Abstract
Mango malformation disease caused by Fusarium spp. is one of the main limiting factors for mango production in Mexico. Fungal isolates obtained from symptomatic tissue from mango trees in Sinaloa, Mexico, were subjected to phylogenetic analysis of the TEF-1α gene, and evaluated for mating type, morphological markers and pathogenicity. Fifteen monosporic Fusarium isolates were identified as Fusarium neocosmosporiellum O’Donnell & Geiser. Phylogenetic analysis confirmed that the sequence obtained in this study corresponds to F. neocosmosporiellum. All 15 isolates were found to be pathogenic, but no differences were observed in aggressiveness among the isolates, which may be a result of low genetic variability within the fungal population. According to the morphological and molecular results, this study confirmed that the causal agent of mango malformation disease was F. neocosmosporiellum. This is the first report of mango malformation disease caused by this fungus in Mexico. This study provides a basis for the development and implementation of an integrated management system for this disease, confirms a new location of this fungus, and highlights that this pathogen is a potential threat to mango cultivation in Mexico.
Résumé
La malformation du manguier, causée par Fusarium spp., est un des principaux facteurs qui limite la production de mangues au Mexique. Des isolats fongiques obtenus de tissus symptomatiques provenant de manguiers poussant dans l’État du Sinaloa, au Mexique, ont été soumis à l’analyse phylogénétique du gène de la TEF-1α et évalués pour en déterminer le type sexuel, les marqueurs morphologiques et la pathogénicité. Quinze isolats monospores ont été identifiés en tant que Fusarium neocosmosporiellum O’Donnell & Geiser. L’analyse phylogénétique a confirmé que la séquence obtenue dans le cadre de cette étude correspondait à F. neocosmosporiellum. Les 15 isolats se sont avérés pathogènes, mais aucune différence n’a été observée quant à l’agressivité parmi ces derniers, ce qui peut émaner d’une faible variabilité génétique au sein de la population fongique. Selon les résultats des analyses morphologiques et moléculaires, cette étude a confirmé que l’agent causal de la malformation du manguier était F. neocosmosporiellum. Il s’agit de la première mention de la malformation du manguier causée par ce champignon au Mexique. Cette étude fournit une base pour élaborer et appliquer un système de gestion intégrée de cette maladie, confirme une nouvelle localization pour ce champignon et démontre que cet agent pathogène constitue une menace possible pour la production de mangues au Mexique.
Introduction
Mango (Mangifera indica L.) is the most economically important fruit in tropical and subtropical regions of the world (FAO Citation2020). In 2018, Mexico ranked sixth worldwide in terms of cultivated area and production, preceded by India, China, Thailand, Indonesia, and Pakistan (FAO Citation2020). However, mango production is affected worldwide by mango malformation disease (MMD), one of the most problematic mango diseases. The disease affects vegetative and flower buds (Ploetz Citation2007). Initially trees exhibit bud swelling and vegetative malformation, followed by eruption of multiple shoots, ending in a bunchy and deformed apex. The disease was first recorded in India in 1891 (Krishnan et al. Citation2009) and is currently distributed in all mango-producing regions in the world (Morales and Rodríguez Citation2009; Qi et al. Citation2013).
The fungus Fusarium mangiferae Britz, Wingfield & Marasas has been reported as a causal agent for the disease (Freeman et al. Citation1999; Bhavna et al. Citation2014), as have other species of the Fusarium genus complex, such as: Fusarium oxysporum Schlecht. emend. Snyder & Hansen (Mora Citation2000), Fusarium decemcellulare (Berk. & Broome) Rossman & Samuels (Castellano and Guanipa Citation2004), Fusarium sterilihyphosum Britz, Wingfield & Marasas (Britz et al. Citation2002), Fusarium proliferatum (Matsush.) Nirenberg ex Gerlach & Nirenberg (Zhan et al. Citation2010), and Fusarium tupiense Lima, Pfenning & Leslie (Crespo et al. Citation2016). In Mexico, species that are reported to cause MMD include Fusarium subglutinans (Wollenweber & Reinking) P. Nelson, Toussoun & Marasas and F. oxysporum in the states of Michoacán, Guerrero, and Morelos (Mora Citation2000). In more recent studies, Fusarium mexicanum Aoki, Freeman, Otero-Colina, Rodríguez-Alvarado, Fernández-Pavía, Ploetz & O’Donnell (Otero-Colina et al. Citation2010; Betancourt et al. Citation2012) and Fusarium pseudocircinatum Nirenberg & O’Donnell (Freeman et al. Citation2014) were identified as causal agents of mango malformation disease.
Many members of the Fusarium solani species complex (FSSC), including Fusarium neocosmosporiellum O’Donnell & Geiserare, are known to cause severe diseases on plants including peas, beans, tomatoes, and tuber roots (Leslie and Summerell Citation2006). The FSSC is constituted by more than 60 phylogenetic species (Schroers et al. Citation2016). Currently, only 23 phylogenetic species have been unambiguously identified and validated as part of the FSSC (Sandoval-Denis and Crous Citation2019), and they are morphologically indistinguishable. (Short et al. Citation2013; Scheel et al. Citation2013; Sandoval-Denis and Crous Citation2019).
Control methods can be very difficult to find when a new disease emerges, particularly if the causal agent is not properly identified. The implementation of appropriate disease management measures requires a reliable identification and detection of the pathogen (Moine et al. Citation2014). In addition to classical morphological identification, the use of molecular biology techniques, especially those involving the polymerase chain reaction (PCR), have provided a rapid and sensitive approach for the detection and identification of plant pathogens (O’Donnell Citation2000; Vincenlli and Tisserat Citation2008). Several DNA regions, particularly, the translation elongation factor 1-α (TEF-1-α) gene, are useful for the identification of Fusarium spp. (O’Donnell et al. Citation1998; Geiser et al. Citation2004).
The objectives of this study were to: identify and characterize the pathogen that causes mango malformation in Sinaloa, Mexico, using morphological and molecular methods, and pathogenicity tests.
Materials and methods
Diseased plant sampling and fungal isolation
In June 2018, Mango trees were observed with symptoms of floral and plant malformation in the municipality of El Rosario (22° 59ʹ 31” N; 105° 51ʹ 4” W), Sinaloa, Mexico. Symptomatic tissues were collected from 15 symptomatic mango trees. Symptomatic trees exhibited bud swelling, vegetative malformation and eruption of multiple shoots ending in a bunchy and deformed apex. Each diseased sample was obtained from a different mango tree. Symptomatic tissues were sampled with a sterile scalpel, transported to the laboratory, and immediately surface-disinfected in 0.5% NaOCl (3 min) and 70% ethanol (3 min), and rinsed twice (5 min) in sterile distilled water. After drying on sterile absorbent paper, the samples were placed in Petri dishes containing potato dextrose agar (PDA). To obtain monosporic cultures, the methodology proposed by Hansen and Smith (Citation1932) was followed. A small fragment of the mycelium from 6-day-old PDA cultures was transferred into 1.0 mL of sterile distilled water, serially diluted, and then plated on a new PDA dish. Two days later, the cultures were observed under a microscope, and individual germinating microspores were transferred to fresh PDA.
Morphological characterization
Morphological characterization was performed following Leslie and Summerell (Citation2006). Isolates were cultivated on PDA at 25 °C in the dark, after which, the colour of the colony and formation of aerial mycelium were observed. On Carnation Leaf Agar (CLA) medium, under the same conditions previously mentioned, microconidial and macroconidial shape, septation, arrangement of conidiogenous cells, and the presence or absence of sporodochia and chlamydospores were recorded and measured (n = 50) under a light microscope at 40× magnification.
DNA extraction and PCR assays
The mycelium of each isolate was collected by scraping the surface of colonies growing on PDA previously incubated for 1 week at 25°C. After grinding 100 mg of fungal mycelia from each isolate in liquid nitrogen, the genomic DNA was extracted using the method described by Ausubel et al. (Citation2003). The DNA concentration and quality were estimated using a Thermo Scientific NanoDrop™ 1000 Spectrophotometer (Fisher Scientific).
The DNA extracted from the Fusarium isolates was analysed by PCR with the primers listed in . The final reaction mixture (25 μL) contained 100 ng DNA templates, an equimolar mixture of dNTPs, 25 mM MgCl2, 1 U Taq DNA polymerase, and 40 pmol of each oligonucleotide (Bioline, TN, USA). The sequences of all the primers and PCR conditions used are provided in . EF-1 and EF-2 are the primers for TEF-1α, MAT-1 and MAT-2 are primers to determine mating type 1 and mating type 2, respectively.
Table 1. Reaction conditions and sequences of primers for PCR analysis of Fusarium spp
The PCR products (TEF-1α DNA) were purified and sequenced by Macrogen Inc. (Seoul, South Korea). The TEF-1α DNA sequences were used to search for sequence similarity against the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) using the BLASTN programme. The molecular identification was confirmed via BLAST on the FUSARIUM ID and Fusarium MLST databases. Species were identified based on 100% sequence identity as the threshold.
Phylogenetic analyses
The TEF-1α sequence from one representative isolate (66VRSIN) was aligned with reference sequences (indicated in ) of the Fusarium solani species complex (FSSC) obtained from GenBank using the multiple alignment in ClustalW of the Geneious R9 software, and phylogenetic relationships among the Fusarium isolates were inferred based on the nucleotide sequence alignment of the gene. Trees were constructed by the neighbour-joining method based on distances determined by the method of Jukes and Cantor (Erickson Citation2010) using 1000 bootstrap replicates.
Pathogenicity studies
All fungal isolates were tested for their ability to cause mango malformation disease by experimental inoculation of healthy 4-month-old mango plants (cv. ‘Kent’) maintained under greenhouse conditions for 12 months. The inoculum was prepared by culturing the isolates on CLA under cool-white fluorescent light for 14 days at 25°C to stimulate sporulation (Freeman et al. Citation1999). The pathogenicity test was conducted during 2018–2020. Inoculation of apical buds was performed by placing a 20 μL drop of conidial suspension (5 × 107 conidia per mL) on dormant buds. Six buds were inoculated per branch, with two branches per plant, and 20 plants inoculated per isolate. Apical bud inoculation on control plants was performed with 20-μL drops of sterile distilled water. Pathogenicity experiments were conducted twice. Isolation of fungi was conducted from developing lesions on inoculated plants, and the isolates were identified to species using morphological characteristics and the sequencing of the TEF1-α gene.
The severity of the disease was assessed using a scale of 0 to 5 as proposed by Iqbal et al. (Citation2006) (), in which 0 is a plant with no symptoms and 5 is a plant with the most severe symptoms.
Table 2. Disease severity scale for mango malformation disease proposed by Iqbal et al. (Citation2006)
Data analysis
The data obtained from the pathogenicity test were subjected to a non-parametric analysis of variance with the Kruskal–Wallis and Dunn test (p ≤ 0.05). All statistical analyses were performed with the XLSTAT software.
Results
Fifteen monosporic isolates of Fusarium spp. were obtained from diseased plant material from 15 mango trees with symptoms of floral and plant malformation in the municipality of El Rosario, Sinaloa, Mexico, in June 2018.
Morphological markers
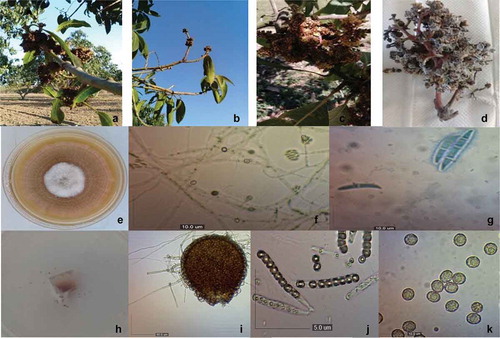
Colonies of the 15 monosporic isolates of Fusarium spp. grown on PDA presented white and brown cottony mycelia (in the centre), with irregular edges (). The microconidia (n = 50) were cylindrical to ellipsoidal oblong and hyaline, with 0–1 septa of 4.2–17.3 × 2.3–7.9 μm (). Macroconidia (n = 50) were hyaline, often almost straight and wider in the middle, from 3 to 4 septa of 41.1–75.7 × 4.9–11.1 μm (). The teleomorphic phase (Neocosmospora vasinfecta E.F. Sm.) was observed. The perithecia (n = 20) were orange to red, pear-shaped, with a short neck, 185.3–294.6 × 126.4–220.7 μm (). The cylindrical asci (n = 30) contained eight ascospores and had short pedicels measuring 78.3–89 × 6.5–8.8 μm (). The ascospores (n = 50) were globose, cylindrical, unicellular, with rough walls of 12.9–29.8 × 11.7–26.5 μm ().
Fig. 1 a–d, (Colour online) Symptoms of mango malformation disease caused by Fusarium neocosmosporiellum from sampled trees. e–k, F. neocosmosporiellum morphological features. e, Top view of a F. neocosmosporiellum colony grown on PDA; f, Microconidia in situ (10 μm bar); g, Macroconidia (10 μm bar); h, Exudation of ascospores; i, Perithecia (50 μm bar); j, Ascus (5 μm bar); k, Ascospores with ornate wall (10 μm bar)
PCR assays and phylogenetic analysis
PCR analysis of the 15 isolates of Fusarium spp. amplified the expected fragment of the TEF-1α gene (683 bp).
The TEF-1α sequence of isolate 66VRSIN was submitted to Genbank (sequence MK932796). The analysis of TEF-1α sequences of these isolates by BLAST in the GenBank, Fusarium ID and Fusarium MLST databases showed that all were affiliated to the species F. neocosmosporiellum with a homology of 99.9 to 100%. The MAT2 gene fragment (800 bp) was amplified for all 15 isolates, indicating they were all mating type 2.
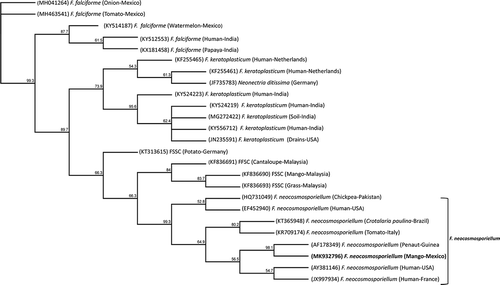
The phylogram generated with the partial sequences of the TEF-1α gene from isolate 66VRSIN (MK932796) and other species from the FSSC obtained from GenBank was divided into groups, aligning each species of Fusarium (). The first group corresponds to F. falciforme, the second group was aligned with isolates of the species Fusarium keratoplasticum. A third small group was formed and included FSSC from different crops including mango. Finally, the fourth group was aligned with isolates of F. neocosmosporiellum isolated from different sources. The TEF-1α sequence (MK932796) from the 66VRSIN isolate was aligned with the group of F. neocosmosporiellum ().
Pathogenicity tests
There were no significant differences between the two assays in terms of symptom severity on mango plants (cv. ‘Kent’) at 270 dpi (P = 0.0782), with an average disease severity of 4.5 in the first assay and 4.6 in the second assay.
The 15 monoconidial F. neocosmosporiellum isolates were pathogenic to mango cv. ‘Kent’. Although plants developed malformation on the vegetative buds following inoculation with each of the 15 isolates, no significant difference in virulence existed amongst the isolates according to the observed symptoms (). The first MMD symptoms were observed at 150 dpi on the inoculated plants in both assays, matching the symptoms originally observed in mango plants grown in commercial fields. Fungi were isolated from symptomatic tissues of inoculated plants and their identity determined to be F. neocosmosporiellum using morphological characteristics and sequencing of the TEF-1α gene, fulfilling Koch’s postulates. No symptoms were observed on the non-inoculated plants after 12 months ().
Table 3. Pooled results of the first and second pathogenicity assays of mango (cv. ‘Kent’) inoculated with 15 monoconidial Fusarium neocosmosporiellum isolates from mango trees
Discussion
This study confirmed the first occurrence in Mexico of mango malformation disease caused by F. neocosmosporiellum. The results provide the basis for the development and implementation of appropriate disease management of this pathogen in Mexico.
The phylogenetic analysis indicated that our TEF-1α sequence (MK932796) grouped closely with a sequence of F. neocosmosporiellum obtained from infected peanut plants. These two sequences were also grouped with other sequences from F. neocosmosporiellum isolates obtained from tomato, chickpea, Crotalaria paulina Schrank, and humans, whereas they were clearly distinguished from species of other clades of the FFSC, such as F. falciforme and F. keratoplasticum. The TEF-1α gene was used in the present study to determine the specific identity of the isolates. It has also been used in other studies, since it provides very useful phylogenetic information at the species level for the genus Fusarium, and has been used as a marker to resolve relationships between species (Geiser et al. Citation2004).
All the isolates of F. neocosmosporiellum were pathogenic on mango cv. ‘Kent’, indicating that F. neocosmosporiellum is able to cause mango malformation disease, which represents an important problem for the production of mango. Fungi were recovered from inoculated symptomatic plants and showed the same morphological characteristics and TEF-1α sequences as those of the originally inoculated isolates, thus confirming their pathogenicity.
In this study, no differences were observed in aggressiveness among the isolates. This may be a result of little genetic variability within the F. neocosmosporiellum populations. The isolates were of the same mating type (MAT 2), which suggests that only asexual reproduction occurs, generating a clonal population of this fungus. The isolates were obtained from different mango trees from different plots. However, more studies are needed to analyse the genetic diversity of this fungus, as well as its virulence.
Fusarium species are among the most challenging fungal species to manage in agricultural crops. Several agroecological techniques are known to impact Fusarium populations (Jarvis Citation1989), including sanitation, organic amendments, biological control, among others. Each of these techniques is only partly effective, necessitating an integrated control program, which further complicates adoption on farms where there is often limited flexibility for the uptake of new practices (Collange et al. Citation2014). Mango is a perennial crop, hence cultivars cannot be easily changed year after year as is the case with annual crops; therefore, it is necessary to implement resistance breeding programmes, biological control and other disease management practices. This study provides the basis for the development of an integrated management system for this disease and confirms a new location of this fungus. It also highlights that this pathogen is a potential threat for mango production in Mexico.
Acknowledgements
Thanks are extended to CONACYT for the scholarship granted to Lorena Molina-Cárdenas to carry out her doctoral studies.
Additional information
Funding
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Scidman JG, Smith JA, Struhl K. 2003. Current protocols in molecular biology. New York: Wiley.
- Betancourt RI, Velázquez MJJ, Montero CJC, Fernández PSP, Lozoya SH, Rodríguez AG. 2012. Fusarium mexicanum agente causal de la malformación del mango en Jalisco, México. Rev Mex Fitopatol. 30:115–127.
- Bhavna J, Wahid AM, Bains G, Chandra PR, Shukla A, Tuteja N, Kumar J. 2014. Fusarium mangiferae associated with mango malformation in the Tarai region of the Uttarakhand state of India. Plant Signal Behav. 9:1–3.
- Britz H, Steenkamp ET, Coutinho TA, Wingfield BD, Marasas WF, Wingfield MJ. 2002. Two new species of Fusarium section Liseola associated with mango malformation. Mycologia. 94(4):722–730. doi:https://doi.org/10.1080/15572536.2003.11833199
- Castellano G, Guanipa N. 2004. Comportamiento de diez cultivares de mango (Mangifera indica L.) a la inoculación con Fusarium decemcellulare Brix. Rev Fac Agron. 21:200–206.
- Collange B, Navarrete M, Montfort F, Mateille T, Tavoillot J, Martiny B, Tchamitchian M. 2014. Alternative cropping systems can have contrasting effects on various soil-borne diseases: relevance of a systemic analysis in vegetable cropping systems. Crop Prot. 55:7–15. doi:https://doi.org/10.1016/j.cropro.2013.10.002
- Crespo M, Arrebola E, Carzola FM, Maymon M, Freeman S, Aoki T, O’Donell K, Torres JA, de Vicente A. 2016. Analysis of Genetic diversity of Fusarium tupiense, the main causal agent of mango malformation disease in southern Spain. Plant Dis. 100:276–286. doi:https://doi.org/10.1094/PDIS-02-15-0153-RE
- Erickson K. 2010. The Jukes-Cantor model of molecular evolution. PRIMUS. 20(5):438–445. doi:https://doi.org/10.1080/10511970903487705
- FAO. 2020. Production statistics by crop. [accessed 2020 Feb]. www.fao.org/faostat/en/#data/QC.
- Freeman S, Maimon M, Pinkas Y. 1999. Use of GUS transformants of Fusarium subglutinans for determining etiology of mango malformation disease. Phytopathology. 89(6):456–461. doi:https://doi.org/10.1094/PHYTO.1999.89.6.456
- Freeman S, Otero-Colina G, Rodríguez-Alvarado G, Fernández-Pavía S, Maymon MR, Ploetz RC, Aoki T, O’Donnell K. 2014. First report of mango malformation disease caused by Fusarium pseudocircinatum in Mexico. Plant Dis. 98(11):1583. doi:https://doi.org/10.1094/PDIS-04-14-0375-PDN
- Geiser D, Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 110:473–479. doi:https://doi.org/10.1023/B:EJPP.0000032386.75915.a0
- Hansen HN, Smith. 1932. The mechanisms of variation in imperfect fungi: Botrytis cinerea. Phytopathology. 37:369–371.
- Iqbal Z, Rehman M, Dasti AA, Saleem A, Zafar Y. 2006. RAPD analysis of Fusarium isolates causing mango malformation disease in Pakistan. World J Microb Biot. 22:1161–1167. doi:https://doi.org/10.1007/s11274-006-9157-x
- Jarvis WR. 1989. Managing diseases in greenhouse crops. Plant Dis. 73:190–194. doi:https://doi.org/10.1094/PD-73-0190
- Krishnan AG, Nailwal TK, Shukla A, Pant RC. 2009. Mango (Mangifera indica. L) malformation an unsolved mystery. Researcher. 1(5):20–36.
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Iowa, USA: Blackwell Publishing Ltd; p. 122–279.
- Moine LM, Labbe C, Louis-Seize G, Seifert KA, Belanger RR. 2014. Identification and detection of Fusarium striatum as a new record of pathogen to greenhouse tomato in northeastern America. Plant Dis. 98:292–298. doi:https://doi.org/10.1094/PDIS-08-13-0844-RE
- Mora AJA. 2000. Patogénesis y epidemiología de la” Escoba de bruja” Fusarium subglutinans (Wollemwed y Reinking) y F. oxysporum (Schlecht.)] del mango (Mangifera indica L.) en Michoacán, México [Tesis de Doctor en Ciencias]. Montecillo (Edo. México): Colegio de Postgraduados; p. 107.
- Morales RV, Rodríguez GM. 2009. Micobiota endofítica asociada al cultivo del mango ‘Haden’ (Mangifera indica L.) en el oriente de Venezuela. Rev UDO Agrícola. 9(2):393–402.
- O’Donnell K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia. 92:919–938. doi:https://doi.org/10.1080/00275514.2000.12061237
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Nat Acad Sci USA. 95:2044–2049. doi:https://doi.org/10.1073/pnas.95.5.2044
- Otero-Colina G, Rodríguez-Alvarado G, Fernández-Pavía S, Maymon M, Ploetz RC, Aoki T, Freeman S. 2010. Identification and characterization of a novel etiological agent of mango malformation disease in Mexico, Fusarium mexicanum sp. nov. Phytopathology. 100(11):1176–1184. doi:https://doi.org/10.1094/PHYTO-01-10-0029
- Ploetz RC. 2007. Diseases of tropical perennial crops: challenging problems in diverse environments. Plant Dis. 91:644–663. doi:https://doi.org/10.1094/PDIS-91-6-0644
- Qi YQ, Pu JJ, Zhang X, Zhang H, Lu Y, Qun-Fang Y, Zhang HQ, Xie YX. 2013. First report of dieback of mango caused by Fusarium decemcellulare in China. J Phytopathol. 161:735–738. doi:https://doi.org/10.1111/jph.12117
- Sandoval-Denis M, Crous PW. 2019. Back to the roots: a reappraisal of Neocosmospora. Persoonia. 43:90–185. doi:https://doi.org/10.3767/persoonia.2019.43.04
- Scheel CM, Hurst SF, Barreiros G, Akiti T, Nucci M, Balajee SA. 2013. Molecular analyses of Fusarium isolates recovered from a cluster of invasive mold infections in a Brazilian hospital. BMC Infect Dis. 13:49. doi:https://doi.org/10.1186/1471-2334-13-49
- Schroers HJ, Samuels GJ, Zhang N, Short DP, Juba J, Geiser DM. 2016. Epitypification of Fusisporium (Fusarium) solani and its assignment to a common phylogenetic species in the Fusarium solani species complex. Mycologia. 108:806–819. doi:https://doi.org/10.3852/15-255
- Short DP, O’Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM. 2013. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet Biol. 53:59–70. doi:https://doi.org/10.1016/j.fgb.2013.01.004
- Vincenlli O, Tisserat N. 2008. Nucleic acid-based pathogen detection in applied plant pathology. Plant Dis. 92:660–669. doi:https://doi.org/10.1094/PDIS-92-5-0660
- Zhan RL, Yang SJ, Ho HH, Liu F, Zhao YL, Chang JM, He YB. 2010. Mango malformation disease in south China caused by Fusarium proliferatum. J Phytopathol. 158:721–725. doi:https://doi.org/10.1111/j.1439-0434.2010.01688.x