basant sur les traits morphologiques de la forme imparfaite. L’identification a été confirmée chez neuf isolats par l’analyse des séquences de l’ITS de l’ADNr. Le champignon correspond à celui décrit dans des rapports précédents provenant d’autres parties du monde où il provoque des symptômes chez le pourpier courant, semblables à ceux décrits ici. Notamment à cause de sa spécificité à l’égard de l’hôte et de son agressivité envers le pourpier courant, G. trianthemae a été considéré comme un mycoherbicide potentiel. La pathogénicité de six isolats fongiques a été démontrée en leur appliquant les postulats de Koch. Notre étude fournit de nouveaux axes de recherche pour déterminer la spécificité de l’hôte, obtenir des souches agressives, établir les taux d’inoculum les plus appropriés et démontrer l’influence des conditions climatiques sur la gravité de la maladie, ce qui contribuera à évaluer l’utilité de l’agent pathogène en tant que mycoherbicide potentiel.
Introduction
Horse purslane (Trianthema portulacastrum L.) is a branched, prostrate annual weed in the family Aizoaceae (Aneja et al., Citation2000), found in Africa and the Neotropics (Balyan and Bhan Citation1986) as well as in the United States (Mitchell Citation1988). Horse purslane is widely distributed in Mexico, where it competes with agricultural crops and grows along watercourses, railways and roadsides, abandoned fields, and urban communities (Calderon-Redzedowski and Redzedowski Citation2004). In Sinaloa, Mexico, the plant grows mainly during the rainy season (August and September).
Several foliar pathogens of horse purslane have been reported around the world, including Drechslera (Exserohilum) indica (J.N. Rai, Wadhwani & J.P. Tewari) Mouch. (Rao and Rao Citation1987), Colletotrichum gloesporoides (Penz.) Penz. & Sacc. ((Darshika and Daniel Citation1992), Alternaria alternata (Fr.) Keissl. (Gupta and Mukherji Citation2001; Bohra et al. Citation2005), and Phoma herbarum sensu Cooke fide Saccoardo (Ray and Vijayachadran Citation2013). Although Curvularia lunata R.R. Nelson & Haasis, C. tuberculata B.L. Jain, and Colletotrichum capsici (Syd.) Sacc. are associated with horse purslane in India, no disease has been induced by these fungal species. However, Bipolaris maydis Nisikado & Miyake, A. alternata and Gibbago trianthemae Simmons cause mild, moderate and severe symptoms, respectively (Gandipilli et al. Citation2017).
In recent years, the incidence of an undescribed foliar disease in horse purslane in Sinaloa, Mexico, has reached 100%, with the diseased leaf area ranging from 35% to 80%. Preliminary observations indicate that the disease is highly destructive during periods of high relative humidity (≥ 90%), especially when there is 12.5 mm to 37.5 mm of precipitation and a temperature range of 27–39°C, during August and September. Symptoms on the stems () appear as pinkish to maroon irregular lesions, mainly on senescent plants. Disease symptoms on the leaves start as maroon lesions of approximately 0.5 mm in diameter, which expand to 10–15 mm in diameter, with a straw colour in the centre surrounded by a maroon border (). Severely affected leaves become chlorotic, with defoliation occurring mainly in the lower parts of plants. Similar symptoms in horse purslane were previously reported in Cuba and Venezuela (Simmons Citation1986), the United States (Mitchell Citation1988), Pakistan (Akhtar et al. Citation2013), and India (Gandipilli et al. Citation2016; Kumar et al. Citation2016; Gandipilli and Ratnakumar Citation2018), and the causal agent was identified as G. trianthemae. As the aetiology of the disease is undescribed in Sinaloa, the aim of this research was to identify the cause of this stem blight and leaf spot of horse purslane based on morphology, molecular techniques, and pathogenicity tests.
Materials and methods
Sample collection and single-spore isolation
Ten symptomatic plants of horse purslane were collected from each one of ten sampling sites, 2 km apart from each other, in the municipality of Ahome, Sinaloa, between August 1st and September 30th, 2019. Since no agricultural crops were planted in the region during the sampling period, the samples were collected from railways and roadsides, abandoned fields, and urban communities. Twenty symptomatic leaves and stems (two leaves and stem pieces from each plant) were collected from each site. The fungus associated with the disease was isolated on 1.6% water agar (WA) following a previously described procedure (Reza and Motlagh Citation2010). The isolates of the unknown fungus were transferred to Petri dishes containing WA with sterilized purslane leaves on top (WAPL; 4 leaves per plate) and incubated for 10 days at 24°C to induce sporulation. To obtain monosporic isolates, three 5-mm mycelial disks with conidia close to the margins on the medium were cut and placed in a test tube with 3.0 mL sterile distilled water and mixed using a vortex for 20 s. Serial dilutions were prepared and spread on 1.6% WA, and individual spores were collected under a stereo microscope (Fisher Scientific model SPTI-ITH; Waltham, MA, USA) and transferred onto WA at 25°C. A few monosporic isolates were obtained per sampling site. However, we selected only one due to morphological similarity among isolates. A total of 10 isolates were kept in slanted WA medium covered with sterile mineral oil at 15°C for subsequent studies.
Presence of the fungus in leaf spots
Symptomatic leaves collected from the same sampling sites as for fungus isolation were wiped down with 70% ethyl alcohol, placed in Petri dishes containing moistened Whatman No. 1 filter paper, and incubated at 25°C for 5 days to induce sporulation of the fungus associated with the disease. After incubation, 20-mm sections of infected tissue were examined for the presence of conidiophores and conidia by direct observation under the stereo microscope (Zeiss Stemi Dv4, Pittsburgh, PA, USA).
Fungal colony morphology
Mycelial growth was determined for 10 monoconidial isolates obtained from horse purslane stems and leaves, cultured on potato dextrose agar (PDA; Bioxon; Cuautitlán Izcalli, Estado de Mexico, Mexico) prepared according to the manufacturer’s directions. Petri dishes were inoculated with a mycelial plug (5 mm in diameter) and incubated on a 14 h light/10 h dark schedule at 25°C. The radial growth in the medium was measured along two axes perpendicular to one another, at 2, 4, 6, 8, and 9 days after inoculation, in order to determine a final value (Kim et al. Citation2005). Colony colour and pigmentation were determined according to colour standards and nomenclature (Ridgway Citation1912) in 10-day-old cultures.
Conidiophore and conidia morphology
Conidiophore and conidial measurements were determined for the 10 isolates. Plugs (5 mm in diameter) from monoconidial cultures were cut from the margins of 5-day-old colonies on WA and transferred onto the PDA. After 10 days of incubation in 14 h light/10 h dark at 25°C, the length and width of 35 conidiophores per isolate were determined at the midpoint. In addition, conidial shape, as well as the number of transverse and longitudinal septa of 35 conidia from each of the isolates, was observed. In order to assure similar developmental stages and uniformity of conidia, samples were taken inside the colony at ~20 mm from the colony margin, mounted in distilled water, and measured at 400X using a compound microscope (Carl Zeiss, Model Axio Imager A2, White Plains, NY, USA). The presence of secondary conidia was determined following previously described procedures (Simmons Citation1986; Gandipilli and Ratnakumar Citation2017). Species identification relied on previous reports dealing with morphological characteristics for this taxonomic level (Simmons Citation1986; Akhtar et al. Citation2013; Kumar et al. Citation2016).
Molecular identification of isolates
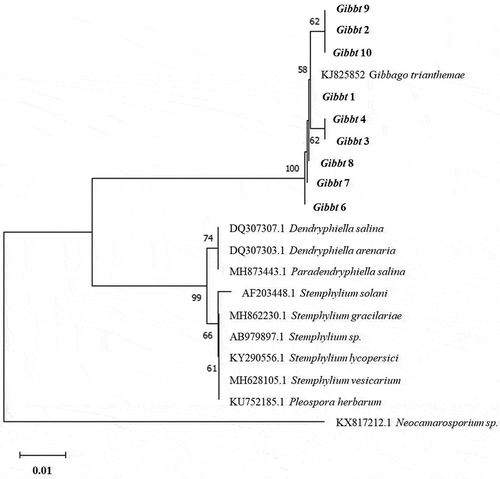
Genomic DNA was extracted from nine isolates in DNAzol® following the manufacturer’s instructions. The ITS rDNA region was amplified using the universal primers ITS1 (5ʹ TCCGTAGGTGAACCTGCGG 3ʹ) and ITS4 (5ʹ TCCTCCGCTTATTGATATGC 3ʹ). PCR reactions were performed in 25-μL volumes containing a 10-ng DNA template, 17.25 μL of ultrapure water, 2.5 μL of 10X PCR reaction buffer (200 mM Tris-HCl pH 8.4, 500 mM KCl), 0.75 μL MgCl2 (50 μM), 1.25 μL of dNTPs (10 mM), 1.0 μL of each primer (10 μM), and 0.25 μL (1 U) of Taq DNA polymerase (Invitrogen Brazil, Cat. No. 11 615–050). The thermocycler program included: an initial denaturation at 95°C for 4 minutes; 34 cycles (each for 1 min) of denaturation at 95°C, annealing at 55°C, and extension at 72°C; and a final extension at 72°C for 5 min. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Cat. No. 28 106) and quantified using a Nanodrop 2000. PCR products were sequenced at the National Laboratory of Genomics and Biodiversity (LANGEBIO, Irapuato, Mexico) and compared to sequences in the NCBI (National Centre for Biotechnology Information) database using the BLAST software and the megablast algorithm. MEGA X was used for alignment and phylogenetic analysis. Partial 314-bp rDNA sequences (partial its1 [106 nt]-5.8S [158 nt]-partial its2 region [50 nt]) were aligned together with Gibbago/Stemphylium reference sequences (Kumar et al. Citation2016) using the ClustalW alignment program. Multiple alignments were subjected to a DNA substitution model analysis in MEGA in order to select the best-fitting model. A phylogenetic tree was constructed using the Kimura 2-parameter (K2P) model and the neighbour joining (NJ) method. Among site rate variation was modelled by a gamma distribution (+G). Tree topology support was assessed by 1000 bootstrap replicates.
Pathogenicity testing
Horse purslane seeds were collected in the field during the last week of September 2019. Seeds were placed in 0.5 NaOCl for 2 min and rinsed 3 times with distilled water, and seeded in a greenhouse in 3 kg plastic pots (five seeds per pot) containing a pasteurized mixture of clay:loam:sand (at a ratio of 28.1:27.0:44:9) at pH 7.0. Upon germination, seedlings were thinned and one plant was left per pot. Plants were fertilized every 2 weeks with Miracle Gro® fertilizer (The Scotts Company LLC; Marysville, Ohio, USA) according to the manufacturer’s recommendations and watered as needed in the greenhouse.
Inoculum was prepared by growing six monoconidial fungal isolates on 1.6% WA with five autoclaved purslane leaves on the surface (WAPL). After a 2-week incubation in 14 h light/10 h dark at 25°C, the mycelium and conidia of each isolate were suspended in water and filtered through two layers of cheesecloth to remove the mycelia (Prieto et al. Citation2016). Conidia were then counted using a hematocytometer, and the suspension density was adjusted to 5 × 105 conidia/mL.
The pathogenicity of the six monoconidial fungal isolates was tested using 1.5-month-old plants grown in 3 kg plastic pots. Each conidial suspension was sprayed separately on four horse purslane plants. Four control plants were sprayed with sterile distilled water. After inoculation, the plants were placed in clear plastic bags to ensure high relative (85%) humidity (RH) for 48 h. Relative humidity was measured using a thermo-hygrometer (Taylor, Model 1523). Inoculated and control plants were arranged in a completely randomized design in the greenhouse with four replications (four pots, each with one plant). Afterwards, plants were subjected to the 85% RH for five days during 12 h per day. The pathogenicity and relative virulence of the isolates were determined 14 days after inoculation. The foliage area diseased (FAD; in cm2) was measured on 10 leaves randomly taken from each inoculated plant, following previously described procedures (Zadoks and Schein Citation1979). The percentage of FAD was determined based on the total leaf area of the replicate plants. In order to standardize the data, percentages of FAD were transformed into arcsine values as previously described (Gomez and Gomez Citation1984). The transformed data were analysed using ANOVA, and the mean separation was achieved following Tukey’s test (Little and Hills Citation1973). The greenhouse experiment was conducted twice. Since the data from the two experiments were qualitatively similar, only the data from one single experiment are presented. In order to fulfil Koch’s postulates the fungus was isolated from lesions on inoculated plants at the end of the experimental periods, and its identity was confirmed using colony, conidiophore and conidial morphometry, and twenty conidia were measured for each isolate. Greenhouse temperatures ranged from 24°C to 36°C in the first experiment, and from 21°C to 32°C in the second experiment.
Results
Ten isolates of an unknown fungus were obtained from symptomatic horse purslane plants. Natural infections on stems occurred mainly on senescent plants, and symptoms in the leaves occurred regardless of the stage. Five isolates were obtained from stems and five from leaves, one isolate each from 10 sampling sites in the municipality of Ahome, Sinaloa, Mexico.
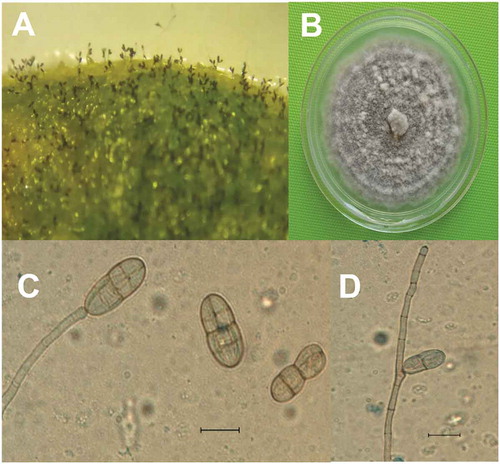
Five days after incubation of infected horse purslane leaf tissue in moist chambers, microscopic observations indicated the presence of conidiophores and conidia on the leaf surface (). Conidiophore and conidia measurements as well as morphology on the leaf surface were similar to those obtained on PDA. On PDA, the colonies of 6-day cultures appeared velvety grey with faint zonation (), and mycelial growth rate ranged from 18.6 mm to 22.3 mm per day. Primary conidiophores were simple, light brown in colour, slightly swollen at the apex (), and geniculated () comprised 1–6 septa. Mature conidia did not present any basal scars, nor did they exhibit any pigmentation halo at this point. Instead, they were muriform; smooth and ranging from broadly ellipsoid to broadly sub-ovate-ellipsoid in shape; light brown in colour; and appearing single or (rarely) in chains of two. Conidia were simple, with 2–5 transepta and constriction at the midpoint, and 1–6 longisepta (). Apical and basal cells of primary conidia germinated producing secondary conidiophores, giving rise to secondary conidia that were morphologically similar to primary conidia. The morphological characteristics of the fungal isolates were similar to those of Gibbago (), as described by Simmons (Citation1986).
Table 1. Morphological characteristics and measurements of asexual structures of ten isolates of Gibbago trianthemae obtained from Trianthema portulacastrum in Sinaloa, Mexico
Fig. 2 Gibbago trianthemae. (a) Conidiophores and conidia on infected tissue; (b) Colony on PDA; (c) Conidiophore with apical swelling bearing conidium and detached conidia; (d) Geniculated conidiophore. Scale bar = 20 µm
Comparisons with the GenBank database revealed an identical sequence coverage (100%) and percentage similarity (99–100%) with G. trianthemae at the partial ITS1-5.8S-ITS2 rDNA region analysed. A phylogenetic tree built with the partial ITS rDNA region showed that all nine isolate sequences used for the analysis were located within the same clade and grouped with G. trianthemae, and that this clade was clearly separated from closely related genera with a similar morphology (). Sequences were submitted to the National Centre for Biotechnology Information (Bethesda, MD, USA) with GenBank accession numbers MN960438-MN960446 (MN960438-MN960442: isolates Gibbt 10 through Gibbt 6 in descending order; and MN960443-MN960446: isolates Gibbt 4 through Gibbt 1 in descending order). These findings support the morphological analysis of the fungal isolates from this work and confirm the identity of the pathogen as G. trianthemae.
Fig. 3 Neighbour joining tree based on the partial internal transcribed spacer region of Gibbago trianthemae specimens. The tree was constructed with MEGA X (bootstrap = 1000), using the Kimura 2-parameter model and a gamma distribution (+G) to model the among-site rate variation. The corresponding sequence of Neocamarosporium sp. was used as an outgroup. Database accession numbers of the sequences are provided in parentheses. Bootstrap values are given as percentages. Nine isolate sequences obtained in this work were included in the tree and are shown in bold. The scale bar indicates the number of nucleotide substitutions per site
All six isolates of G. trianthemae (Gibbt 1 through Gibbt 6) were pathogenic on horse purslane. Variation in virulence was detected among the isolates. The first symptoms (pinpoint maroon lesions 0.5–1.0 mm in diameter) were observed five days after inoculation. At day 14, lesions on leaves were circular (8–10 mm in diameter) and surrounded by a maroon halo (), similar to those observed under field conditions. In experiment 1, isolates Gibbt 5 and Gibbt 6 were significantly (F = 65.08; P < 0.0001; CV = 24.08) more virulent than the other isolates. Gibbt 1 and Gibbt 3 showed moderate virulence, and isolates Gibbt 2 and Gibbt 4 were the least virulent. Qualitative similar results (data not shown) were obtained in experiment 2 which was confirmed with one-way ANOVA (F = 35.34; P < 0.0001; CV = 30.44). Control plants sprayed with sterile distilled water remained asymptomatic during the study (, ). In order to fulfil Koch’s postulates, the fungal pathogen was isolated from inoculated plants and its identity was confirmed by comparing colony conidiophores as well as conidial morphology with the original isolates.
Table 2. Pathogenicity of Gibbago trianthemae in horse purslane plants
Discussion
Conidiophore and conidia morphology of the fungal isolates obtained from symptomatic horse purslane stems and leaves were similar to those in the genera Alternaria, Embellisia, Stemphylium, and Ulocladium (Simmons Citation1986). However, the lack of a basal pigmented halo and a conspicuous scar around the conidium’s detachment point from the conidiophore as well as the unique pattern of secondary sporulation, allowed us to identify the pathogen as G. trianthemae (Simmons Citation1986). Furthermore, conidiophore and conidial measurements and colony morphology are within the range for this fungal species (Simmons Citation1986; Kumar et al. Citation2016; Gandipilli et al. Citation2017). Identification of the pathogen was complemented by pathogenicity tests in which the isolates caused symptoms in horse purslane similar to those observed in natural infections under field conditions. These observations coincide with symptoms caused by the same fungus in the same plant species in other parts of the world (Akhtar et al. Citation2013; Kumar et al. Citation2016). Although perithecia have been found associated with infected tissue in India (Gandipilli and Ratnakumar Citation2017), the teleomorph of the fungus was not observed during the sampling period in Sinaloa, Mexico. Phylogenetic analysis comparing the G. trianthemae isolate sequences with previous sequences of this pathogen (Kumar et al. Citation2016) and species belonging to Stemphylium, Dendryphiella, and Pleospora confirmed morphological observations, and the identity of the sequenced isolates was established as G. trianthemae.
In this study, isolates Gibbt 5 and Gibbt 6 were the most virulent, with FAD values of 58% and 53.8%, respectively, 14 days after inoculation. Aneja et al. (Citation2000) reported that isolates of G. trianthemae from India could cause a FAD of 84% at day 30 post-inoculation showing how severe this pathogen can be on horse purslane.
This is the first report of G. trianthemae causing the disease in horse purslane in Mexico. As the fungus has a specific pathogenicity to this host (Mitchell Citation1988; Kumar et al. Citation2016), this characteristic makes it a potential candidate as a mycoherbicide, as stated in previous reports (Kumar et al. Citation2016; Gandipilli et al. Citation2016). Future research should focus on exploring the role for G. trianthemae in agriculture. In order to evaluate this pathogen as a potential mycoherbicide (Aneja et al., Citation2000), several lines of work will be required, including: the selection of the most virulent strains of the pathogen in Sinaloa optimization of sporulation (Gandipilli and Ratnakumar Citation2018); determining proper inoculation techniques in the field; environmental conditions influencing disease severity; and the confirmation of host specificity.
Acknowledgements
We thank Dr. Brandon Loveall from Improvence editing services for English proofreading of the manuscript.
Additional information
Funding
References
- Akhtar KP, Sarwar N, Saleem K, Ali S. 2013. Gibbago trianthemae causes Trianthema portulacastrum (horse purslane) blight in Pakistan. Australas Plant Dis Notes. 8(1):109–110. doi:10.1007/s13314-013-0108-8
- Aneja KR, Khan SA, Kaushal S 2000. Management of horse purslane (Trianthema portulacastrum L.) with Gibbago trianthemae Simmons in India. Proceedings of the X international symposium on biological control of weeds. 1999 July 4–14; Montana, United States; p. 27–33.
- Balyan R, Bhan V. 1986. Emergence, growth, and reproduction of horse purslane (Trianthema portulacastrum) as influenced by environmental conditions. Weed Sci. 34(4):516–519. doi:10.1017/S0043174500067345
- Bohra B, Vyas BN, Godrej NB, Mistry KB. 2005. Evaluation of Alternaria alternata (Fr.) Keissler for biological control of Trianthema portulacastrum L. Indian Phytopathol. 58:184–188.
- Calderon-Redzedowski G, Redzedowski J. 2004. Flora del Bajío y de regiones adyacentes. Manual de malezas de la región de Salvatierra, Guanajuato, México. Fascículo complementario XX. Diciembre. Xalapa, Mexico: Instituto de Ecología, A.C.; p. 315.
- Darshika P, Daniel M. 1992. Changes in chemical content of Adhatoda and Trianthema due to fungal diseases. Indian J Pharmaceut Sci. 54:73–77.
- Gandipilli G, Bonthu S, Geddada ER. 2017. Mycoflora of Trianthema portulacastrum L., an invasive weed of Andra Pradesh. Int J Curr Adv Res. 6:4244–4248.
- Gandipilli G, Kumar PKR, Pilaka B. 2016. Selection of some fungal pathogens for biological control of Trianthema portulacastrum L., a common weed of vegetable crops. J Appl Biol Biotechnol. 4:90–96.
- Gandipilli G, Ratnakumar PK. 2017. In vitro screening of foliar pathogens for biological control of horse purslane weed. Indian J Exp Biol. 55:389–395.
- Gandipilli G, Ratnakumar PK. 2018. Factors influencing growth and sporulation of Gibbago trianthemae, a prospective biocontrol agent. Indian J Exp Biol. 56:847–852.
- Gomez KA, Gomez AA. 1984. Statistical procedures for agricultural research. 2nd ed. New York: John Wiley and Sons; p. 704.
- Gupta R, Mukherji KG. 2001. Environmental effect on the occurrence of Alternaria alternata on Trianthema portulacastrum. J Env Biol. 22:83–86.
- Kim YK, Xiao CL, Rogers JD. 2005. Influence of culture media and environmental factors on mycelial growth and pycnidial production of Sphaeropsis pyriputrescens. Mycologia. 97(1):25–32. doi:10.1080/15572536.2006.11832835
- Kumar V, Kumar N, Aneja KR, Kaur M. 2016. Gibbago trianthemae, phaeodictyoconidial genus, cause leaf spot disease of Trianthema portulacastrum. Arch Phytopathol Pflanzenschutz. 49(1–4):48–58. doi:10.1080/03235408.2016.1152066
- Little TM, Hills FJ. 1973. Agricultural experimentation: design and analysis. New York: John Wiley and Sons; p. 368.
- Mitchell JK. 1988. Gibbago trianthemae, a recently described hyphomycete with bioherbicide potential for control of horse purslane (Trianthema portulacastrum). Plant Dis. 72(4):354–355. doi:10.1094/PD-72-0354
- Prieto KR, de Maderos LS, Isidoro MM, Toffano L, da Silva MFGF, Fernández JB, Vieira PC, Forim MR, Rodrigues-Filho E, Stuart RM, et al. 2016. Rapid determination of ACTG-and AK-toxins in Alternaria alternata by LC-ESI-MS/MS analysis and antifungal properties of citrus compounds. J Braz Chem Soc. 27:1493–1505.
- Rao AP, Rao AS. 1987. New fungal diseases of some weeds. Indian Bot Rep. 6:38.
- Ray P, Vijayachadran LS. 2013. Evaluation of indigenous fungal pathogens from horse purslane (Trianthema portulacastrum) for their relative virulence and host range assessments to select a potential mycoherbicidal agent. Weed Sci. 61(4):580–585. doi:10.1614/WS-D-12-00076.1
- Reza M, Motlagh S. 2010. Isolation and characterization of some important fungi from Echinochloa spp. the potential agents to control rice weeds. Aust J Crop Sci. 4:457–460.
- Ridgway R. 1912. Color standards and color nomenclature. Baltimore: A. Hoens and Company; p. 42.
- Simmons EG. 1986. Gibbago, a new phaeodyctioconidial genus of hyphomycetes. Mycotaxon. 27:107–111.
- Zadoks JC, Schein RD. 1979. Epidemiology and plant disease management. New York: Oxford University Press; p. 427.

