Abstract
Alternaria panax (sect. Panax) is reported to be the main causal agent of alternaria blight on ginseng (Panax ginseng) in Asia. However, the species distribution causing ginseng alternaria blight in China has not been examined since 1964. Two hundred fifty-seven Alternaria isolates were obtained from 2015 to 2019 from 13 ginseng-producing areas in Jilin province, China. Based on morphological characteristics and sequence analysis of the internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), RNA polymerase second largest subunit (RPB2), translation elongation factor (TEF), and Alternaria major allergen (Alt a 1) genes, 248 (96.5%) of the isolates were identified as A. alternata and A. tenuissima (sect. Alternaria) and nine (3.5%) isolates were classified as A. panax (sect. Panax). To our knowledge, this is the first report of A. alternata (sect. Alternaria) as the primary cause of alternaria blight on ginseng in Jilin province of China.
Résumé
On rapporte qu’Alternaria panax (sect. Panax) est le principal agent causal de l’alternariose chez le ginseng (Panax ginseng) en Asie. Toutefois, la distribution de l’espèce causant l’alternariose chez le ginseng en Chine n’a pas été revue depuis 1964. De 2015 à 2019, 257 isolats d’Alternaria ont été obtenus de 13 régions productrices de ginseng de la province du Jilin, en Chine. En se basant sur les caractéristiques morphologiques et l’analyse de la séquence des régions de l’espaceur transcrit interne (ITS) de l’ADN ribosomique (ADNr), sur la glycéraldéhyde-3-phosphate déshydrogénase (GAPDH), la deuxième plus grande sous-unité de l’ARN polymérase (RPB2), le facteur d’élongation de la transcription (TEF) et les principaux gènes de l’allergène d’Alternaria (Alt a 1), 248 (96,5%) isolats ont été identifiés en tant qu’A. alternata et A. tenuissima (sect. Alternaria), et 9 isolats (3,5%) ont été classifiés en tant qu’A. panax (sect. Panax). À notre connaissance, il s’agit de la première mention d’A. alternata (sect. Alternaria) comme cause principale de l’alternariose chez le ginseng dans la province du Jilin, en Chine.
Introduction
Ginseng, Panax ginseng C. A. Meyer, has been used for thousands of years as an important medicinal plant in East Asia (Park et al. Citation2012). It is a perennial plant of the Araliaceae family, and its roots are highly valued for medicinal and nutritional uses (Kim et al. Citation2010). Ginseng is widely used as a traditional medicine to improve human immune and metabolic systems. In China, P. ginseng is planted mainly in the northeast of the country, especially in Jilin province (Wang et al. Citation2018). Ginseng crops may suffer from significant biotic and abiotic stresses, as noted by Kim et al. (Citation2019), of which fungal diseases are the main source of biotic stresses, and the effects of these stresses can be exacerbated by the plant’s long cultivation period (four to six years). The major fungal and fungal-like diseases of ginseng limiting production in China and Korea are alternaria blight (Alternaria panax Whetzel), anthracnose (Colletotrichum gloeosporioides Penz.), damping-off (Pythium debaryanum Hesse, Pythium ultimum Trow, and Rhizoctonia solani Kühn), root rot (Fusarium redolens Wollenw, F. cerealis (Cke.) Sacc. and Cylindrocarpon destructans (Zinsm.) Scholten), grey mould (Botrytis cinerea Pers.: Fr. and B. pelargonii Roed.), and phytophthora blight (Phytophthora cactorum (Leb. et Cohn.) Schrot.) (Reeleder Citation2003; Kim et al. Citation2010; Gao et al. Citation2014; Guan et al. Citation2014; Lu et al. Citation2019).
Species of Alternaria are ubiquitous fungal pathogens and saprophytes occurring worldwide in different habitats. The genus Alternaria contains important pathogens of several vegetable, oil, cereal and medicinal plants, including both ginseng (P. ginseng) and American ginseng (P. quinquefolius L.). Disease caused by A. panax was first found on P. quinquefolius in the early 1900s (Nakata and Takimoto Citation1922), and on P. ginseng in 2013 (Deng et al. Citation2013).
Alternaria blight normally displays symptoms of circular or irregular necrotic lesions surrounded by chlorotic zones. Infection of the host can occur on leaves. However, the fungus can affect all parts of the plant through stems, fruits and roots, and has the potential to spread rapidly because of the production of large numbers of conidia resulting in secondary infection (Deng et al. Citation2013).
Alternaria blights are among the most commonly occurring and serious diseases in ginseng cultivation, with a typical incidence of 20–30%, and reports of 100% from Korea (Deng et al. Citation2013; Wei et al. Citation2018). It is generally believed that the causal agent of alternaria blight of ginseng is A. panax, but research into the possible involvement of other Alternaria species in causing this disease in Jilin province, China, has not been conducted. The purpose of this study was to determine the species and pathogenicity of isolates of Alternaria spp. associated with ginseng in Jilin province of northeastern China.
Materials and methods
Collection of samples, isolation, and purification
Over 400 samples comprised seeds, leaves, stems, and roots showing disease symptoms were collected from the cities of Baishan, Changchun, Dunhua, Jiaohe, and Tonghua, the main ginseng-producing areas in Jilin province (, and S1) during 2015 to 2019 (). Symptomatic leaf, stem, seed or root tissues were cut into small pieces (5 × 5 mm) and surface-disinfected with 70% ethanol for 40 s, disinfected with 1% sodium hypochlorite (NaClO) for 1 min, then rinsed three times with sterile distilled water and placed into Petri dishes containing potato dextrose agar (PDA) (Liu et al. Citation2017). The PDA dishes were then incubated at 25 °C without light for 7 days. Pure cultures were obtained from single spores (Al-Nadabi et al. Citation2018).
Table 1. Lists of 26 representative isolates of three Alternaria spp. collected from ginseng in Jilin province in China with details about hosts, isolates source, date, origins, pathogenicity and Genbank accession numbers
Table 2. Sequences of Alternaria isolates used in the phylogenetic study
Table 3. Summary of morphological comparison of three Alternaria spp
Table 4. Geographical distribution of Alternaria spp
Morphological identification
The preliminary screening for Alternaria-like fungi was conducted following Basim et al. (Citation2017), and other fungi were omitted. A total of 257 putative Alternaria cultures were obtained and maintained on PDA at 25°C. To characterize the sporulation pattern of Alternaria species, isolates were incubated on potato carrot agar (PCA) at 25°C for 7 days. The colony morphology was then examined, including colony colour, margin, and texture (Deng et al. Citation2013). The size and morphological characteristics of conidia (n = 50) and the sporulation patterns on PCA were examined by light microscopy (Nikon, Tokyo, Japan) and photographed. The images were captured with a Digital Sight DS-5 M digital camera (Nikon, Tokyo, Japan) using the NIS-elements F Package version 3.0.
DNA extraction, PCR, and sequencing
Pure cultures of 26 selected isolates representing different tentative species and different locations () were incubated on PDA at 25°C for 7 days. To extract total genomic DNA, approximately 15 mg (fresh weight) of mycelia from each isolate was harvested by scraping the colony surface with an inoculation loop. The mycelia were ground in liquid nitrogen, and transferred to a 1.5 mL microcentrifuge tube containing 800 μL cetyltrimethylammonium bromide (CTAB) extraction buffer [2% (w/v) CTAB, 200 mM Tris-HCl, pH 8.0, 20 mM EDTA, pH 8.0, 1.4 M NaCl, 1% (w/v) polyvinylpyrrolidone, 1% (v/v) β-mercaptoethanol]. The extraction was performed following Adachi et al. (Citation1993). Finally, the DNA pellet was dissolved in 50 μL of Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) and stored at −20°C until further use.
Identification of Alternaria species was based on a combination of five different genomic regions. The internal transcribed spacer (ITS) region of ribosomal DNA was amplified using ITS4 and ITS5 primers (White et al. Citation1990); the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) region was amplified using gpd 1 and gpd 2 primers (Berbee et al. Citation1999); the RNA polymerase second largest subunit (RPB2) region was amplified using RPB2-5F2 and RPB2-7cR primers (Liu et al. Citation1999); the translation elongation factor (TEF) region was amplified using EF1-728 F and EF1-986 R primers (Carbone et al. Citation1999); and the Alternaria major allergen gene (Alt a 1) was amplified using Alt-for and Alt-rev primers (Hong et al. Citation2005). All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). PCR amplification was performed in a 25 μL volume containing 10.5 μL ddH2O, 12.5 μL Premix Ex Taq (Version 2.0, TaKaRa, containing 0.625 U DNA polymerase, 200 μM dNTP mixture, and 1.5 mM Mg2+), 0.5 μL each of the two primers (10 μM), and 1.0 μL DNA template (100 μg/mL). Negative controls containing the same reagents, but without the DNA template were also included in all PCR reaction sets. The amplification conditions for the ITS gene consisted of a denaturation step at 95°C for 5 min, followed by 40 cycles of 30 s at 94°C, 30 s at 51°C and 45 s at 72°C with a final elongation step of 7 min at 72°C. The PCR conditions for the TEF and GAPDH genes consisted of a denaturation step at 95°C for 5 min, followed by 40 cycles of 30 s at 94°C, 30 s at 54°C and 45 s at 72°C with a final elongation step of 7 min at 72°C. The cycle for the partial RPB2 gene consisted of a denaturation step at 95°C for 5 min, followed by 40 cycles of 30 s at 94°C, 30 s at 58°C and 45 s at 72°C with a final elongation step of 7 min at 72°C (Wang et al. Citation2019). The cycle for the Alt a 1 gene consisted of a denaturation step at 95°C for 5 min, followed by 40 cycles of 30 s at 94°C, 30 s at 57°C and 60 s at 72°C with a final elongation step of 7 min at 72°C (Woudenberg et al. Citation2014). The amplified products were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China).
Phylogenetic analysis
Phylogenetic analyses using MEGA 5.0 were based on Maximum Likelihood analyses of concatenated sequences of five genomic regions (ITS, TEF, GAPDH, RPB2 and Alt a 1) for 26 isolates obtained in this work () plus concatenated sequences of 45 other strains of Alternaria spp. obtained from the NCBI website (; ), included in Woudenberg et al. (Citation2015) and Deng et al. (Citation2013). These sequences were aligned by MUSCLE (Edgar Citation2004), and the alignments were manually adjusted. Maximum likelihood (ML) analysis was used to construct the phylogenetic trees based on the multi-locus (ITS, GAPDH, RPB2, TEF, and Alt a 1) sequences. Phylogenetic trees were constructed using MEGA v.5 (Park et al. Citation2012). Percentage bootstrap support >50% (1,000 replications) by the ML analysis is shown on the respective branch. The resulting trees were visualized in FigTree v. 1.4.2 (Rambaut Citation2014).
Pathogenicity tests
The wounded detached and attached leaves were initially used to provide a rapid pre-screening. Detached leaves of 3-year-old ginseng plants were placed on wet filter paper in 9-cm diameter Petri dishes. Hyphal plugs (8 mm diameter) were inoculated, mycelium side down, on detached leaves of 3-year-old ginseng plants. Isolates were propagated on PDA for 7 days at 25°C without light. The leaves were lightly wounded with a fine sterile needle before inoculation. Mock-inoculated leaves were wounded, and clean agar plugs were used. Attached leaves of 3-year-old ginseng plants were also inoculated with hyphal plugs for 26 of the isolates in a similar manner. Pathogenicity tests were conducted on nine leaves per isolate (one set for attached and one set for detached). Each of the leaves was inoculated with four hyphal plugs, and kept in a greenhouse with a temperature range from 22°C to 25°C, a 12 h photoperiod of fluorescent light and relative humidity of 90%. At 7 days, the symptoms were recorded. The pathogen was re-isolated from symptomatic leaf tissues and identified using both morphological and molecular techniques. Each pathogenicity experiment was repeated three times.
Conidial inoculation assays were also conducted. Isolates were propagated as above, and conidia were harvested with sterile distilled water and passed through two layers of cheesecloth to remove debris and mycelium. The conidial suspension was adjusted to a concentration of 106 spores/ml with sterile deionized water (Liu et al. Citation2017). Detached stems, seeds or roots from 3 year old ginseng plants and intact plants were used to assess the pathogenicity of Alternaria species. Twenty-six isolates of Alternaria species in sect. Alternaria and sect. Panax (A. alternata, sect. Alternaria: JH3-4, DH2-6, DH2-12, FS3-5, FS3-10, BSGJ2, SJHJ3, SJHJ5, YC1, YC8, YC9, YC10, BQSZ6, BQSZ9; A. tenuissima, sect. Alternaria: JY25, JY30, YC3, YC6, JLCB1, JLCB2, JLCB3, JLCB4, 3JP9; A. panax, sect. Panax: CPA, CPB, CR2) were used in pathogenicity tests (). Controls were mock inoculated with sterile distilled water. Six detached stems, seeds, and roots, and six whole plants were inoculated with each isolate by spraying the conidial suspension following Liu et al. (Citation2020). The detached stems, seeds and roots were placed on wet filter paper in 9-cm diameter Petri dishes and were inoculated as above. The whole plants were covered with plastic bags for 7 days and kept in a greenhouse with a temperature range from 22°C to 25°C, a 12 h photoperiod of fluorescent light and relative humidity of 90%. The symptoms were recorded and photographed after 13 days. The pathogen was re-isolated from the symptomatic tissues and identified using both morphological and molecular techniques. Each pathogenicity experiment was repeated two times.
Results
Alternaria species associated with ginseng
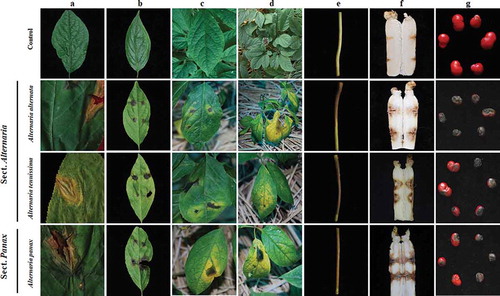
Symptoms of alternaria blight were seen on most of the leaves as circular or irregular necrotic lesions surrounded by chlorotic zones (), while on stems the lesions were fusiform-shaped and brown with chlorotic zones (). Seeds displayed a brown and black discoloration of the seed coat (), in contrast to healthy seeds, which were red (). Symptoms on roots were black brown rots ().
Fig. 2 (Colour online) Morphological characteristics of species of Alternaria isolated from ginseng. a, g, m, Colony morphology on PDA incubated for 7 days at 25°C. b, h, n, Colony morphology on PCA incubated for 7 days at 25°C. c, i, o–p conidia; d, j, q–r conidiophores; e, f, k, l, s, t conidia chains. a–f, Alternaria alternata. g–l, A. tenuissima. m–t, A. panax. Scale bars: 20 μm
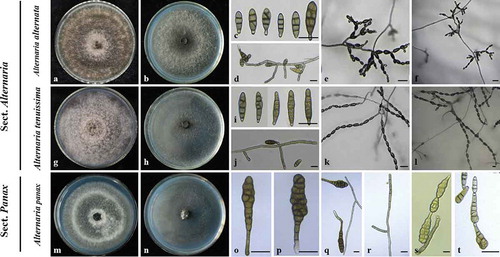
Fig. 3 (Colour online) Pathogenicity of representative isolates of three Alternaria species on ginseng. a, Natural symptoms of Alternaria spp. on the leaves of ginseng. b–c, Symptom development on detached and attached Panax ginseng leaves wounded with a fine sterile needle and inoculated with agar plugs of Alternaria mycelium at 7 days post inoculation. d, Symptom development on attached P. ginseng leaves, which were droplet-inoculated with Alternaria species conidial suspension at 13 days post inoculation. e–g, Symptom development on detached P. ginseng stems, roots and seeds at 7 days post inoculation. The first line showed the different parts of ginseng were inoculated with sterile water control. The second to the last line shows the different parts of ginseng inoculated with Alternaria alternata, A. tenuissima, and A. panax.
The 257 cultures were initially identified as putative Alternaria spp. based on morphological and cultural characteristics, following descriptions of Deng et al. (Citation2013). Alternaria spp. represented 61% of the fungal isolations from affected plants with typical symptoms of alternaria blight.
Morphological characterization and identification
The Alternaria isolates were separated into two main groups, based on morphological characteristics and the size of conidia. The first group (the small-spored Alternaria) contained two morphotypes, and the second group (the large-spored Alternaria) contained one morphotype. In the first group, one morphotype consisted of 181 isolates, and these produced cottony, olive brown colonies with slight concentric growth rings on PDA after incubation at 25°C for 7 days without light ( and ). On PCA, these isolates produced conidial chains of 5‒10 conidia with numerous secondary chains branching on short conidiophores like short trees (). Conidia were obpyriform to ellipsoid (27.1 ± 6.6 × 9.7 ± 1.9 µm). The number of transverse septa and longitudinal septa of conidia varied from 1 to 4 and from 0 to 3, respectively (). Based on these features, isolates in this group were identified as A. alternata (Fries) Keissler ( and ), and these characteristics were consistent with those described by Nishikawa and Nakashima (Citation2019).
There were 67 isolates for the second morphotype in the first group, which were loosely cottony and greyish-green to olive brown colonies on PDA ( and ). These isolates produced chains of 8–12 conidia with occasional secondary and tertiary chains branching from apical and median cells on PCA (–l). Conidia were ovoid to obclavate (31.6 ± 6.8 × 9.9 ± 1.6 μm), with 2–7 transverse and 0–3 longitudinal septa (–j and ). These characteristics corresponded to A. tenuissima (Nees & Nees: Fries) Wiltshire as described by Wang et al. (Citation2019). Alternaria alternata and A. tenuissima were placed in sect. Alternaria-(the first group), which contains most of the small-spored Alternaria species with concatenated conidia (Woudenberg et al. Citation2015).
In the second group, cultures of the 9 isolates were pale grey, normally loose and cottony with smooth, round margins and did not secrete pigment on PDA after 7 days at 25 °C ( and ). On PCA, these isolates produced chains of 2–3 conidia from apical and median cells (–t). Conidia were obclavate with ture beaks (95.8 ± 17.5 × 27.3 ± 5.6 μm), with 1–2 longitudinal and 5–13 transverse septa (–q and ). These isolates were classified as A. panax in agreement with descriptions by Deng et al. (Citation2013) and Hill (Citation2009), placed inside sect. Panax (Woudenberg et al. Citation2015).
Alternaria alternata (sect. Alternaria) was the most prevalent (70.4%), followed by A. tenuissima (sect. Alternaria) (26.1%), but both were detected in almost all surveyed regions (). Alternaria panax (sect. Panax) (3.5%) was less common () and was found in just three regions: Fusong, Jingyu, and Changchun ().
Phylogenetic analyses
Among the 257 Alternaria isolates, 26 purified isolates were further identified based on sequence analysis of the ITS, TEF, GAPDH, RPB2, and Alt a 1 genes. These were selected to represent different morphotypes and locations ().
Fourteen strains (SJHJ5, YC10, FS3-5, DH2-12, BQSZ6, BSGJ2, FS3-10, SJHJ3, BQSZ9, DH2-6, JH3-4, YC1, YC8, YC9) obtained from ginseng clustered in the A. alternata and formed a distinct clade (ML bootstrap support value 99) as sister to A. tenuissima (). Nine isolates (3JP9, JLCB1, JLCB2, JLCB3, JLCB4, JY25, JY30, YC3, YC6) were recognized as A. tenuissima, and three isolates (CPA, CPB, CR2) clustered with representative strains of A. panax (). The phylogenetic analysis indicated that the 23 isolates were A. tenuissima and A. alternata in sect. Alternaria (). Three isolates (CPA, CPB, CR2) obtained from ginseng clustered in the A. panax clade (ML bootstrap support value 100), A. panax sect. Panax ().
Fig. 4 (Colour online) Maximum Likelihood tree obtained from the combined ITS, GAPDH, RPB2, TEF, and Alt a 1 sequence alignment analysis of 45 Alternaria species. Bootstrap support values above 50% are shown at the nodes and the bootstrap values are given at the nodes. The isolates of Alternaria alternata and A. tenuissima are highlighted in the green boxes which are under sect. Alternaria. The isolates of A. panax are highlighted in yellow box which are inside sect. Panax. The 26 strains from ginseng are highlighted in bold. The tree was rooted to A. infectoria CBS 210.86 (Woudenberg et al. Citation2015). T: ex-type isolate; R: representative isolate; Species names between parentheses refer to the former species name
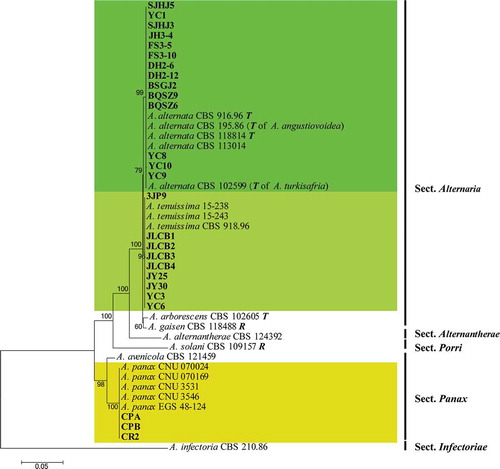
Fig. 5 (Colour online) Maximum Likelihood tree obtained from the combined ITS, GAPDH, RPB2, TEF, and Alt a 1 sequences alignment analysis of 59 Alternaria species. Bootstrap support values above 50% are shown at the nodes and the bootstrap values are given at the nodes. The species Alternaria alternata and A. tenuissima are highlighted in differently green boxes. The 23 strains from ginseng are highlighted in bold. The tree was rooted to A. alternantherae CBS 124392 (Woudenberg et al. Citation2015). T: ex-type isolate; R: representative isolate; Species names between parentheses refer to the former species name

Pathogenicity of the Alternaria isolates
Wounded detached leaves provided a rapid prescreening of Alternaria isolates for pathogenicity on ginseng. After preliminary screening, putatively virulent Alternaria isolates were confirmed by intact plant assays. Amongst 257 Alternaria isolates, 177 (68.9%) isolates were pathogenic, and 80 (31.1%) were found to be non-pathogenic (Table S1). The symptoms on detached leaves (), in an agar plug test with wounding, consisted of circular and black necrotic lesions without yellow zones. The non-pathogenic isolates contained 78 isolates of sect. Alternaria and two isolates of sect. Panax.
In the conidial inoculation assays, various disease symptoms were expressed (). The first type of symptom on leaves () with a conidial inoculation test was circular or irregular necrotic lesions surrounded by chlorotic zones. The second type of symptom was on stems (), which became brown at infection sites, with the browning spreading. The third type of symptom was on roots, which became black-brown or black at infection sites, and abundant hyphae were produced on the lesions (). The fourth type of symptom occurred on seeds (), which developed black spots and became withered. A fungus similar to the original inoculated fungus was consistently re-isolated from lesions on inoculated plant material (–d), but never from the control leaves (). The hyphal plug inoculation test and the conidial inoculation test gave identical results (pathogenicity or non-pathogenicity), but the different symptoms from the two tests were that the areas of chlorotic zones on unwounded plant material inoculated with conidia were large () compared with the hyphal plug inoculation of wounded host material ( and c).
Discussion
Alternaria species have been reported as important crop pathogens (Gur et al. Citation2016). In this study, two sections, sect. Alternaria and sect. Panax, were associated with leaf, stem, seed, and root disease on ginseng. Alternaria alternata, in sect. Alternaria has been reported as an important pathogen causing leaf spot disease on many crops such as wheat and date palms in Oman (Al-Nadabi et al. Citation2018), alfalfa in Canada (Abbasi et al. Citation2018), sunflower in South Africa (Kgatle et al. Citation2018), apple leaf spot (Gur et al. Citation2016), and black spot of carob (Basim et al. Citation2017). Furthermore, it has been reported to be associated with leaf spot of strawberry (Nishikawa and Nakashima Citation2019). However, until recently, there have been few reports of A. alternata causing disease on ginseng (Li et al. Citation2020). This study showed that A. alternata was the most common Alternaria species associated with alternaria blight in Jilin province in China.
Alternaria tenuissima, in sect. Alternaria is highly adaptable to different environments, and has been identified as the causal agent of many kinds of diseases on other plants, including black chokeberry (Aronia melanocarpa (Michx.) Elliot) (Wee et al. Citation2016), rhizoma atractylodis (Atractylodes lancea (Thunb.) DC.) (Wang et al. Citation2007), sunflower (Helianthus annus L.) (Khodaei and Arzanlou Citation2013; Wang et al. Citation2014, Citation2019), eggplant (Solanum melongena L.) (Raja et al. Citation2006), potato (S. tuberosum L.) (Zheng et al. Citation2015; Zhao et al. Citation2018), blueberry (Vaccinium corymbosum L.) (Fernández et al. Citation2015), and broad bean (Vicia faba L.) (Mohamed et al. Citation2002). However, we did not find any reports of A. tenuissima associated with Panax ginseng. Over 26% of the isolates found in this study from ginseng in Jilin province were associated with A. tenuissima.
Alternaria panax-sect. Panax has been reported as the main pathogen on many important araliaceous plants (Deng et al. Citation2013), such as P. ginseng (Zhao et al. Citation1993), P. notoginseng (Chen et al. Citation2005) and P. quinquefolius (Zhao et al. Citation1993). However, we found that only nine isolates from P. ginseng were A. panax out of 257 Alternaria isolates obtained. This species was detected only in three regions. More research needs to be done in other ginseng growing regions in China to see if this same pattern holds, and whether this pattern truly differs from other areas of the world where A. panax is the main cause of alternaria blight of P. ginseng.
The use of the ITS sequence can identify A. panax, but could not distinguish A. alternata from A. tenuissima (White et al. Citation1990; Berbee et al. Citation1999; Liu et al. Citation1999; Carbone et al. Citation1999). Characterization of the Alternaria isolates based on the combined sequences of the ITS, TEF, GAPDH, RPB2 and Alt a 1 genes identified the 26 Alternaria isolates to the species level. Among the two sections of Alternaria spp., A. alternata and A. tenuissima in sect. Alternaria were found in the 13 surveyed regions, and the second section in sect. Panax was only found in three surveyed regions (). The distribution of Alternaria spp. on P. ginseng in Jilin province differs from that of previous studies (Nakata and Takimoto Citation1922; Zhao et al. Citation1993). This study showed that among 257 Alternaria isolates, 177 (68.9%) isolates were pathogenic, and 80 (31.1%) isolates were non-pathogenic. The causal agent of alternaria blight of P. ginseng was reported as A. panax in previous studies (Nakata and Takimoto Citation1922; Deng et al. Citation2013; Wei et al. Citation2018). However, A. alternata was the prevalent species in this study (70.4%), followed by isolates belonging to A. tenuissima (26.1%). Our study also found P. ginseng as a new host for A. tenuissima in China, and is also the first report of disease on the roots and stems of ginseng caused by Alternaria species. Future studies should include more isolates from ginseng in other provinces of China in order to investigate the distribution and host specialization of the Alternaria species on ginseng.
Table S1. Geographic origins and numbers of Alternaria isolates obtained from infected ginseng.
Download MS Word (25.5 KB)Acknowledgements
The technical assistance of Shiran Wang, Lin Zhang, and Xiuhua Wang is gratefully acknowledged.
Supplementary material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2020.1858167.
Additional information
Funding
References
- Abbasi PA, Ali S, Renderos W, Naeem HA, Papadopoulos Y. 2018. First report of Alternaria alternata causing leaf spot and blight symptoms on alfalfa in Canada. Can J Plant Pathol. 40:451–455. doi:10.1080/07060661.2018.1470111.
- Adachi Y, Watanabe H, Tanabe K, Doke N, Nishimura S, Tsuge T. 1993. Nuclear ribosomal DNA as a probe for genetic variability in the Japanese pear pathotype of Alternaria alternata. Appl Environ Microb. 59:3197–3205. doi:10.1128/AEM.59.10.3197-3205.1993.
- Al-Nadabi HH, Maharachchikumbura SSN, Agrama H, Al-Azri M, Nasehi A, Al-Sadi AM. 2018. Molecular characterization and pathogenicity of Alternaria species on wheat and date palms in Oman. Eur J Plant Pathol. 152:577–588. doi:10.1007/s10658-018-1550-4.
- Basim H, Basim E, Baki E, Abdulai M, Öztürk N, Balkic R. 2017. Identification and characterization of Alternaria alternata (Fr.) Keissler causing Ceratonia Blight disease of carob (Ceratonia siliqua L.) in Turkey. Eur J Plant Pathol. 151:73–86.
- Berbee ML, Pirseyedi M, Hubbard S. 1999. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 91:964–977. doi:10.1080/00275514.1999.12061106.
- Carbone I, Anderson JB, Kohn LM. 1999. Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. Evolution. 53:11–21. doi:10.1111/j.1558-5646.1999.tb05329.x.
- Chen YQ, Wang Y, Feng GQ, Liu YZ. 2005. Biological characteristics of Alternaria panax Whetz isolated from Panax notoginseng. Acta Phytopathologica Sinica. 35:267–269.
- Deng JX, Paul NC, Park MS, Yu SH. 2013. Molecular characterization, morphology, and pathogenicity of Alternaria panax from araliaceous plants in Korea. Mycol Progress. 12:383–396. doi:10.1007/s11557-012-0844-8.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi:10.1093/nar/gkh340.
- Fernández RL, Rivera MC, Varsallona B, Wright ER. 2015. Disease prevalence and symptoms caused by Alternaria tenuissima and Pestalotiopsis guepinii on blueberry in Entre Ríos and Buenos Aires, Argentina. AJPS. 6:3082–3090. doi:10.4236/ajps.2015.619301.
- Gao J, Wang Y, Guan YM, Chen CQ. 2014. Fusarim cerealis, a new pathogen causing ginseng (Panax ginseng) root rot in China. Plant Dis. 98:1433. doi:10.1094/PDIS-03-14-0328-PDN.
- Guan YM, Lu BH, Wang Y, Gao J, Wu LJ. 2014. First report of root rot caused by Fusarim redolens on ginseng (Panax ginseng) in Jilin Province of China. Plant Dis. 98:844. doi:10.1094/PDIS-09-13-0922-PDN.
- Gur L, Reuveni M, Cohen Y. 2016. Occurrence and etiology of Alternaria leaf blotch and fruit spot of apple caused by Alternaria alternata f. sp. mali on cv. Pink lady in Israel. Eur J Plant Pathol. 147:695–708. doi:10.1007/s10658-016-1037-0.
- Hill SN. 2009. Epidemiology of Alternaria panax on American ginseng and evaluation of a disease forecaster. Ann Arbor, MI: Michigan State University. ProQuest LLC.
- Hong SG, Cramer RA, Lawrence CB, Pryor BM. 2005. Alt a 1 allergen homologs from Alternaria and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genet Biol. 42:119–129. doi:10.1016/j.fgb.2004.10.009.
- Kgatle MG, Truter M, Ramusi TM, Flett B, Aveling TAS. 2018. Alternaria alternata, the causal agent of leaf blight of sunflower in South Africa. Eur J Plant Pathol. 151:667–688. doi:10.1007/s10658-017-1402-7.
- Khodaei S, Arzanlou M. 2013. Morphology, phylogeny and pathogenicity of Alternaria species, involved in leaf spot disease of sunflower in northern Iran. Arch Phytopathol Plant Protect. 46:2224–2234. doi:10.1080/03235408.2013.790259.
- Kim H, Mohanta TK, Park Y-H, Park SC, Shanmugam G, Park JS, Jeon J, Bae H. 2019. Complete genome sequence of the mountain-cultivated ginseng endophyte Burkholderia stabilis and its antimicrobial compounds against ginseng root rot disease. Biol Control. 140:104–126.
- Kim YC, Lee JH, Bae YS, Sohn BK, Park SK. 2010. Development of effective environmentally-friendly approaches to control Alternaria blight and anthracnose disease of Korean ginseng. Eur J Plant Pathol. 127:443–450. doi:10.1007/s10658-010-9610-4.
- Li MJ, Pan X, Wang Q, Liu Z, Sun H, Shao C, Guan YM, Zhang Y. 2020. First report of leaf spot caused by Alternaria alternata on ginseng (Panax ginseng) in China. Plant Dis. 104:571. doi:10.1094/PDIS-09-19-1897-PDN.
- Liu LP, Liu YN, Yang LY, Lu BH, Yang LN, Wang X, Li Y, Gao J, Hsiang T. 2017. First report of anthracnose caused by Colletotrichum destructivum on curly dock in China. Plant Dis. 101:256–257. doi:10.1094/PDIS-08-16-1084-PDN.
- Liu LP, Zhang L, Qiu PL, Wang Y, Liu YN, Li Y, Gao J, Hsiang T. 2020. Leaf spot of Polygonatum odoratum caused by Colletotrichum spaethianum. J Gen Plant Pathol. 86:157–161. doi:10.1007/s10327-019-00903-4.
- Liu YJ, Whelen SR, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 16:1799–1808. doi:10.1093/oxfordjournals.molbev.a026092.
- Lu BH, Wang XH, Wang R, Wang X, Yang LN, Liu LP, Yang C, Gao J, Liu XN. 2019. First report of Botrytis pelargonii causing postharvest gray mold on fresh ginseng roots in China. Plant Dis. 103(1):149. doi:10.1094/PDIS-01-17-0031-PDN.
- Mohamed ZR, Honda Y, Islam SZ, Muroguchi N, Arase S. 2002. Leaf spot disease of broad bean (Vicia faba L.) caused by Alternaria tenuissima–A new disease in Japan. J Gen Plant Pathol. 68:31–37. doi:10.1007/PL00013049.
- Nakata K, Takimoto S. 1922. Studies on ginseng diseases in Korea. Bull.Agric. Exper. Stat. Chosen. 1–18.
- Nishikawa J, Nakashima C. 2019. The Mycological Society of Japan. 2019. Morpholoical and molecular characterization of the strawberry black leaf spot pathogen referred to as the strawberry pathotype of Alternaria alternata. Mycoscience. 60:1–9. doi:10.1016/j.myc.2018.05.003.
- Park Y-H, Lee S-G, Ahn DJ, Kwon TR, Park SU, Lim H-S, Bae H. 2012. Diversity of fungal endophytes in various tissues of Panax ginseng Meyer cultivated in Korea. J Gins Res. 36:211–217. doi:10.5142/jgr.2012.36.2.211.
- Raja P, Reddy AVR, Allam US. 2006. First report of Alternaria tenuissima causing leaf spot and fruit rot on eggplant (Solanum melongena) in India. Plant Pathol. 55:579–579. doi:10.1111/j.1365-3059.2006.01379.x.
- Rambaut A. 2014. FigTree v. 1.4.2. Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
- Reeleder RD. 2003. The ginseng root pathogens Cylindrocarpon destructans and Phytophthora cactorum are not pathogenic to the medicinal herbs Hydrastis canadensis and Acataea racemosa. Can J Plant Pathol. 25:218–221. doi:10.1080/07060660309507072.
- Wang D, Wang SR, Yang MJ, Lu BH, Liu LP, Gao J. 2018. Detection of the resistance of Alternaria panax and cross-resistance on ginseng in Jilin Province. Agrochemicals. 57:603–605.
- Wang TY, Zhao J, Ma GP, Bao SW, Wu XH. 2019. Leaf blight of sunflower caused by Alternaria tenuissima and A. alternata in Beijing, China. Can J Plant Pathol. 41:372–378. doi:10.1080/07060661.2019.1583288.
- Wang TY, Zhao J, Sun P, Wu XH. 2014. Characterization of Alternaria species associated with leaf blight of sunflower in China. Eur J Plant Pathol. 140:301–315. doi:10.1007/s10658-014-0464-z.
- Wang WX, Jia FJ, Li Y, Huang JB. 2007. Biological characteristics of the pathogen causing dark leaf spot on Atractylodes lancea. Plant Protection. 33:107–110.
- Wee J-I, Park J-H, Back C-G, You Y-H, Chang T. 2016. First report of leaf spot caused by Alternaria tenuissima on black chokeberry (Aronia melanocarpa) in Korea. Mycobiology. 44:187–190. doi:10.5941/MYCO.2016.44.3.187.
- Wei SQ, Sun Y, Xi G, Zhang H, Xiao M, Yin R. 2018. Development of a single-tube nested PCR-lateral flow biosensor assay for rapid and accurate detection of Alternaria panax Whetz. PloS One. 13:e0206462. doi:10.1371/journal.pone.0206462.
- White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand GH, Sninsky JJ, White TJ, editors. PCR protocols–A guide to methods and applications. New York (NY): Academic Press; p. 315–322.
- Woudenberg JHC, Seidl MF, Groenewald JZ, de Vries M, Stielow JB, Thomma BPHJ, Crous PW. 2015. Alternaria section Alternaria: species, formae speciales or pathotypes? Stud Mycol. 82:1–21. doi:10.1016/j.simyco.2015.07.001.
- Woudenberg JHC, Truter M, Groenewald JZ, Crous PW. 2014. Large-spored Alternaria pathogens in section Porri disentangled. Stud Mycol. 79:1–47. doi:10.1016/j.simyco.2014.07.003.
- Zhao J, Ma GP, Liu YY, Wu XH. 2018. Alternaria species infecting potato in southern China. Can J Plant Pathol. 40:312–317. doi:10.1080/07060661.2018.1459850.
- Zhao RF, Chen WQ, Zhang TY. 1993. Research progress on black spot disease of Panax ginseng and Panax quinquefolius in China. Plant Protection. 1:31–32.
- Zheng HH, Zhao J, Wang TY, Wu XH. 2015. Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathol. 64:425–433. doi:10.1111/ppa.12274.

