Abstract
Pseudopyrenochaeta lycopersici is a soil-borne fungus causing corky root of tomato. The slow growth and poor sporulation of the pathogen make its isolation difficult. Isolates of the fungus are classified into types 1 and 2, with many different physiological and molecular characteristics between both types. In general, mating type genes enable mating ability and sexual development. The type 1 strains CBS267.59 and Oha3-6 of P. lycopersici were utilized to amplify DNA binding domains (HMG box). The MAT1-1-1 gene of CBS267.59 encodes a predicted protein including an alpha box DNA binding domain, while MAT1-2-1 of Oha3-6 encodes a predicted protein including an HMG DNA binding domain. The sequences of the putative P. lycopersici α and HMG boxes were very similar to established α and HMG boxes. ORF1 and the DNA lyase gene were found in the 5ʹ and 3ʹ regions, respectively, of the MAT1-1 locus. Comparison of the mating type genes and flanking regions was carried out between the strains CBS267.59 and Oha3-6 and other Loculoascomycetes, Pyrenomycetes and Discomycetes. The assembled sequences were 2932 bp (nucleotide positions 426–3358) for the MAT1-1 idiomorph and 3410 bp (nucleotide positions 438–3848) for the MAT1-2 idiomorph. Multiplex polymerase chain reaction amplified 432 bp and 786 bp bands in the MAT1-1 and MAT1-2 strains, respectively. The isolation and characterization of the MAT genes of P. lycopersici improve our knowledge and may help to explain the taxonomic differences in types 1 and 2 of this fungus.
Résumé
Pseudopyrenochaeta lycopersici est un champignon terricole qui cause la racine liégeuse de la tomate. La lente croissance et la médiocre sporulation de l’agent pathogène le rendent difficile à isoler. Les isolats du champignon sont classifiés en types 1 et 2, les deux types affichant plusieurs caractéristiques physiologiques et moléculaires différentes. Généralement, les gènes du type sexuel permettent la reproduction et le développement sexuel. Les souches CBS267.59 et Oha3-6 du type 1 de P. lycopersici ont été utilisées pour amplifier les domaines de liaison à l’ADN (boîte HMG). Le gène MAT1-1-1 de CBS267.59 code une protéine prédite, incluant une boîte alpha du domaine de liaison à l’ADN, tandis que le gène MAT1-2-1 d’Oha3-6 code une protéine prédite, incluant un domaine de liaison à l’ADN de type HMG. Les séquences des boîtes putatives α et HMG de P. lycopersici étaient très semblables aux boîtes α et HMG établies. Le gène ORF1 et le gène d’ADN lyase ont été trouvés dans les régions 5ʹ et 3ʹ, respectivement, du locus MAT1-1. Une comparaison des gènes du type sexuel et des régions flanquantes a été effectuée entre les souches CBS267.59 et Oha3-6 ainsi qu’entre d’autres loculoascomycètes, pyrenomycètes et discomycètes. Les séquences assemblées étaient de 2 932 bp (aux positions 426-3358 des nucléotides) pour l’idiomorphe de MAT1-1 et de 3 410 bp (aux positions 438-3848 des nucléotides) pour l’idiomorphe de MAT1-2. La PCR multiplex a amplifié des bandes de 432 bp et de 786 bp des souches MAT1-1 et MAT1-2, respectivement. L’isolement et la caractérisation des gènes du type sexuel de P. lycopersici améliorent nos connaissances et pourraient nous aider à expliquer les différences taxinomiques entre les types 1 et 2 de ce champignon.
Introduction
Corky root rot is a soil-borne disease of Solanaceous crops caused by Pseudopyrenochaeta lycopersici R.W. Schneid. and Gerlach (Aragona et al. Citation2014). Two days after infection, disease symptoms appear, including brown lesions, necrosis and rotting of small roots (Valente et al. Citation2011). Yield and economic losses due to pathogen infection are reported under field and protected conditions (Golzar Citation2009). Both microsclerotia and mycelium of P. lycopersici remain viable for a prolonged time on the infected roots or on plant residues in the soil. Based on physiological and molecular traits, P. lycopersici is categorized into types 1 and 2 (Bayraktar and Oksal Citation2011; Hieno et al. Citation2015).
The sex cycle in heterothallic fungi begins when opposite mating types interact and continues with the production of progeny that are genetically different from the parents. Sexual reproduction in ascomycetes is controlled by a single regulatory locus known as a mating type (MAT) (Coppin et al. Citation1997; Klix et al. Citation2010). Cloning of MAT loci has been performed for different heterothallic ascomycetes, such as Schizosaccharomyces pombe, Saccharomyces cerevisiae and Neurospora crassa (Hicks et al. Citation1979; Kelly et al. Citation1988; Staben and Yanofsky Citation1990; Tsong et al. Citation2003; Debuchy et al. Citation2010).
The MAT locus has two alternate forms (MAT1-1/MAT1-2 idiomorphs); the MAT1-1 idiomorph has the gene MAT1-1-1, which encodes a DNA-binding protein with an alpha box motif, while MAT1-2 idiomorph has the MAT1-2-1 gene, which encodes a DNA-binding protein with a high mobility group (HMG) box motif. It appears that MAT genes encode transcriptional regulators and control the expression of certain genes needed for sexual reproduction (Turgeon Citation1998; Bardwell Citation2005; Kim and Borkovich Citation2006; Kothe Citation2008).
Pseudopyrenochaeta lycopersici is a mitosporic fungus that produces solitary dark pycnidia, including single-celled conidia on simple, septate branched conidiophores (Infantino and Pucci Citation2005). However, in pure culture the isolated fungus rarely sporulates and the pycnidia have never been found on naturally infected roots (Hockey and Javes Citation1984).
Sequencing and functional characterization of the P. lycopersici genome has been performed to understand the virulence mechanisms in the pathogen, but the mating type genes were not explored (Aragona et al. Citation2014). Therefore, this study aims to isolate and characterize the mating type genes in P. lycopersici to expand ecological and molecular knowledge of the species.
Materials and methods
Strains and culture conditions
The strains of P. lycopersici type 1 used in the experiments are listed in . Strains CBS267.59 and Oha3-6 were used for idiomorph determination. Other strains and species in were used for the confirmation of mating type. The strains were maintained on 1/4 strength potato dextrose agar (PDA) medium at 25°C.
Table 1. Strains of Pseudopyrenochaeta lycopersici examined in this study
DNA and RNA extraction and RNA cleanup
DNA and RNA were extracted from all strains to conduct polymerase chain reaction (PCR) and reverse transcription (RT)-PCR. Genomic DNA was extracted from frozen mycelia grown on potato dextrose broth (PDB) after 1 week of incubation at 25°C. For RNA extraction, mycelial tissues were harvested from 5 mL of Czapek-dox (CD), homogenized at 15 000 rpm for 1 min in 9 mL PDB or CD, and the homogenate (4 mL) was transferred to 100 mL of PDB or CD in a 500 mL baffle flask and cultured on a rotary shaker (120 rpm) at 25°C for 48 h. Total RNA was prepared from frozen mycelia using RNAiso (TaKaRa-Bio, Japan) total RNA extraction reagent following the manufacturer’s instructions. Total RNA cleanup was performed with an RNeasy Mini Kit (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions.
PCR and sequencing
PCR amplification of and subsequent sequencing of the complete mating type genes were performed to compare the strains CBS267.59 and Oha3-6 with other Loculoascomycetes. The PCR primers are listed in . The DNA binding domains (HMG box) were amplified using the degenerate Sn-HMG1 and Sn-HMG2 primers (Bennett et al. Citation2003). For amplification of the alpha box, Sn-ab1 and a nested degenerate primer (Pl-alpha-2) were used following primary PCR using Sn-ab1 and Sn-ab2 (Bennett et al. Citation2003). Pl-alpha-2 was designed from amino acid sequences of the alpha box of five species (Phaeosphaeria nodorum, Leptosphaeria maculans, Cochliobolus heterostrophus, Ascochyta lentis and Ascochyta rabiei). The GenBank accession numbers are AY212018, AY174048, AF029913, DQ341314 and DQ341313, respectively.
Table 2. Primers used in this study
The PCR mixture (final volume 30 μL) contained 1 × PCR buffer, dNTP mixture (containing 2.5 mM of dNTP), 1 μM of each primer, 1.25 U TaKaRa Taq (TaKaRa-Bio) and 50 ng of fungal genomic DNA. PCR conditions were initiated by denaturation for 2 min at 94°C, followed by 30 cycles (30 s at 94°C, 1 min at 42°C and 30 s at 72°C) with a final extension for 5 min at 72°C. The nested PCR for the alpha box utilizes a PCR mixture (final volume 30 μL) containing the same components as the first PCR with 1 μM of each primer (Sn-ab1 and Pl-alpha-2), and the template was 1 μL of a 1:50 dilution of the first PCR mixture. The PCR conditions were similar to the previous PCR conditions.
Thermal asymmetric interlaced (TAIL)-PCR (Liu and Whittier Citation1995) was used to amplify mating type idiomorphs. The primary TAIL-PCR mixture (final volume 30 μL) contained 1 × PCR buffer, dNTP mixture (containing 2.5 mM of dNTP), 0.2 μM of specific primer, 4 μM arbitrary (AD) degenerate primer, 1.25 U TaKaRa Taq and 20 ng of genomic DNA. The secondary TAIL-PCR mixture (final volume 30 μL) contained the same components as the primary reaction with 0.2 μM nested specific primer, 4 μM AD primer, and the template was 1 μL of a 1:50 dilution of the primary reaction mixture.
RT-PCR was performed with TaKaRa PrimeScript RT-PCR Kit (TaKaRa-Bio). The denaturation mixture (final volume 10 μL), containing 5 μg of total RNA, dNTP mixture (containing 1 mM of dNTP) and 0.25 μM of Oligo dT primer, was denatured and annealed at 65°C for 5 min. The RT-reaction mixture (final volume 20 μL) containing 1 × PrimeScript buffer, 20 U of RNase inhibitor, 0.5 μL of PrimeScript RTase (for 2step) and denaturation mixture, was incubated at 42°C for 30 min and the enzyme was inactivated at 70°C for 15 min. The PCR mixture (final volume 50 μL) contained 1 × PCR buffer, dNTP mixture (containing 400 μM of dNTP), 0.2 μM appropriate sense primer, 0.2 μM appropriate anti-sense primer and 2.5 U of TaKaRa Ex Taq HS (TaKaRa-Bio). The PCR conditions included denaturation for 1 min at 94°C, followed by 35 cycles (30 s at 94°C, 30 s at 53°C and 1 min at 72°C), with a final extension for 5 min at 72°C.
The 3ʹ-untranslated regions (UTRs) of mating type genes were obtained by rapid amplification of cDNA ends (RACE)-PCR using T7 primer and polyT-T7 primer with TaKaRa PrimeScript RT-PCR Kit. The denaturation mixture (final volume 10 μL), containing 5 μg of total RNA, dNTP mixture (containing 1 mM of dNTP) and 0.25 μM of polyT-T7 primer, was denatured and annealed at 65°C for 5 min. The RT-reaction mixture (final volume 20 μL), containing 1 × PrimeScript buffer, 20 U of RNase inhibitor, 0.5 μl of PrimeScript RTase (for two steps) and whole denaturation mixture, was incubated at 42°C for 30 min. The enzymes were inactivated at 70°C for 15 min. The primary PCR mixture (final volume 50 μL) contained 1 × PCR buffer, dNTP mixture (containing 400 μM of dNTP), 0.2 μM sense primer, 0.2 μM T7 primer and 2.5 U of TaKaRa Ex Taq HS (TaKaRa-Bio). The PCR conditions included denaturation for 1 min at 94°C, followed by 30 cycles (30 s at 94°C, 30 s at 45°C and 2 min at 72°C), with a final extension for 5 min at 72°C. The secondary TAIL-PCR mixture (final volume 50 μl) contained the same components as the primary reaction with 0.2 μM nested sense primer, 0.2 μM T7 primer, and the template was 1 μL of a 1:50 dilution of the primary reaction mixture. PCR conditions consisted of 1 min at 94°C, followed by 30 cycles (30 s at 94°C, 30 s at 54°C and 1.5 min at 72°C), with a final extension for 5 min at 72°C.
Multiplex PCR was carried out to determine the mating type rapidly. Multiplex PCR mixtures (final volume 30 μL) contained 1 × PCR buffer, dNTP mixture (containing 2.5 mM of dNTP), 1 μM of four RT primers (PlT1MAT1-RT-F: 6, PlT1MAT1-RT-R: 7, PlT1MAT2-RT-F: 8 and PlT1MAT2-RT-R: 9), 1.25 U TaKaRa Taq and 50 ng of fungal genomic DNA. PCR conditions included 2 min at 94°C, followed by 30 cycles (30 s at 94°C, 30 s at 53°C and 1 min at 72°C), and a final extension for 5 min at 72°C. All PCR products were extracted using a QIAquick Gel Extraction Kit (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. Sequence analysis was conducted on an Applied Biosystems 3100 automated DNA sequencer (Applied Biosystems, Foster City, CA) using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) following the manufacturer’s instructions. All the sequences obtained in this study were deposited in the DDBJ/EMBL/GenBank databases. Dot-plots were generated using DOTTUP from EMBOSS explore.
Results
The fungal isolates included in this study were collected from different locations. This study is the first to explore the mating type idiomorphs of strains CBS267.59 and Oha3-6 of P. lycopersici.
Amplification of DNA binding domain and MAT idiomorphs
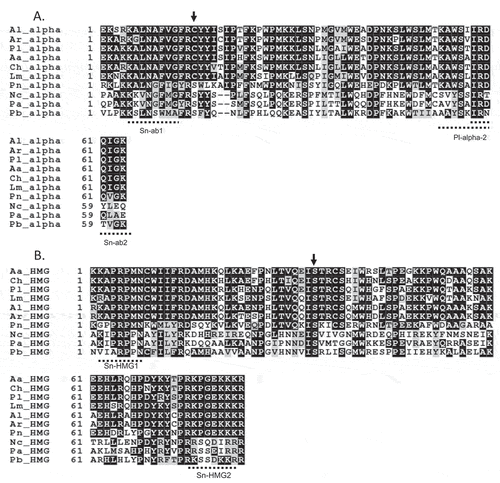
Degenerate primer pairs designed for mating type genes in P. nodorum () were used to amplify the DNA binding domains of P. lycopersici type 1 strains (CBS267.59 and Oha3-6). Phaeosphaeria nodorum is a member of the Loculoascomycetes and is phylogenetically close to P. lycopersici. A PCR product of approximately 250 bp was amplified using Sn-ab1 and Pl-alpha-2 from genomic DNA of strain CBS267.59, while a product of approximately 300 bp was obtained using Sn-HMG1 and Sn-HMG2 from genomic DNA of strain Oha3-6 (). Sequencing of the putative P. lycopersici alpha and HMG box and BLAST searches of the GenBank database with these sequences confirmed that they are closely related to the known alpha and HMG boxes ().
Fig. 1 Alignment of deduced amino acid sequences of the (a) alpha box, (b) HMG box and amino acid identity and similarity to Pseudopyrenochaeta lycopersici (Pl) of Ascochyta lentis (Al), A. rabiei (Ar), Alternaria alternata (Aa), Cochliobolus heterostrophus (Ch), Leptosphaeria maculans (Lm), Phaeosphaeria nodorum (Pn), Neurospora crassa (Nc), Podospora anserina (Pa) and Pyrenopeziza brassicae (Pb). Amino acid residues shaded in black are identical, while those in grey are similar. The primers Sn-ab1, Sn-ab2, Pl-alpha-2, Sn-HMG1 and Sn-HMG2 are indicated as dotted lines. The positions of introns are shown as vertical arrows
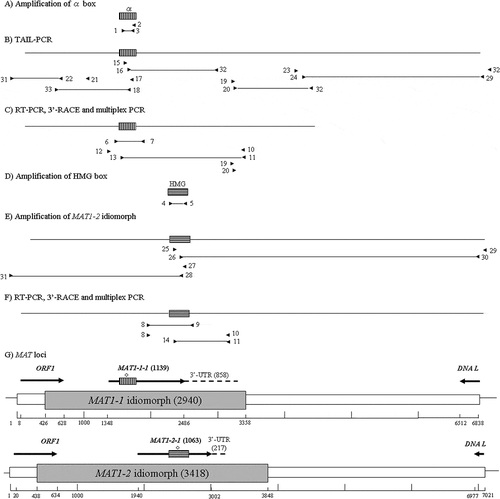
Confirmation of the idiomorph region was inferred based on the use of DOTTUP with default parameters (). The MAT1-1 idiomorph was obtained by TAIL-PCR using alpha box-specific primers and arbitrary degenerate primers. Approximately 1.6 kb was extended from the alpha box to the 5ʹ region using two TAIL-PCRs and approximately 5.2 kb was extended 3ʹ from the alpha box by three TAIL-PCR reactions (). The ORF1 and DNA lyase genes were observed in the 5ʹ and 3ʹ regions of the MAT1-1 locus, respectively. The primers ORF1-1 and GSP-F4-2 R were designed for ORF1 and DNA L sequences of the MAT1-1 strain, respectively. The MAT1-2 idiomorph was obtained using the primer pairs ORF1-1 and T1-GSP2-1 and T1-GSP1-1 and GSP-F4-2 R (). A fragment, approximately 2.5 kb in length, was amplified using ORF1-1 and T1-GSP2-1, and an approximately 4.6 kb fragment was obtained using T1-GSP1-1 and GSP-F4-2 R ().
Structure of the MAT loci
Full sequences of the mating type genes and flanking regions were obtained from P. lycopersici type 1 strains CBS267.59 and Oha3-6 and were compared with mating type genes of other Loculoascomycetes (Cochloibilus heterostrophus, Alternaria alternata, Ascochyta rabiei, Ascochyta lentis, Leptosphaeria maculans and Phaeosphaeria nodorum), Pyrenomycetes (Neurospora crassa and Podospora anserina) and Discomycetes (Pyrenopeziza brassicae). Analysis of the assembled sequences identified a MAT1-1 idiomorph (strain CBS267.59) of 2932 bp (nucleotide positions 426–3358) and a MAT1-2 idiomorph (strain Oha3-6) of 3410 bp (nucleotide positions 438–3848) (). The MAT1-1-1 is 1139 bp (1348–2486) in length, encoding a predicted protein of 361 amino acids. Sequencing of the cDNA fragments amplified by RT-PCR indicated an intron-sized 56 bp at a similar position to the alpha box of other Dothideomycetes. MAT1-2-1 is 1063 bp (1940–3002) in length, encoding a predicted protein of 336 amino acids. Sequencing of the cDNA fragments amplified by RT-PCR indicates an intron-sized 55 bp at a similar position to the HMG box of other Dothideomycetes. The P. lycopersici type 1 alpha box domain shared 87%, 87%, 87%, 82%, 81%, 57%, 42%, 37% and 37% amino acid sequence similarity, respectively, with the alpha box domains of C. heterostrophus, A. alternata, A. lentis, A. rabiei, L. maculans, P. nodorum, N. crassa, P. anserina and P. brassicae (). The P. lycopersici type 1 HMG box domain amino acid sequence shared 85%, 84%, 83%, 81%, 81%, 55%, 39%, 34% and 31% similarity, respectively, with L. maculans GenBank accession nos. AY174048 and AY174049, A. lentis accession nos. DQ341314 and DQ341315, A. rabiei accession nos. DQ341313 and DQ341312, C. heterostrophus accession nos. AF029913 and AF027687, A. alternata accession nos. AB009451 and AB009452, P. nodorum accession nos. AY212018 and AY212019, P. brassicae accession nos. AJ006073 and AJ006072, P. anserina accession nos. X64194 and X64195, and N. crassa accession nos. M33876 and M54787 ().
The ORF1 and DNA lyase were obtained from the 5ʹ region and 3ʹ region of the P. lycopersici type 1 MAT locus, respectively. ORF1 was observed in other Loculoascomycetes: C. heterostrophus, L. maculans and P. nodorum (Cozijnsen and Howlett Citation2003). Pseudopyrenochaeta lycopersici ORF1 overlapped in the MAT idiomorphs () and was similar to ORF1 in C. heterostrophus, L. maculans and P. nodorum ().
Fig. 2 Alignment of the deduced amino acid sequence of ORF1 genes in MAT1-1 and MAT1-2 isolates of Cochliobolus heterostrophus (Ch) GenBank accession nos. AF029913 (MAT1-1) and AF027687 (MAT1-2), Phaeosphaeria nodorum (Pn) GenBank accession nos. AY212018 (MAT1-1) and AY212019 (MAT1-2), Leptosphaeria maculans (Lm) GenBank accession nos. AY174048 (MAT1-1) and AY174049 (MAT1-2) and Pseudopyrenochaeta lycopersici (Pl). The arrow indicates the beginning of idiomorph
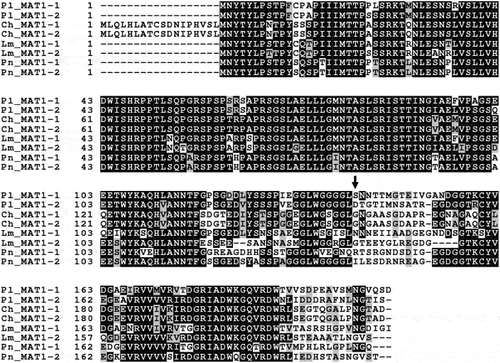
Fig. 3 Dot-plot comparison of MAT loci. The MAT1-1 idiomorph starts from 355 bp and ends at 3295 bp. The MAT1-2 idiomorph starts from 133 bp and ends at 3551 bp. Each ORF is indicated as arrows
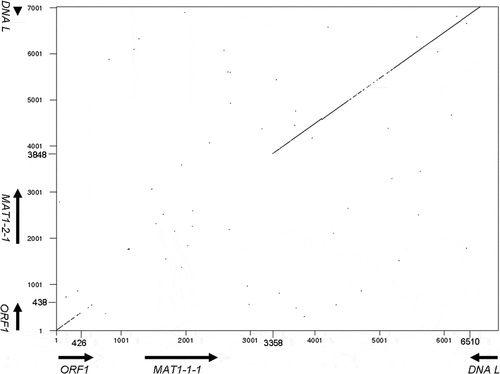
Fig. 4 PCR cloning of MAT loci from Pseudopyrenochaeta lycopersici type 1. (a) Amplification of the sequence corresponding to the alpha box using degenerate primers 1, 2 and 3 from strain CBS267.59 of the fungus. (b) TAIL-PCR to obtain the 5ʹ and 3ʹ region sequence from the alpha box. (c) RT-PCR with primers 6 and 7, 3ʹ-RACE with primers 10, 11, 12 and 13 and multiplex PCR with primers 6 and 7. (d) Amplification of the sequence corresponding to the HMG box from strain Oha3-6 using degenerate primers 4 and 5. (e) Amplification of the MAT1-2 idiomorph using primers 25, 26, 27 and 28 designed based on the sequence of the HMG box and primers 29, 30 and 31 designed based on the sequences of DNA lyase and ORF1 gene. (f) RT-PCR with primers 8 and 9, 3ʹ-RACE with primers 8, 10, 11 and 14 and multiplex PCR with primers 8 and 9. (g) Structural organization of the P. lycopersici type 1 MAT loci. Grey boxes indicate MAT idiomorphs. Scale bar is marked to denote the entire region sequenced. Direction of transcription and limits of the ORFs is indicated by arrows, with a horizontally striped and a vertically striped box representing the alpha and HMG boxes in MAT1-1-1 and MAT1-2-1, respectively. 3ʹ-UTR are shown as dotted lines. Introns are indicated by diamonds on the striped boxes. Primers are indicated as arrowheads
Transcription of MAT genes
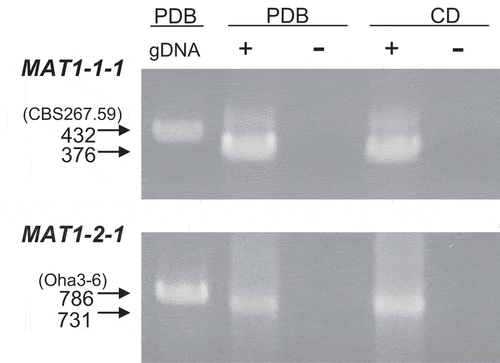
Transcription of the MAT genes was examined in P. lycopersici type 1 strains CBS267.59 (MAT1-1) and Oha3-6 (MAT1-2) by RT-PCR. Mycelia cultured in PDB (nutrient-rich medium) and CD (nutrient minimum medium) were used for RNA extraction. The PlT1MAT1-RT-F and PlT1MAT1-RT-R primer pair and PlT1MAT2-RT-F and PlT1MAT2-RT-R primer pair ( and ) were used for amplification of total RNA of CBS267.59 and Oha3-6, with 376 and 731 bp cDNA fragments expected, respectively (). Additionally, 3ʹ-UTRs were determined by 3ʹ-RACE using the PlT1MAT1-RT-F1 or PlT1MAT2-RT-F and polyT-T7 primer pair (primary PCR) and the RTN-Plalpha-1 or RTN-PlHMG-1 and T7 primer pair (secondary PCR) ( and ). A fragment (858 bp) was obtained in CBS267.59 (MAT1-1) and sequenced, with the polyadenylation site detected 749 bp downstream from the stop codon. A fragment (217 bp) was also obtained from strain Oha3-6 (MAT1-2) and sequenced, with the polyadenylation site detected 171 bp downstream from the stop codon.
Fig. 5 Expression of the mating type genes in Pseudopyrenochaeta lycopersici. gDNA, genomic DNA of strain CBS267.59 (MAT1-1) and Oha3-6 (MAT1-2) as the template. + = RT-PCR with reverse transcriptase; – = without reverse transcriptase. Genomic DNA was extracted from each strain grown on PDB (potato dextrose broth). Total RNA was extracted from each strain grown on PDB or CD (Czapek-dox)
Multiplex PCR for determination of mating type
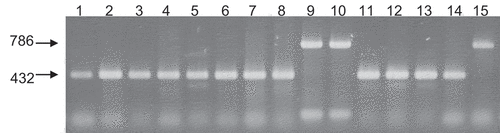
Multiplex PCR was performed with the genomic DNA of 33 strains of P. lycopersici using RT-PCR primers ( and and ). The strains CBS267.59 (GenBank accession no. AB979660.1) and MAFF712040 (GenBank accession no. AB971217.1) were found to be MAT1-1 while the strain Oha3-6 (GenBank accession no. AB979661.1) was determined to be MAT1-2. In general, 432 bp and 786 bp bands were detected in the MAT1-1 and MAT1-2 strains, respectively, and the results from 15 strains are shown in . Most type 1 strains were found to be MAT1-1 while only three were determined to be MAT1-2. The assembled genome sequence (GenBank assembly accession GCA_003313425.1) was 80%, 96% and 99% identical to CBS267.59, MAFF712040 and Oha3-6, respectively (Supplementary ).
Fig. 6 Multiplex PCR with primers 6, 7, 8 and 9. From left to light: lane 1, CBS267.59; lane 2, MAFF712039; lane 3, Au3-10; lane 4, Au4-3; lane 5, Ty2-7; lane 6, Lt2-12; lane 7, GR2-1; lane 8, GR3-3; lane 9, Y502-H1; lane 10, Y502-M33; lane 11, Y519-31; lane 12, 24-C; lane 13, PrF2; lane 14, Oha3-5; lane 15, Oha3-6. Lanes 1–8 and 11–14 are MAT1-1 and lanes 9, 10 and 15 are MAT1-2.
Discussion
The term ‘mating types’ was regularly used following the discovery of heterothallism in fungi. Cloning and sequencing analyses elucidated the role of mating types in the sexual development of Ascomycetes (Glass et al. Citation1988). Afterwards, several genetic and physiological investigations were carried out in numerous species. Despite this, no information is available regarding the mating types of P. lycopersici. Therefore, in this study the mating type genes of P. lycopersici were isolated and characterized to increase ecological and molecular biological knowledge of the species. So far, sexual reproduction has not been observed in this fungus.
PCR-based methods enabled the isolation of mating type genes in P. lycopersici type 1 using degenerate primers designed based on the DNA binding domain of the related fungus, P. nodorum. As a result, MAT1-1-1 and MAT1-2-1 were identified in strains CBS267.59 and MAFF712040 (type 1) and Oha3-6 (type 1), respectively, of P. lycopersici. Three rounds of BLAST were carried out to compare nucleotide sequences of the mating type genes from these strains with the assembled genome sequence of the fungus (Aragona et al. Citation2014). Based on the BLAST searches, a high similarity was detected. This means that the mating genes were not annotated by Aragona et al. (Citation2014). They considered P. lycopersici to be an imperfect fungus. In this study, MAT1-1-1 derived from strain CBS267.59 encodes a predicted protein including an alpha box DNA binding domain, while MAT1-2-1 derived from strain Oha3-6 encodes a predicted protein including an HMG DNA binding domain. The amino acid sequences in each domain were similar to those of other asexual and heterothallic fungi in the Dothideomycetes, and the introns were observed at the same position as the mating type genes of the fungi mentioned above.
Heterologous expression of the MAT genes of presumed asexual fungi has been performed in C. heterostrophus (Sharon et al. Citation1996; Arie et al. Citation2000). Arie et al. (Citation2000) examined whether the A. alternata MAT genes were functional in C. heterostrophus and found that C. heterostrophus strains with the A. alternata MAT genes produced pseudothecia that were indistinguishable from those produced in wild-type mating, but which were less abundant. This indicates that the MAT genes of A. alternata are functional, suggesting that the MAT genes of other asexual fungi could also be functional. In this study, the mating type genes of P. lycopersici were transcribed when the fungus was cultured in both nutrient-rich and nutrient-poor conditions. It was not, however, determined whether or not sexual reproduction can occur between MAT1-1 and MAT1-2 isolates. From the results, the MAT1-1 strains were predominantly found in type 1 (30 strains were MAT1-1 and three strains were MAT1-2), and the mating type ratio in type 1 strains was not 1:1. The reason for asexuality in P. lycopersici may be the absence of compatible mating partners in the field. In type 2 of P. lycopersici, MAT1-2 was obtained, but not MAT1-1 (data not shown).
Traditionally, the taxonomy of fungal species was based mainly on morphological and biological features, but recently, phylogenetic analysis using more than one gene has been utilized to determine more precisely taxonomy. As a result, some species may be subdivided further as varieties, forma speciales, anastomosis groups, or even as different species (Geiser et al. Citation1998; Nirenberg and O’Donnell Citation1998; O’Donnell et al. Citation1998; Kasuga et al. Citation1999; Vigot and Wostemeyer Citation2000). Pseudopyrenochaeta lycopersici type 1 and type 2 are recognized to be distinct based on morphological, physiological and molecular biological characteristics. Each could be reclassified as a subspecies (form or mating population) or as a phylogenetically closely related species within the genus Pseudopyrenochaeta.
While MAT1-1-1 and MAT1-2-1 from P. lycopersici type 1 were identified in this study, we are now attempting to find these two genes in type 2. Furthermore, the morphology of the sexual structures (pseudothecia) of these types could be compared using isolates with different mating type genes. Such research may help to clarify the taxonomic positions of P. lycopersici type 1 and type 2.
Supplemental Figures 1, 2 and 3
Download MS Word (116.5 KB)Supplemental data
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2021.1934733
Additional information
Funding
References
- Aragona M, Minio A, Ferrarini A, Valente M, Bagnaresi P, Orrù L, Tononi P, Zamperin G, Infantino A, Valè G, et al. 2014. De novo genome assembly of the soil-borne fungus and tomato pathogen Pyrenochaeta lycopersici. BMC Genomics. 15:313. doi:https://doi.org/10.1186/1471-2164-15-313.
- Arie T, Kan`eko I, Yoshida T, Noguchi M, Nomura Y, Yamaguchi I. 2000. Mating type genes from asexual phytopathogenic ascomycetes, Fusarium oxysporum and Alternaria alternata. Mol Plant-Microbe Interact. 13:1330–1339. doi:https://doi.org/10.1094/MPMI.2000.13.12.1330.
- Bardwell L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides. 26:339–350. doi:https://doi.org/10.1016/j.peptides.2004.10.002.
- Bayraktar H, Oksal E. 2011. Molecular, physiological and pathogenic variability of Pyrenochaeta lycopersici associated with corky rot disease of tomato plants in Turkey. Phytoparasitica. 39:165–174. doi:https://doi.org/10.1007/s12600-011-0150-z.
- Bennett RS, Yun S-H, Lee TY, Turgeon BG, Arseniuk E, Cunfer BM, Bergstrom GC. 2003. Identity and conservation of mating type genes in geographically diverse isolates of Phaeosphaeria nodorum. Fungal Genet Biol. 40:25–37. doi:https://doi.org/10.1016/S1087-1845(03)00062-8.
- Coppin E, Debuchy R, Arnaise S, Picard M. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev. 61:411–428. doi:https://doi.org/10.1128/.61.4.411-428.1997.
- Cozijnsen AJ, Howlett BJ. 2003. Characterization of the mating-type locus of the plant pathogenic ascomycete Leptosphaeria maculans. Curr Genet. 43:351–357. doi:https://doi.org/10.1007/s00294-003-0391-6.
- Debuchy R, Berteaux-Leceleir V, Silar P. 2010. Mating systems and sexual morphogenesis in ascomycetes. In: Borkovich KA, Ebbole DJ, editors. Cellular and molecular biology of filamentous fungi. Washington (DC): ASM Press; p. 501–535.
- Geiser DM, Pitt JI, Taylor JW. 1998. Cryptic speciation and recombination in the aflatoxin producing fungus Aspergillus flavus. Proc Natl Acad Sci USA. 95:388–393. doi:https://doi.org/10.1073/pnas.95.1.388.
- Glass NL, Vollmer SJ, Staben C, Grotelueschen J, Metzenberg RL, Yanofsky C. 1988. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science. 241:570–573. doi:https://doi.org/10.1126/science.2840740.
- Golzar H. 2009. First report of Pyrenochaeta lycopersici, causal agent of tomato corky root rot in Australia. Australas Plant Dis Notes. 4:126–128.
- Hicks J, Strahern JN, Klar AJS. 1979. Transposable mating type genes in Saccharomyces cerevisiae. Nature. 282:478–483. doi:https://doi.org/10.1038/282478a0.
- Hieno A, Naznin HA, Suga H, Yamamoto YY, Hyakumachi M. 2015. Specific detection of Type 1 and Type 2 isolates of Pyrenochaeta lycopersici by loop-mediated isothermal amplification reaction. Acta Agric Scand B – Soil Plant Sci. 66:353–358.
- Hockey AG, Javes TM. 1984. Isolation and identification of Pyrenochaeta lycopersici, causal agent of tomato brown root rot. Trans Brit Mycol Soc. 82:151–152. doi:https://doi.org/10.1016/S0007-1536(84)80220-X.
- Infantino A, Pucci N. 2005. A PCR-based assay for the detection and identification of Pyrenochaeta lycopersici. Eur J Plant Pathol. 112:337–347. doi:https://doi.org/10.1007/s10658-005-6605-7.
- Kasuga T, Taylor JW, White TJ. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus, Histoplasma capsulatum Darling. J Clin Microbiol. 37:653–663. doi:https://doi.org/10.1128/JCM.37.3.653-663.1999.
- Kelly M, Burke J, Smith M, Klar A, Beach D. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7:1537–1548. doi:https://doi.org/10.1002/j.1460-2075.1988.tb02973.x.
- Kim H, Borkovich KA. 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot Cell. 5:544–554. doi:https://doi.org/10.1128/EC.5.3.544-554.2006.
- Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Pöggeler S. 2010. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and non-essential sexual regulators. Eukaryot Cell. 9(6):894–905. doi:https://doi.org/10.1128/EC.00019-10.
- Kothe E. 2008. Sexual attraction: on the role of fungal pheromone/receptor systems. Acta Microbiol Immunol Hung. 55:125–143. doi:https://doi.org/10.1556/AMicr.55.2008.2.5.
- Liu YG, Whittier F. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragment from P1 and YAC clones for chromosome walking. Genomics. 25:674–681. doi:https://doi.org/10.1016/0888-7543(95)80010-J.
- Nirenberg HI, O’Donnell K. 1998. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia. 90:434–458. doi:https://doi.org/10.1080/00275514.1998.12026929.
- O’Donnell K, Cigelink E, Nirenberg HI. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 90:465–493. doi:https://doi.org/10.1080/00275514.1998.12026933.
- Sharon A, Yamaguchi K, Christiansen S, Horwitz BA, Yoder OC, Turgeon BG. 1996. An asexual fungus has the potential for sexual development. Mil Gen Genet. 251:60–68. doi:https://doi.org/10.1007/BF02174345.
- Staben C, Yanofsky C. 1990. Neurospora crassa mt a mating-type region. Proc Natl Acad Sci USA. 87:4917–4921. doi:https://doi.org/10.1073/pnas.87.13.4917.
- Tsong AE, Miller MG, Raisner RM, Johnson AD. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 115:389–399. doi:https://doi.org/10.1016/S0092-8674(03)00885-7.
- Turgeon BG. 1998. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol. 36:115–137. doi:https://doi.org/10.1146/annurev.phyto.36.1.115.
- Valente MT, Infantino A, Aragona M. 2011. Molecular and functional characterization of an endoglucanase in the phytopathogenic fungus Pyrenochaeta lycopersici. Curr Genet. 57:241–251. doi:https://doi.org/10.1007/s00294-011-0343-5.
- Vigot K, Wostemeyer J. 2000. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiol Res. 155:179–195. doi:https://doi.org/10.1016/S0944-5013(00)80031-2.
