?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Several species of Pythium cause seed rot and damping-off of soybean in the United States and Canada. The aggressiveness of 14 isolates representing five Pythium species was evaluated on six North American soybean (Glycine max) cultivars based on seed disease in a Petri plate assay and on seedling emergence, plant and root weights, and root rot severity in temperature-controlled greenhouse experiments. In the in vitro assay, the P. aphanidermatum, P. ultimum var. ultimum and P. spinosum isolates caused higher disease severity on germinating seeds than the P. sylvaticum and P. irregulare isolates. In the greenhouse inoculum layer assays, all isolates reduced emergence and tissue weights of at least some cultivars, with significant isolate × cultivar interactions. The aggressiveness of isolates within each species varied significantly based on all or most of the disease parameters measured. At least one isolate from each species reduced emergence by at least 50% on some cultivars, but Pythium ultimum var. ultimum isolate PU 350 caused the most damping-off, stunting and root rot overall. There was a significant negative correlation between root rot severity or seedling emergence (r = −0.93, P < 0.001). ‘Archer’ and ‘Maple Glen’ were more resistant overall than ‘Conrad’, ‘Maple Isle’, ‘Sloan’, ‘Williams’, but some isolates were not very aggressive on any of the cultivars. This study revealed significant variation in the relative ability of the 14 isolates to cause disease and illustrated the importance of identifying aggressive isolates of Pythium species for applications like resistance screening or genetic mapping studies.
Résumé: Aux États-Unis et au Canada, certaines espèces de Pythium causent la pourriture des semences et la fonte des semis chez le soya (Glycine max). L’agressivité de 14 isolats représentant 5 espèces de Pythium a été évaluée chez 6 cultivars de soya en se basant sur le développement de la maladie des semences dans une boîte de Petri ainsi que sur l’émergence des semis, le poids des plants et des racines, de même que sur la gravité du pourridié dans le cadre d’expériences menées en serre, dans des conditions de température contrôlées. Au cours de l’épreuve biologique in vitro, les isolats de P. aphanidermatum, de P. ultimum var. ultimum et de P. spinosum ont infecté plus gravement les semences en voie de germination que les isolats de P. sylvaticum et de P. irregulare. Au cours des épreuves biologiques d’inoculation en couche menées en serre, tous les isolats ont retardé l’émergence et réduit le poids des tissus d’au moins quelques cultivars, et ce, avec des interactions importantes entre l’isolat et le cultivar. En se basant sur tous ou la plupart des paramètres de la maladie mesurés, l’agressivité des isolats au sein de chaque espèce a varié considérablement. Au moins un isolat de chaque espèce a retardé l’émergence d’au moins 50% chez certains cultivars, mais l’isolat PU 350 de Pythium ultimum var. ultimum a causé généralement la majorité des cas de fonte des semis, de rabougrissement et de pourridié. Il y avait une corrélation négative significative entre la gravité du pourridié et l’émergence des semis (r = −0,93, P < 0,001). Les cultivars ‘Archer’ et ‘Maple Glen’ étaient généralement plus résistants que ‘Conrad’, ‘Maple Isle’, ‘Sloan’ et ‘Williams’, mais certains isolats n’étaient pas très agressifs à l’égard d’aucun cultivar. Cette étude a révélé une importante variation quant à la capacité relative des 14 isolats à causer la maladie et a illustré l’importance d’identifier les isolats agressifs des espèces de Pythium pour des applications telles que le criblage en vue de la résistance ou des études sur la cartographie génétique.
Introduction
The genus Pythium contains some of the most important soilborne pathogens of soybean grown in the United States and in southern Ontario, Canada (Rojas et al. Citation2017). Pythium spp. are necrotrophic oomycetes that often have a wide host range, including maize (Zea mays), which is often grown in rotation with soybean. Pythium pathogens in soybean fields are typically part of a complex of soilborne pathogens that may include Phytophthora sojae and Rhizoctonia spp. in addition to multiple Pythium spp. (Rizvi and Yang Citation1996; Dorrance et al. Citation2004; Broders et al. Citation2007). These pathogens cause seed rot, pre- and post-emergence damping-off, and root injury on surviving plants (Rupe et al. Citation2011; Rothrock et al. Citation2015). As cool, damp soils are favourable for promoting infection by many oomycete pathogens, the potential for disease and economic losses has increased because of a trend towards earlier planting dates (Hu and Wiatrak Citation2012). Serrano and Robertson (Citation2018) found that periods of cold stress for 24 hours or more reduced emergence in the presence of P. sylvaticum. Moreover, cool temperatures also slow the rate of seed germination. Abundant precipitation before and after planting reduced emergence and increased post-emergence damping-off caused by Pythium spp. (Kirkpatrick et al. Citation2006b). Volatile compounds from germinating seeds induce the germination of Pythium sporangia, and saturated soils facilitate infection of seeds and seedlings (Nelson Citation1987). Damage to seeds may increase infection and root rot (Rupe et al. Citation2011).
At least 17 species of Pythium have been recovered from diseased soybean plants in North America (Rupe et al. Citation2011; Rojas et al. Citation2017). A 2011–2012 survey of Oomycete pathogens recovered from diseased soybean seedlings in Ontario and several U.S. states indicated that P. sylvaticum, P. oopapilum, P. irregulare, P. heterothalicum, P. ultimum var. ultimum, and P. perplexum were among the most common pathogens identified (Rojas et al. Citation2017). Pythium sylvaticum and P. irregulare were among the most common Pythium species found in Ohio fields in 2004 and 2005, with isolates recovered from both maize and soybean (Broders et al. Citation2007). Pythium ultimum var. ultimum was less common in Ohio, but was one of the main species isolated from infected soybean plants.
Seed treatment with fungicide products containing metalaxyl, mefenoxam, ethaboxam, or a strobilurin is widely used in North America to protect soybean seeds and seedlings from Pythium and other soilborne pathogens (Dorrance et al. Citation2009; Urrea et al. Citation2013; Noel et al. Citation2020). This exerts selection pressure on soil pathogens, and some field populations have evolved strains with increased tolerance to metalaxyl and mefenoxam (Broders et al. Citation2007; Urrea et al. Citation2017; Scott et al. Citation2020). Moreover, as seedling roots grow beyond the zone with concentrated fungicide/oomicide, they may become more vulnerable to infection if soil moisture and/or temperature remain favourable for oomycete pathogens (Dorrance et al. Citation2009). Noel et al. (Citation2020) were able to isolate more than 20 Pythium spp. from soybean seedlings that developed from seeds treated with mefenoxam and/or ethaboxam. Soybean resistance to the most common and most aggressive Pythium spp. would therefore be a valuable trait.
Resistance to one or more Pythium spp. has been reported in several soybean cultivars, breeding lines and plant introductions (PIs) (Kirkpatrick et al. Citation2006a, Citation2006b; Rod et al. Citation2018; Klepadlo et al. Citation2019; Lerch-Olson et al. Citation2020). Soybean resistance to Pythium spp. typically involves quantitative differences in damping-off and injury to the root systems of infected seedlings. Pythium spp. do not have a complex race structure like Phythophthora sojae, and relatively little has been reported about the extent of variation in aggressiveness within different species. Kirkpatrick et al. (Citation2006a, Citation2006b) reported that the soybean cultivar ‘Archer’ had resistance to P. ultimum populations in the state of Arkansas, and that it reduced the severity of both seed rot and root rot. Rosso et al. (Citation2008) identified a P. aphanidermatum resistance gene on Chromosome 13 (Linkage Group F) of ‘Archer’ that was assigned the name Rpa1. Although Rpa1 appeared to segregate as a single dominant gene in the mapping population, no other major R genes with a large effect on Pythium resistance have been reported to the best of our knowledge. ‘Magellan’ is another U.S. cultivar with moderate resistance to some isolates of P. ultimum var. ultimum (Klepadlo et al. Citation2019), and ‘Maverick’ has resistance to four species of Pythium (Lerch-Olson et al. Citation2020). Rod et al. (Citation2018) reported that PI 84637, the Canadian cultivar ‘Maple Isle’, and the Swedish lines Fiskeby III and Fiskeby 840-7-3 have resistance to Illinois isolates of P. ultimum var. ultimum, P. irregulare and P. sylvaticum.
Many of the studies conducted to date have focused on either the reactions of one or two soybean lines to a wide range of Pythium isolates, or the reactions of a wider range of soybean genotypes to one or a few Pythium spp. isolates. The objectives of this study were to evaluate the relative aggressiveness of 14 isolates representing five Pythium spp. based on seed rot and root rot severity on seedlings of six North American soybean cultivars. In this report, we follow the definitions used by Matthiesen et al. (Citation2016) of aggressiveness as the quantitative degree of disease injury caused by a pathogen species or isolate, and pathogenicity as the ability of a pathogen to cause disease on a specific host.
Materials and methods
Soybean cultivars
Six soybean cultivars were chosen on the basis of information about their levels of resistance to certain Pythium species and isolates. ‘Williams’ (Bernard and Lindahl Citation1972) and ‘Sloan’ (Bahrenfuss and Fehr Citation1980) were chosen as susceptible checks because they have been used in that role in several previous studies of Pythium resistance (Broders et al. Citation2007; Ellis et al. Citation2013; Klepadlo et al. Citation2019). ‘Archer’ was included because it is known to be resistant or moderately resistant to Arksansas isolates of at least five Pythium spp. (Cianzio et al. Citation1991; Bates et al. Citation2008), though it was moderately susceptible to a P. irregulare isolate from Ohio (Ellis et al. Citation2013). ‘Conrad’ had moderate resistance to an Ohio isolate of P. irregulare in a study by Ellis et al. (Citation2013) but seemed less resistant in another study (Stasko et al. Citation2016). The Canadian cultivar ‘Maple Isle’ was included because Rod et al. (Citation2018) found that it had resistance to Illinois isolates of three different Pythium species. Subsequent assays showed that another Canadian cultivar, ‘Maple Glen’, was at least as resistant to P. ultimum var. ultimum (unpublished data), so it was also included. Seeds used in the experiments were obtained from the USDA Soybean Germplasm Collection in Urbana, IL, USA, or were harvested from plants grown from seeds obtained from the Collection.
Pythium isolates
Fourteen isolates representing five species of Pythium were used in this study (). The species were P. aphanidermatum (three isolates), P. irregulare (two isolates), P. spinosum (two isolates), P. sylvaticum (two isolates), and P. ultimum var. ultimum (three isolates). All of the isolates were recovered from infected soybean plants and were therefore known to be pathogenic on soybean. The isolates originated from three states, but the majority were collected in Arkansas. All isolates were obtained from sources listed in and had previously been identified to the species level based on morphology and molecular analyses. The P. ultimum var. ultimum 12Py391 isolate was previously used in Pythium resistance assays conducted by Rod et al. (Citation2018).
Table 1. Names of Pythium spp. isolates used in this study and their origins and sources.
Petri plate radial growth assay
The inherent vigour of the Pythium spp. isolates was compared in a Petri plate assay to determine growth rates on potato dextrose agar (PDA). For each isolate, a 5-mm diameter mycelial plug was punched from the edge of a fully colonized PDA plate that had been incubated at room temperature (RT; 23 ± 2°C) for 7 days. The plug was transferred to the centre of a 90-mm Petri plate containing PDA and the plate was incubated at RT for 4 days. The plates were covered with aluminium foil to keep them dark during the incubation period (Groenewald et al. Citation2006), and were arranged in a completely random design, with three replications of each isolate. Each day the diameter of the colonized area in each plate was measured in two perpendicular directions, and the mean of those two measurements was regarded as the colony size after 5 mm had been subtracted out.
Petri plate aggressiveness assay
A separate in vitro assay was conducted with seeds from the six cultivars to assess the aggressiveness of the 14 isolates based on seed germination and early root disease. The procedures used were similar to those described in Broders et al. (Citation2007) and Zhang and Yang (Citation2000). A 5-mm plug of each Pythium isolate was transferred from a 7-day-old culture grown on cornmeal agar (CMA) to the centre of each 90-mm wide Petri plate containing 1.5% water agar (WA). The plates were then incubated at RT in the dark for 2–3 days. In the meantime, seeds were disinfested in a solution containing 0.5% sodium hypochlorite (NaOCl) for 3 min, washed with tap water for 3 min, and then rinsed in sterilized water for 3 min. Eight evenly spaced seeds were placed in each plate near the outer edge of the mycelial growth. Non-inoculated control plates with eight seeds were also set up to determine seed viability and vigour in the absence of the pathogen. The plates were then incubated at RT for 7 days in the dark, after which disease ratings were made. Each isolate was considered a treatment and was replicated in three plates. A disease severity scale from Rojas et al. (Citation2017) was used in which 0 = healthy germinated seed; 1 = delayed development, with minimal or no discolouration on roots; 2 = germination, with isolated lesions and discoloured roots; 3 = germination, but with coalesced lesions; and 4 = no germination due to colonization of seed. A disease severity index (DSI) for each isolate was then calculated as:
Preparation of inoculum for greenhouse assays
Inoculum was prepared using methods similar to those described by Broders et al. (Citation2007) and Ellis et al. (Citation2013). A soil-based substrate mixture was prepared in a ratio of 950 mL of steamed sandy loam soil (2 parts sand: 1 part soil), 500 mL of medium coarse vermiculite (Perlite, Inc., Canada), and 50 mL of cornmeal. After being mixed thoroughly, 1-litre portions of the mix were transferred to autoclave-safe plastic bags (Worldwide Life Sciences Division, Inc., Bristol, PA). The mixture was autoclaved twice at 121°C for 60 min on successive days, with a 24-hour cooling period in between. Subcultures of each isolate grown on cornmeal agar medium were transferred to potato dextrose agar (PDA) medium in Petri dishes and were incubated at room temperature (23 ± 2°C) for 6–7 days. Cultures from two plates of each isolate were then diced into small pieces and aseptically transferred to each bag of the autoclaved soil substrate and mixed thoroughly with 200–250 mL of double-distilled water. The inoculated bags were covered with aluminium foil to keep them dark and were incubated for 7 days at room temperature to promote colonization of the substrate. The bags were periodically shaken to enhance uniform distribution.
Greenhouse assays
Aggressiveness of each isolate on the six soybean genotypes was assessed in factorial experiments with the 14 Pythium isolates. The experiments were arranged in randomized complete blocks with five replications, and each experiment was repeated once. Each pot was considered an experimental unit, and in each assay, there were three non-inoculated control pots for each soybean/isolate combination. Plastic pots with 10-cm diameter and depth, and a volume of 370 mL were filled with approximately 240 mL of a steamed sandy loam soil mix, which was then saturated with water for 24 hours. Assay inoculum was prepared by thoroughly mixing a 1:1 ratio of sandy loam soil with the inoculum mixture. A 40-mL layer of this mixture was then added to each pot and covered with another 40 mL of the steamed sandy loam soil. Eight seeds of the same genotype were placed on this layer of soil, approximately 1.5-cm apart, and were then covered with 50 mL of coarse vermiculite. The seeds used in the assays were disinfested and rinsed with water. Pots for the non-inoculated checks were prepared the same way as the inoculated pots, except that the layer containing inoculum was omitted and was substituted with sterilized soil mix. Approximately 1 g of Osmocote 17-6-12 slow-release fertilizer (The Scotts Company, Marysville, OH) was added to each pot and the pots were watered daily to maintain a high level of moisture in the soil substrate. Following emergence of seedlings, the pots were kept in trays flooded with water to a depth of 3–4 cm until 2 days before the end of each assay to keep the potting medium moist. The experiment was repeated once.
The assays were conducted in a greenhouse in the Plant Care Facility at the University of Illinois in Urbana-Champaign. Central air conditioning and shade curtains controlled by a computer monitoring system were deployed on sunny days to help keep the temperatures between 18° and 22°C. This temperature range was intermediate between the ideal temperatures for the Pythium spp. used in the experiments. Pythium ultimum and P. aphanidermatum are highly pathogenic at 23°C (Zitnick-Anderson and Nelson Citation2015), whereas the optimal temperatures for some of the other Pythium spp. are cooler (Wei et al. Citation2011). Natural sunlight was supplemented with light from cool-white fluorescent bulbs for 10 hours each day to reduce seedling etiolation.
Disease evaluation in greenhouse assays
Isolate aggressiveness was assessed on the basis of seedling emergence (i.e., plant stands) 10 days after planting and on whole plant and root weights 17 days after planting. A seedling was considered to have emerged if its cotyledons were above the soil surface. Surviving plants were separated from the soil mix 17–18 days after planting by gently washing the soil from the roots, after which the plants were placed on paper towels to dry before being weighed. The roots of plants in non-inoculated control pots were also washed and used for comparison. Data were collected for fresh plant weight (FPW), fresh root weight (FRW), dry root weight (DRW) and root rot severity when plants were in the V2 growth stage (Fehr et al. Citation1971). The methods were similar to those described in Broders et al. (Citation2007).
Root rot severity was rated using the 1 to 5 scale of Klepadlo et al. (Citation2019), in which 1 = a healthy root system with no visible lesions; 2 = small lesions on the lateral roots, with visible symptoms on 1 to 20% of the roots; 3 = rot on lateral roots and lesions beginning on the tap root, with visible discolouration on 21 to 75% of the roots; 4 = visible symptoms on main tap root and lateral roots and discolouration of 76 to 100% of the root system; and 5 = no germination due to seed rot. The root rot rating for a pot was the mean for the surviving plants in that pot. After root disease severity was rated, the roots were placed in an oven and dried at 50°C for 72 hours to obtain the root dry weights. Standardized plant and root weights were calculated for each cultivar using an equation from Ellis et al. (Citation2013) to adjust for genotype-related variability among non-inoculated cultivars for seed germination and seedling vigour. Weights for each cultivar were adjusted as:
Data analyses
Data for the radial growth of the isolates on PDA were log-transformed and Levene’s test of homogeneity of variance was performed to ensure that the assumptions for analysis of variance (ANOVA) were met using PROC GLM of SAS version 9.4 (SAS Institute, Inc., Cary, NC). If the variances in the two runs of experiments were not significantly different, these data were combined for the analysis. Variation in radial growth across species and among isolates of a species was also analyzed using PROC GLM. Significant differences among means were determined using Fisher’s protected least significant difference test (α = 0.05).
For the in vitro pathogenicity tests, disease severity index (DSI) in a range of 0 to 1 was calculated and arcsine-transformed to meet the assumptions required for ANOVA. Levene’s test of homogeneity of variances was then used to compare the two experimental runs using PROC GLM in SAS version 9.4. If there was no significant difference, data from the two runs were combined before ANOVA was carried out in PROC GLIMMIX. The isolate and soybean cultivar were treated as fixed effects and DSI as a response variable. Significant differences among the means were tested using Tukey’s grouping test at α = 0.05.
In the greenhouse pot assays, seedling emergence was calculated as the proportion of seeds that emerged in inoculated pots compared to in non-inoculated pots with the same soybean cultivar. The emergence data were arcsine-transformed to normalize the distribution for proportion of seeds that emerged in a range of 0 to 100% (Sokal and Rohlf Citation1995; Warton and Hui Citation2011). Standardized data for plant fresh plant weight (FPW), fresh root weight (FRW) and dry root weight (DRW) were arcsine-transformed to meet the normal distribution assumption for ANOVA. The two runs were tested for homogeneity of error variance using PROC GLM of SAS Version 9.4 (SAS Institute, Inc., Cary, NC). If no heterogeneity was detected between different runs of the experiment, data were pooled and then analyzed using PROC UNIVARIATE of SAS to examine the distribution of the data. Dunnett’s contrast was carried out using PROC MIXED to identify isolates within species that caused significantly different levels of disease. The confidence intervals were obtained and plotted for each Pythium isolate. Least square means of data for standardized plant and root weight were calculated and were tested using Tukey’s multiple comparison test at α = 0.05, with 95% confidence intervals. The ordinal rating means for root rot were analyzed using a nonparametric approach recommended by Shah and Madden (Citation2004) using PROC MIXED of SAS to determine the effects of experimental factors on ratings (Sokal and Rohlf Citation1995). Tukey’s multiple comparison test was used to identify significant differences among Pythium spp. and among isolates within a species. Linear correlation coefficients were calculated for each replication for seed emergence, root rot severity and other disease parameters using the PROC CORR statement in SAS. Regression analysis was used to assess the relationship between root rot severity rating and the proportion of emerged seedlings. The determination coefficient of regression (R2) between the two disease assessments was calculated and tested for significance using a t test.
Results
Radial growth assay
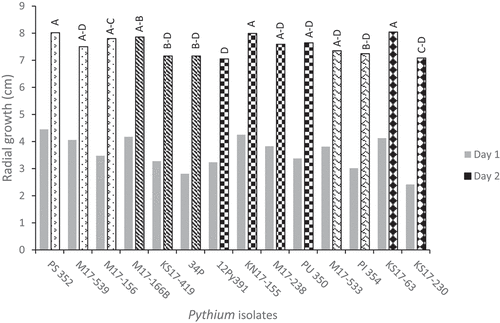
There were significant differences in the radial growth of the 14 isolates on PDA after 2 days (P < 0.0001), though the differences were not large, with colony diameters ranging between 7.1 and 8.0 cm, with an average of 7.5 ± 0.2 cm (). PS 352 (P. sylvaticum), KN17-155 (P. ultimum var. ultimum) and KS17-63 (P. spinosum) had the most rapid growth and P. ultimum var. ultimum isolate 12Py391 had the slowest. The Pearson correlation coefficient for radial growth after one and after two days was significant (r = 0.70; P < 0.0001), so we focused on differences in the diameters of the colonies at the end of the second day. By the third day, the plates had become completely colonized by all of the isolates. No obvious species differences for radial growth rates were evident, and colony diameter after 2 days appeared to depend primarily on the isolate. Each of the species had at least one isolate with a growth rate that was not significantly slower than that of PS 352 or KN17-155, and another isolate with growth that was not significantly faster than that of 12Py391 ().
Fig. 1 Radial growth of 14 isolates of five Pythium species on potato dextrose agar (PDA) after 1 and 2 days. Different letters above bars indicated significant differences based on Tukey grouping analysis of day 2 means at α = 0.05. Means with same letters were not significantly different (SE = 0.21). Species of isolates: Pythium sylvaticum (1–3) = PS 352, M17-539, M17-156; P. aphanidermatum (4–6) = M17-166B, KS17-419, 34P; P. ultimum var. ultimum (7–10) = PU 350, M17-238, KN17-155, 12Py391; P. irregulare (11–12) = M17-533, PI 354; P. spinosum (13–14) = KS17-63, KS17-230.
In vitro pathogenicity assay
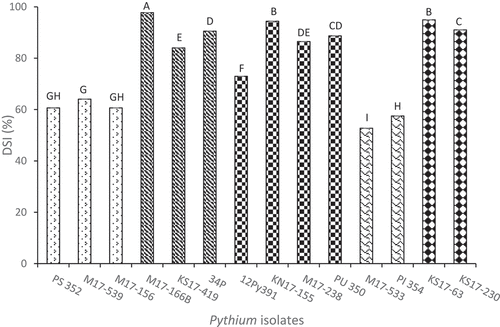
As no problems with heterogeneity of DSI data were detected between the two experimental runs of the Petri plate seed assays (P = 0.33), the DSI data from the two runs were combined for ANOVA. There were significant differences among Pythium isolates, soybean cultivars, and isolate x cultivar interactions (P < 0.0001) ( & ). Averaged across the five cultivars, the mean DSI ratings ranged from < 60% for the two P. irregulare isolates to > 80% for the P. aphanidermatum, P. spinosum and P. ultimum var. ultimum isolates, except for 12Py391 (). The isolates that caused the highest average DSI ratings were M17-166B (P. aphanidermatum), KN17-155 (P. ultimum var ultimum) and KS17-63 (P. spinosum) (). The mean DSI values for ‘Sloan’, ‘Williams’, ‘Conrad’, and ‘Archer’ were significantly higher than those for ‘Maple Glen’ and ‘Maple Isle’, but even for those two cultivars, the DSI ratings were above 60% (Data not shown). ‘Conrad’ and ‘Sloan’ seeds were the most susceptible, with poor germination frequencies and a tendency to become completely colonized by the pathogen. The mean DSI ratings for ‘Williams’ (85.8%) and ‘Archer’ (84.6%) were nearly identical.
Table 2. Variance in seed and seedling disease parameters contributed by six soybean cultivars, 14 Pythium isolates and cultivar × isolate interactions. Disease parameters were seed disease severity index (DSI) from Petri plate assays and seedling fresh plant weight (FPW), fresh root weight (FRW), dry root weight (DRW) and root rot severity from greenhouse pot assays. Statistical significance is based on analysis of variance of transformed data, as described in Materials and Methods.
Fig. 2 Mean disease severity index (DSI) for 14 isolates of five Pythium species, averaged across six soybean cultivars in Petri plate germination assays. Significant differences are indicated by different letters or letter ranges from Tukey’s grouping of least square means (α = 0.05). Species of isolates: Pythium sylvaticum (1–3) = PS 352, M17-539, M17-156; P. aphanidermatum (4–6) = M17-166B, KS17-419, 34P; P. ultimum var. ultimum (7–10) = KN17-155, M17-238, PU 350, 12Py391; P. irregulare (11–12) = M17-533, PI 354; P. spinosum (13–14) = KS17-63, KS17-230.
Inoculum layer greenhouse assays
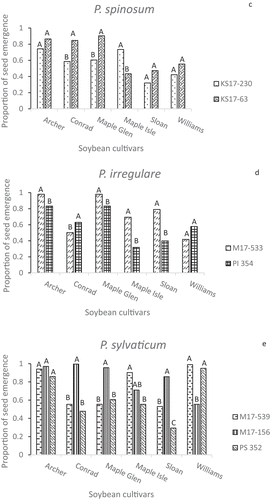
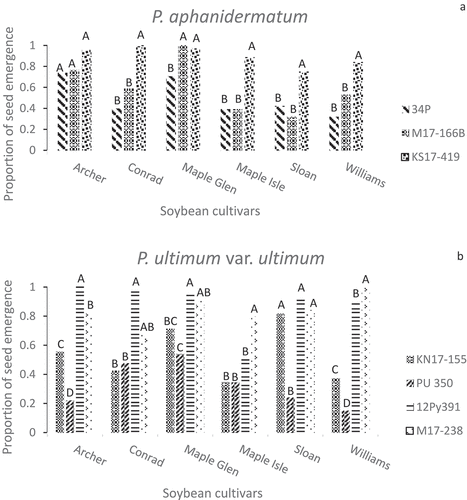
Data from the two inoculum layer greenhouse experiments were pooled after no significant interactions were found between experiment and standardized fresh plant weight, fresh root weight (P = 0.992) or root rot severity score (P = 0.934). Analysis of the different disease parameters assessed in the greenhouse assays detected significant differences among Pythium isolates and soybean cultivars, as well as highly significant isolate × cultivar interactions for emergence and fresh tissue weights (). The 14 isolates varied in their ability to reduce emergence compared to plant stands in non-inoculated pots with the same soybean cultivar (). This variation existed in all five Pythium spp., but P. ultimum var. ultimum was the only species in which all of the isolates tested showed significant differences in aggressiveness on all five cultivars (). Pythium aphanidermatum isolates 34P and M17-166B, P. ultimum var. ultimum isolates KN17-155 and PU 350, P. spinosum KS17-230, P. irregulare M17-533 and PI 354, and P. sylvaticum PS 352 each reduced emergence of one or more cultivars by more than 50% (). Isolates 34P and M17-166B reduced emergence of all cultivars, except ‘Maple Glen’, more than isolate KS17-419 (). PU 350 suppressed emergence of all the cultivars more uniformly than KN17-155, which had a more cultivar-specific effect on emergence (). The two P. spinosum isolates reduced emergence to a similar extent on ‘Sloan’ and ‘Williams’ (). KS17-63 barely affected emergence of ‘Archer’, ‘Conrad’ and ‘Maple Glen’, but suppressed ‘Maple Isle’ emergence to a greater extent than KS17-230. The two P. irregulare isolates suppressed ‘Williams’ emergence to a similar degree, but PI 354 was significantly more aggressive towards ‘Sloan’ (). ‘Archer’ showed a high level of resistance to all of the P. aphanidermatum and P. sylvaticum isolates, in contrast to some of the other cultivars that were affected more by some isolates than by others (). Isolates like KS17-419 and 12Py391 seldom caused much damping-off, while KS17-230 (P. spinosum) and the P. sylvaticum isolates M17-539 and PS 352 affected plant stands of some cultivars more than others ().
Fig. 3 Mean proportion of seed emergence in greenhouse inoculum layer assays with 14 Pythium isolates and six soybean cultivars. Different letters indicate significantly different effects of isolates on emergence of individual cultivars based on Tukey’s multiple comparison of arcsine-transformed data (α = 0.05), with a confidence interval of 95%. Back-transformed data are presented in figure. Pythium spp. represented are (a) P. aphanidermatum, (b) P. ultimum var. ultimum, (c) P. spinosum, (d) P. irregulare, and (e) P. sylvaticum.
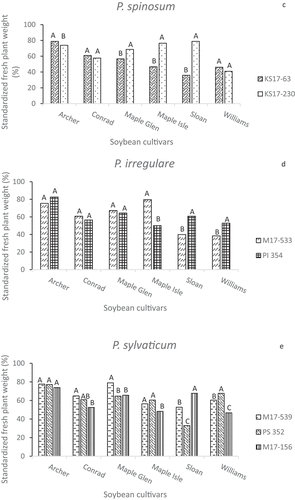
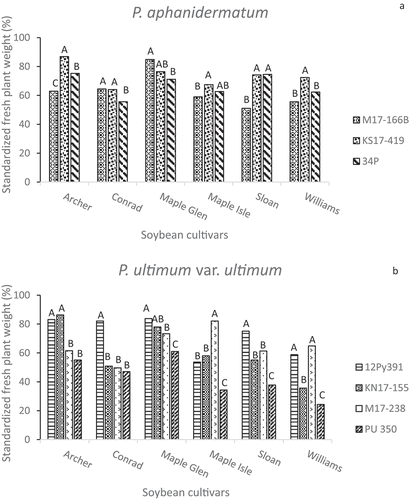
All of the isolates caused reductions in standardized fresh plant weights compared to seedlings from non-inoculated pots (). Some isolates of P. aphanidermatum and P. ultimum var. ultimum reduced FPWs to a different degree on different cultivars, while isolates of P. spinosum and P. irregulare had similar effects on the five cultivars. Although ‘Sloan’ and ‘Williams’ were both considered to be susceptible checks, some isolates were more aggressive towards one than to the other. KS17-230 (P. spinosum) caused a 50% reduction in the FPW of ‘Williams’ plants but only a 20% reduction for ‘Sloan’ (), but the P. sylvaticum isolates PS 352 and M17-156 had opposite effects on these two cultivars (). Pythium irregulare isolates, M17-533 and PI 354 had very similar effects on seedlings of ‘Archer’, ‘Conrad’ and ‘Maple Glen’, but differed in the extent that they reduced the FPWs of ‘Maple Isle’, ‘Sloan’ and ‘Williams’ plants (). The greatest range in aggressiveness within a species based on FPW was once again among the four P. ultimum var. ultimum isolates ().
Fig. 4 Standardized fresh plant weights (%) in greenhouse inoculum layer assays with 14 Pythium isolates and six soybean cultivars. Different letters indicate significantly different effects of isolates on individual cultivars based on Tukey’s multiple comparison tests (α = 0.05) of log-transformed data. Back-transformed data are presented. Pythium spp. represented are (a) P. aphanidermatum, (b) P. ultimum var. ultimum, (c) P. spinosum, (d) P. irregulare, and (e) P. sylvaticum.
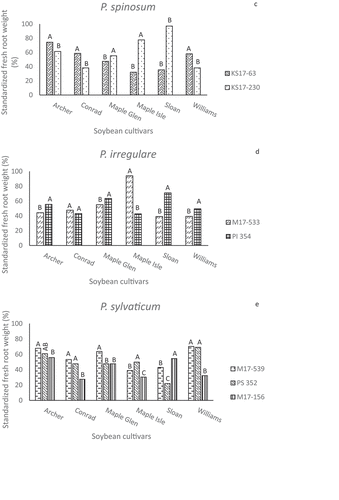
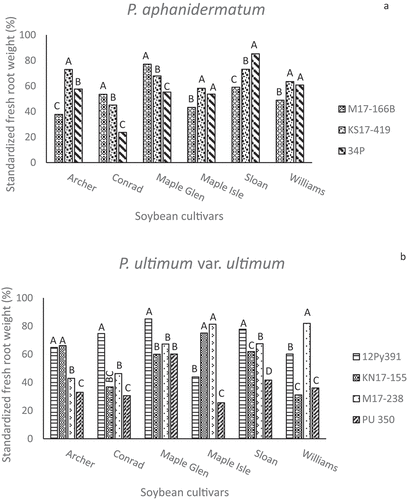
The relative effects of the isolates on standardized fresh root weights generally resembled the FPW results, with significant variation among or between isolates of the same species (). Most isolates reduced mean FRW 20 to 60% compared to the root weights of non-inoculated plants of the same cultivar. Overall, PU 350 was the most aggressive of the four P. ultimum var. ultimum isolates (). ‘Maple Isle’ and ‘Sloan’ were less susceptible to the KS17-230 isolate of P. spinosum than some of the other cultivars or had similar reductions in FRW. ‘Sloan’ root weights were least affected by the KS17-230 isolate, which was unexpected for a susceptible check. ‘Maple Isle’ was also more resistant to the P. irregulare M17-533 isolate than the other cultivars based on FRW (). As dry root weights were highly correlated with FRW, those data are not presented.
Fig. 5 Standardized fresh root weights (%) in greenhouse inoculum layer assays with 14 Pythium species and six soybean cultivars. Different letters indicate significantly different effects of isolates on individual cultivars based on Tukey’s multiple comparison tests (α = 0.05). Pythium spp. represented are (a) Pythium aphanidermatum, (b) P. ultimum var. ultimum, (c) P. spinosum, (d) P. irregulare, and (e) P. sylvaticum.
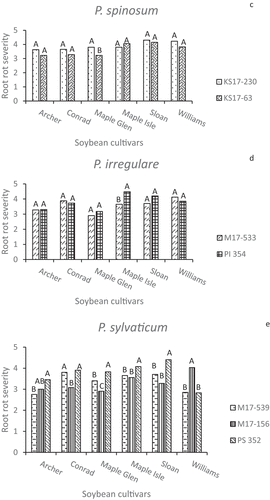
Severe or moderately severe root disease symptoms were caused by all of the isolates on surviving seedlings, with most severity ratings between 3 and 5 (). Differences in severity ratings among isolates of P. aphanidermatum, P. ultimum var. ultimum, and P. sylvaticum were often significant (P < 0.05), but were seldom large, whereas visible symptoms caused by the P. spinosum and P. irregulare isolates were usually not significantly different (P > 0.05). There were no obvious or consistent differences between root disease severity on ‘Archer’ and ‘Maple Isle’ compared to the susceptible checks ‘Sloan’ and ‘Williams’. When the overall mean root rot severity ratings for each cultivar were analyzed, ‘Maple Isle’ and ‘Sloan’ had the highest means, though the mean ratings for all cultivars averaged across isolates were between 3.0 and 4.0 on a scale of 1 to 5 (). The Pearson correlation coefficient indicated a significant negative linear correlation between seedling emergence and root severity score (r = −0.90, P < 0.0001) (). Linear regression showed a strong relationship between these disease parameters (P < 0.0001), with R2 = 0.88 at the 95% confidence level ().
Fig. 6 Visual root rot severity ratings (1–5) on seedlings in greenhouse inoculum layer assays with 14 Pythium isolates and six soybean cultivars. Different letters indicate differences in aggressiveness between isolates on the same cultivar based on Tukey’s multiple comparison tests at α = 0.05 and a confidence level of 95%. Pythium spp. represented are (a) P. aphanidermatum (b) P. ultimum var. ultimum (c) P. spinosum (d) P. irregulare, and (e) P. sylvaticum.
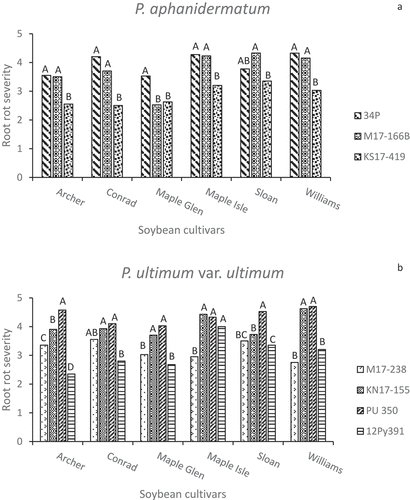
Fig. 7 Regression of root rot severity (scale 1–5) on proportion of emerged seedlings for Pythium isolates in the greenhouse layer inoculum assays. Emergence data were arcsine-transformed prior to the analysis.
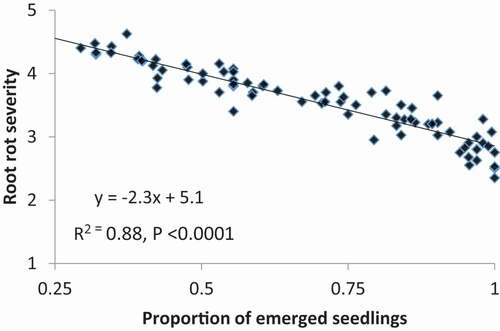
Table 3. Pearson correlation coefficients between root rot severity and seedling emergence, fresh plant weight (FPW), fresh root weight (FRW), and dry root weight (DRW) from the greenhouse pot assays. Correlations are followed by probabilities in parentheses.
Overall, P. ultimum var. ultimum isolate PU 350 from Illinois, USA, was the most aggressive of the Pythium isolates evaluated in this study. Detrimental effects caused by some other isolates, such as 34P (P. aphanidermatum) from Missouri, KS17-63 (P. spinosum) from Arkansas and M17-156 (P. sylvaticum) from Illinois were sometimes considerable but varied depending on the host cultivar. All isolates reduced the fresh plant weight of susceptible cultivars, while even isolates that had negligible effects on seedling emergence reduced the growth rates of most of the cultivars and caused visible lesions and root rot.
Cultivar reactions
Although ‘Archer’ and ‘Maple Isle’ are known to have resistance to certain isolates of Pythium, their responses to several isolates used in this study were similar to those of ‘Sloan’ and ‘Williams’, which are commonly used as susceptible checks in Pythium resistance assays. ‘Archer’, which has resistance to some P. aphanidermatum isolates from Arkansas, did have better emergence than ‘Conrad’, ‘Sloan’ and ‘Williams’ seedlings when inoculated with the 34P and M17-166B isolates of that species (). Based on the FPW () and FRW (), however, ‘Archer’ seedlings had no more resistance to the P. aphanidermatum isolates than any of the other cultivars, and ‘Archer’ emergence in pots inoculated with the KN17-155 isolate of P. ultimum var. ultimum was significantly less than that of ‘Sloan’ and only marginally better than emergence of ‘Conrad’ (). For mean plant fresh weight from KN-155 pots, however, that of Archer was higher than Sloan (), and the fresh root weights for the two cultivars were nearly identical ().
Pythium ultimum var. ultimum isolate 12Py391 had little effect on emergence of the six cultivars cultivars (). Emergence and fresh weights of ‘Maple Isle’ seedlings in pots inoculated with isolate 12Py391 were similar to those of other cultivars, even though ‘Maple Isle’ had been included in the present study because of its high level of resistance to 12Py391 in previous experiments (–). Based on seedling emergence, ‘Archer’ and ‘Maple Glen’ had less damping-off than ‘Maple Isle’ when infected with P. irregulare isolate M17-533 (), but ‘Maple Glen’ FPW and FRW were less affected by that isolate ().
Discussion
The northern and westward expansion of soybean production in the United States and Canada, in addition to a risk of cool, damp soils at planting time in many production regions, has increased the incidence of disease caused by Pythium spp. Until recently, little was known about pathogenic diversity within and among Pythium species. This study investigated the pathogenicity of isolates representing five Pythium species using six soybean cultivars previously reported to vary in their susceptibility to Pythium species (Kirkpatrick et al. Citation2006a; Rosso et al. Citation2008; Rod et al. Citation2018). It thus differed from previous studies that investigated the reactions of just one or two soybean genotypes to larger numbers of Pythium isolates (Broders et al. Citation2007; Matthiesen et al. Citation2016; Matthiesen and Robertson Citation2021), or that tested the reactions of a large number of germplasm accessions to Pythium spp. represented by a single isolate (Rod et al. Citation2018; Lerch-Olson et al. Citation2020). Our study should be of interest to soybean breeders and pathologists who want to learn more about potential host resistance × pathotype interactions. The results support the hypothesis that significant variation in aggressiveness exists among isolates of the same Pythium spp. To effectively screen breeding lines for resistance, soybean breeders should therefore identify aggressive isolates of Pythium spp. that are common in the targeted production area. Our data also revealed significant cultivar × isolate interactions that should be investigated further to optimize selection methods.
All of the disease parameters measured in this study indicated there were differences in aggressiveness among the isolates of each of the five Pythium species, as shown by their effects on seedling survival and growth. Although the rate of radial growth differed among isolates (P < 0.05), the differences were not numerically large, indicating that isolates had similar growth vigour on PDA. In the seed disease assay, differences in aggressiveness associated with Pythium spp. were detected. The DSI percentage was higher for the P. aphanidermatum, P. spinosum, and P. ultimum var. ultimum isolates than it was for the P. sylvaticum and P. irregulare isolates.
In the greenhouse seedling assay, differences in aggressiveness related to species of Pythium were less evident. Based on emergence and the fresh weights of whole plants and roots of each of the five cultivars, variation in aggressiveness was often as high among isolates of the same species as it was between Pythium spp. Certain isolates from each of the five species caused substantial reductions in seedling stands, and the weights of surviving plants and root systems of all or most of the cultivars, while other isolates were less aggressive. The range of aggressiveness was greater within some species, but that may have been biased by the unequal numbers of isolates from each species. Although the cultivars ‘Sloan’ and ‘Williams’ have been used in a number of Pythium resistance or aggressiveness studies as susceptible checks, they developed little disease when challenged with some of the isolates. In fact, their emergence frequencies and fresh weights were sometimes equal to the cultivars ‘Archer’ and ‘Maple Isle’, which have previously shown resistance to certain Pythium species and isolates (Kirkpatrick et al. Citation2006a; Rod et al. Citation2018).
The five species of Pythium included in the present study are among the most common and/or most pathogenic on soybean grown in the U.S. Midwest and south-central soybean production regions. From a regional survey of Pythium spp. and related oomycetes in Ontario and in 11 soybean-producing U.S. states in 2011 and 2012, Rojas et al. (Citation2017) reported that P. sylvaticum and P. ultimum var. ultimum were common in both years. Pythium spinosum was recovered from diseased seedlings at a moderate frequency in 2011, but not at all in 2012, while P. aphanidermatum was relatively uncommon in both years. Although P. ultimum var. ultimum was only moderately common among the species of Pythium collected from Ohio fields, Broders et al. (Citation2007) reported that it was an important soybean pathogen. In the Ohio study, P. sylvaticum and P. irregulare were more common in the same fields but were isolated primarily from diseased maize seedlings. Although P. aphanidermatum may not be common in the northern United States and Canada, it is an important pathogen in eastern Arkansas (Rosso et al. Citation2008), and it was one of the most aggressive Pythium species at 20°C in a root-rot assay conducted by Rojas et al. (Citation2017). Isolates of P. ultimum var. ultimum and P. irregulare also caused high levels of disease on dry bean (Phaseolus vulgaris) in seedling assays conducted by Rossman et al. (Citation2017).
The aggressiveness of Pythium species and isolates on soybean plants depends on environmental as well as genetic factors, and the aggressiveness of some species of Pythium is influenced by temperature (Thomson et al. Citation1971; Wei et al. Citation2011; Rupe et al. Citation2011; Matthiesen et al. Citation2016; Rojas et al. Citation2017). Pythium ultimum is among the species that cause more damage at temperatures below 20°C, while P. aphanidermatum and P. sylvaticum tend to be more aggressive when temperatures are 23 to 30°C or higher (Thomson et al. Citation1971; Rupe et al. Citation2011; Matthiesen and Robertson Citation2021). Growth of P. irregulare is optimal at 27°C (Huzar-Novakowiski and Dorrance Citation2018). Radmer et al. (Citation2017) examined the relative aggressiveness of Minnesota isolates of 18 different Pythium spp. on soybean and maize seedlings at three temperatures. Although relative aggressiveness of some species differed significantly at 20 and 25°C, the numerical differences were relatively small, and very few species were more aggressive at 15°C than at the higher temperatures. In addition, cooler temperatures may slow seed germination and seedling emergence, even in the absence of soil pathogens. For our experiments we maintained temperatures in a moderate range (i.e., 18 to 23°C) that was expected to be favourable, though not necessarily optimal, for all of the Pythium species tested. This range is also typical of soil temperatures that soybean seeds and seedlings are likely to encounter after being planted.
Contrary to the results of an earlier study by Rod et al. (Citation2018), isolate 12Py391 was not very aggressive in the assays reported here. Rod et al. (Citation2018) had found it to be the most aggressive of six P. ultimum var. ultimum isolates collected in Illinois. In other inoculum layer assays that we have conducted using inoculum grown on millet seeds, 12Py391 caused severe damping-off in ‘Sloan’ and ‘Williams’, reducing plant stands to less than 10%, whereas ‘Maple Isle’ emergence and seedling growth differed little from the non-inoculated check pots (Rod et al. Citation2018; Zhang and Walker, unpublished data). Two possible explanations for the discrepancies are diminished aggressiveness in our laboratory strain of 12Py391 and/or differences in the layer inoculation assays. It is possible that the strain of 12Py391 that we have maintained in our laboratory for several years has become less aggressive over time. A reduction in the aggressiveness of Pythium isolates has been observed before by other researchers (Anne Dorrance, personal communication), and the data from the two Petri plate assays hint at this possibility. 12Py391 had the slowest radial growth of the 14 isolates on PDA (), and it was the least aggressive of the four P. ultimum var. ultimum isolates based on DSI ratings in the second Petri plate assay (). Although differences in the protocols of inoculum layer assays would be expected to have a similar influence on all pathogenic Pythium isolates, it is possible that some isolates might be affected more than others. In the Rod et al. (Citation2018) study, inoculum was increased on autoclaved millet seeds, whereas in the present study a medium consisting of a large amount of sandy loam soil and a much smaller amount of cornmeal was used. The nutrient-rich millet seeds may have favoured rapid growth of inoculum, and the soil/cornmeal mix used in the present study may have simply been a poor medium for increasing the 12Py391 isolate. Other potentially important differences between the two assays were the volume and height of the pots used in each, and possibly the watering methods used. In the present study, the pots had a usable volume of 350 mL, whereas Rod et al. (Citation2018) used much larger pots with a volume of 1600 mL. The larger pots were also several cm taller, leaving more vertical space for root growth. Moreover, in the Rod et al. (Citation2018) assay the pots were watered to saturation daily, but were allowed to drain, whereas the lower third of the pots used in the present study remained submerged in flooded trays; 12Py391 may be more sensitive to the differences in edaphic conditions than some other isolates. Neither of these possibilities offers a satisfactory explanation of why ‘Maple Isle’ appeared to be more susceptible to the 12Py391 isolate than ‘Sloan’, however, so further experiments will be needed to determine the cause(s).
In a study of the aggressiveness of 118 isolates of P. sylvaticum, P. lutarium, P. oopapilum and P. torulosum, Matthiesen and Robertson (Citation2021) concluded that Arkansas isolates of each species tended to be more aggressive on the cultivar ‘Sloan’ than isolates from 10 other states. Although some of the most aggressive isolates in the present study were from Arkansas, so were some of the mildest ones. In any case, the relatively small number of isolates and unbalance representation from different states did not permit a reliable assessment. Matthiesen et al. (Citation2016) speculated that the greater aggressiveness of the Arkansas isolates might be related to the fact that the isolates were collected from fields with poor soil drainage and a history of rice-soybean rotations. Some of the most aggressive isolates in our study, such as P. ultimum var. ultimum PU 350 and P. irregulare PI 354, were from fields the U.S. Midwest that have never been used to grow rice.
Some researchers have speculated that seedling resistance to Pythium pathogens may involve some genes that do not also contribute to seed resistance (Rosso et al. Citation2008; Ellis et al. Citation2013; Urrea et al. Citation2017). Certain Pythium spp. may be more likely to infect seeds before or immediately after germination, while others may initiate infection later in the development of seedlings. Of 18 species of Pythium studied by Radmer et al. (Citation2017), the five most aggressive on soybean seedlings were P. ultimum var. ultimum, P. sylvaticum, P. irregulare, P. ultimum var. sporangiferum, and P. intermedium. In an interesting examination of the primary disease effects of various Pythium spp. on dry bean (Phaseolus vulgaris L.), Rossman et al. (Citation2017) described P. aphanidermatum and P. ultimum var. ultimum as being primarily ‘seed rot pathogens’, P. sylvaticum as a ‘root rot pathogen’, and P. irregulare and P. spinosum as being both. Our results do not help to resolve this issue, but it is possible that a phenomenon like this could have contributed to some of the isolate × cultivar interactions observed for different disease parameters.
The cultivar ‘Sloan’ has been widely used as a susceptible check in Pythium resistance studies and was included in this study for that reason. In some of our assays, however, it appeared to have as much resistance as ‘Archer’ towards certain isolates of P. aphanidermatum, P. irregulare, P. spinosum and P. sylvaticum. While this level of resistance was unexpected, ‘Sloan’ has been reported to have at least two quantitative trait loci (QTL) associated with resistance to the P. irregulare isolate Brown 2-3-5, an Ohio isolate that is highly aggressive on soybean (Stasko et al. Citation2016). It is possible that these QTL or other unreported minor genes in ‘Sloan’ provided resistance to some of the isolates in the present study. ‘Archer’ resistance to P. aphanidermatum and some other Pythium pathogens has been investigated and confirmed in previous studies (Rosso et al. Citation2008). It our assays, this resistance appeared to provide germinating seeds with some protection against the P. aphanidermatum isolates based on the emergence data ().
‘Maple Glen’ and ‘Maple Isle’ were both developed by the Agriculture and Agri-Food Canada breeding programme in Ottawa, Ontario, Canada (Voldeng et al. Citation1985, Citation1996) and appeared to have resistance to many of the isolates used in this study. Their differential reactions to some isolates suggested that they might not carry the same Pythium resistance genes. For example, ‘Maple Glen’ had a much higher standardized fresh root weight than ‘Maple Isle’ when challenged with P. aphanidermatum isolate M17-166B and P. ultimum var. ultimum isolate 12Py391, although ‘Maple Isle’ has had greater resistance to the latter isolate in some other studies (Rod et al. Citation2018). These two lines have different ancestors (Cober and Voldeng Citation2012), but it is conceivable that they each inherited Pythium resistance from a different source through indirect selection for cold tolerance.
Evaluation of seedling emergence or plant stands is much easier and quicker than collecting plant or root weights or rating the extent of disease on washed roots. As there was a strong negative correlation between emergence and tissue weights, selection based on emergence should therefore be effective in a plant-breeding programme. In addition, while surviving seedlings in pots with low stands would presumably remain susceptible to a Pythium isolate or species, the reduced competition for light, water and nutrients might counteract the effects of disease to some extent. For phenotypic selection in a breeding programme, the results of this study indicate that seedling emergence should be an effective criterion for selecting resistant plants or lines.
In conclusion, this study revealed substantial within-species variation for aggressiveness among isolates of five Pythium species that are important soybean pathogens in North America, and we identified aggressive isolates within each species. One implication of the results is that soybean breeders or pathologists evaluating or studying soybean resistance to Pythium spp. should assess the aggressiveness of the isolates available to them before assuming that pathotype variation within Pythium species is negligible. As all five of the cultivars, including the ones that were expected to be more susceptible (‘Sloan’ and ‘Williams’) exhibited varying levels of resistance to different isolates. The considerable number of isolate × cultivar interactions in this study suggests that soybean resistance to different Pythium species and isolates is likely to be a complex trait, and that resistance to one isolate does not ensure that a host will be resistant to other Pythium isolates or species. Some of the 14 isolates were aggressive against the cultivars Archer and Maple Isle, which were previously known to have Pythium resistance, while other isolates caused very little damping-off or disease symptoms on the susceptible checks ‘Sloan’ and ‘Williams’. Although seed treatment with oomicides has been effective in reducing disease and damping-off of soybean seedlings, resistance to Pythium spp. would help to protect cultivars when oomicides applied to seed coats become degraded or diluted over time, and in the event that some Pythium populations evolve tolerance to protectants. The development of new soybean cultivars with broad and effective resistance to Pythium pathogens will be challenging, but could be important in production regions where soybean seeds are commonly planted in cool, damp soils.
Acknowledgements
The authors sincerely thank Dr. John Rupe (Dept. of Plant Pathology, University of Arkansas), Dr. Carl Bradley (currently in Dept. of Plant Pathology, University of Kentucky), Dr. Nathan Kleczewski (formerly in Dept. of Crop Sciences, University of Illinois), and Clayton Rushford and Dr. Jeanne Mihail (University of Missouri) for sharing the Pythium isolates used in this study.
Mention of trade names or commercial products in this publication is solely for the purposes of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, colour, national origin, age, or disability, and where applicable, sex, marital status, familial status, parental status, religion, sexual orientation, genetic information, political beliefs, reprisal, or because all or part of an individual’s income is derived from any public assistance program. The USDA is an equal opportunity provider and employer.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bahrenfuss JB, Fehr WR. 1980. Registration of ‘Sloan’ soybean. Crop Sci. 20:673. doi:https://doi.org/10.2135/cropsci1980.0011183X002000050045x.
- Bates GB, Rothrock CS, and Rupe JC. 2008. Resistance of the soybean cultivar Archer to Pythium damping-off and root rot caused by several Pythium spp. Plant Dis. 92:763–766. doi:https://doi.org/10.1094/PDIS-92-5-0763.
- Bernard RL, Lindahl DA. 1972. Registration of ‘Williams’ soybean. Crop Sci. 12:716. doi:https://doi.org/10.2135/cropsci1972.0011183X001200050067x.
- Broders KD, Lipps PE, Paul PA, Dorrance AE. 2007. Characterization of Pythium spp. associated with corn and soybean seed and seedling disease in Ohio. Plant Dis. 91:727–735. doi:https://doi.org/10.1094/PDIS-91-6-0727.
- Cianzio SR, Shultz SP, Fehr WR, Tachibana H. 1991. Registration of ‘Archer’ soybean. Crop Sci. 31:1707. doi:https://doi.org/10.2135/cropsci1991.0011183X003100060081x.
- Cober ER, Voldeng H. 2012. A retrospective look at short-season soybean cultivar development in Canada. Can J Plant Sci. 92:1239–1243. doi:https://doi.org/10.4141/CJPS2012-032.
- Dorrance AE, Berry SA, Bowen P, Lipps PE. 2004. Characterization of Pythium spp. from three Ohio fields for pathogenicity on corn and soybean and matalaxyl sensitivity. Plant Health Prog. 2:1–7.
- Dorrance AE, Robertson AE, Cianzio S, Giesler LJ, Grau CR, Draper CR, Draper MA, Tenuta AU, Anderson TR. 2009. Integrated management strategies for Phythophthora sojae combining host resistance and seed treatments. Plant Dis. 93:875–882. doi:https://doi.org/10.1094/PDIS-93-9-0875.
- Ellis ML, McHale LK, Paul PA, St. Martin SK, Dorrance AE. 2013. Soybean germplasm resistant to Pythium irregulare and molecular mapping of resistance quantitative trait loci derived from the soybean accession PI 424354. Crop Sci. 53:1008–1021. doi:https://doi.org/10.2135/cropsci2012.08.0461.
- Fehr WR, Caviness CE, Burmood DT, Pennington JS. 1971. Stage of development descriptions of soybeans, Glycine max (L.) Merrill. Crop Sci. 11:929–931. doi:https://doi.org/10.2135/cropsci1971.0011183X001100060051x.
- Groenewald S, van den Berg N, Marasas WFO, Viljoen A. 2006. Biological, physiological and pathogenic variation in a genetically homogeneous population of Fusarium oxysporum f.sp. cubense. Australas Plant Pathol. 35:401–409. doi:https://doi.org/10.1071/AP06041.
- Hu M, Wiatrak P. 2012. Effect of planting date on soybean growth, yield, and grain quality: review. Agron J. 104:785–790. doi:https://doi.org/10.2134/agronj2011.0382.
- Huzar-Novakowiski J, Dorrance AE. 2018. Genetic diversity and population structure of Pythium irregulare from soybean and corn production fields in Ohio. Plant Dis. 102:1989–2000. doi:https://doi.org/10.1094/PDIS-11-17-1725-RE.
- Kirkpatrick MT, Rothrock CS, Rupe JC, Gbur EE. 2006a. The effect of Pythium ultimum and soil flooding on two soybean cultivars. Plant Dis. 90:597–602. doi:https://doi.org/10.1094/PD-90-0597.
- Kirkpatrick MT, Rupe JC, Rothrock CS. 2006b. Soybean response to flooded soil conditions and the association with soilborne plant pathogen genera. Plant Dis. 90:592–597. doi:https://doi.org/10.1094/PD-90-0592.
- Klepadlo M, Balk CS, Vuong TD, Dorrance AE, Nguyen HT. 2019. Molecular characterization of genomic regions for resistance to Pythium ultimum var. ultimum in the soybean cultivar Magellan. Theor Appl Genet. 132:405–417. doi:https://doi.org/10.1007/s00122-018-3228-x.
- Lerch-Olson ER, Dorrance AE, Robertson AE. 2020. Resistance of the SoyNAM parents to seed and root rot caused by four Pythium species. Plant Dis. 104:2489–2497. doi:https://doi.org/10.1094/PDIS-10-19-2237-RE.
- Matthiesen RL, Ahmad AA, Robertson AE. 2016. Temperature affects aggressiveness and fungicide sensitivity of four Pythium spp. that cause soybean and corn damping-off in Iowa. Plant Dis. 100:538–591. doi:https://doi.org/10.1094/PDIS-04-15-0487-RE.
- Matthiesen RL, Robertson AE. 2021. Comparison of aggressiveness and fungicide sensitivity of four Pythium spp. that cause damping-off of soybean in the United States. Can J Plant Pathol. 43:769–782. doi:https://doi.org/10.1080/07060661.2021.1881162.
- Nelson EB. 1987. Rapid germination sporangia of Pythium species in response to volatiles from germinating seeds. Phytopathology. 77:1108–1112. doi:https://doi.org/10.1094/Phyto-77-1108.
- Noel ZA, McDuffie D, Chilvers MI. 2020. Influence of soybean tissue and oomicide seed treatments on oomycete isolation. Plant Dis. 105:1281–1288. doi:https://doi.org/10.1094/PDIS-03-20-0642-RE.
- Radmer L, Anderson G, Malvick DM, Kurle JE. 2017. Pythium, Phytophthora, and Phytopythium spp. associated with soybean in Minnesota, their relative aggressiveness on soybean and corn, and their sensitivity to seed treatment fungicides. Plant Dis. 101:62–72. doi:https://doi.org/10.1094/PDIS-02-16-0196-RE.
- Rizvi SSA, Yang XB. 1996. Fungi associated with soybean seedling disease in Iowa. Plant Dis. 80 57–60.
- Rod KS, Walker DR, Bradley CA. 2018. Evaluation of major ancestors of North American soybean cultivars for resistance to three Pythium species that cause seedling blight. Plant Dis. 102:2241–2252. doi:https://doi.org/10.1094/PDIS-09-17-1341-RE.
- Rojas JA, Jacobs JL, Napieralski S, Karaj B, Bradley CA, Chase T, Esker PD, Giesler LJ, Jardine DJ, Malvik DK, et al. 2017. Oomycete species associated with soybean seedlings in North America Part I: identification and pathogenicity characterization. Phytopathology. 107:280–292. doi:https://doi.org/10.1094/PHYTO-04-16-0177-R.
- Rossman DR, Rojas A, Jacobs JL, Mukankusi C, Kelly JD, Chilvers MI. 2017. Pathogenicity and virulence of soilborne Oomycetes on Phaseolus vulgaris. Plant Dis. 101:1851–1859. doi:https://doi.org/10.1094/PDIS-02-17-0178-RE.
- Rosso ML, Rupe JC, Chen P, Mozzoni LA. 2008. Inheritance and genetic mapping of resistance to Pythium damping-off caused by Pythium aphanidermatum in ‘Archer’ soybean. Crop Sci. 48:2215–2222. doi:https://doi.org/10.2135/cropsci2008.01.0035.
- Rothrock CS, Avanzato MV, Rupe JC. 2015. Pythium seed rot, damping-off, and root rot. In: Hartman GL, Rupe JC, Sikora EJ, Domier LL, Davis JA, Steffey KL, editors. Compendium of soybean diseases and pests. 5th ed. St. Paul (MN): APS Press; p. 76–79.
- Rupe JC, Rothrock CS, Bates G, Rosso ML, Avanzato MV, and Chen P. 2011. Resistance to Pythium seedling disease in soybean. In: Sudaric A, editor. Molecular aspects of breeding. 9789533072401, . 9789533072401, IntechOpen Ltd., London, U.K: InTech. p. 261–276. Accessed Feb 16, 2022. http://www.intechopen.com/books/soybean-molecular-aspects-of-breeding/resistance-to-pythium-seedling-disease-in-soybean
- Scott K, Eyre M, McDuffee D, Dorrance AE. 2020. The efficacy of ethaboxam as a seed treatment toward Phytophthora, Phytopythium, and Pythium in Ohio. Plant Dis. 59:605–623. doi:https://doi.org/10.1094/PDIS-09-19-1818-RE.
- Serrano M, Robertson AE. 2018. The effect of cold stress on damping-off of soybean caused by Pythium sylvaticum. Plant Dis. 102:2194–2200. doi:https://doi.org/10.1094/PDIS-12-17-1963-RE.
- Shah DA, Madden LV. 2004. Nonparametric analysis of ordinal data in designed factorial experiments. Phytopathology. 94:33–43. doi:https://doi.org/10.1094/PHYTO.2004.94.1.33.
- Sokal RR, Rohlf FJ. 1995. Introduction to biostatistics. 3rd ed. New York (NY): W.H. Freeman & Co.
- Stasko AK, Wickramasinghe D, Nauth BJ, Acharya B, Ellis ML, Taylor CG, McHale LK, Dorrance AE. 2016. High-density mapping of resistance QTL toward Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Sci. 56:2476–2492. doi:https://doi.org/10.2135/cropsci2015.12.0749.
- Thomson TB, Athow KL, Laviolette FA. 1971. The effect of temperature on the pathogenicity of Pythium aphanidermatum, P. debaryanum, and P. ultimum on soybean. Phytopathology. 61:933–935. doi:https://doi.org/10.1094/Phyto-61-933.
- Urrea K, Rupe J, Chen P, Rothrock CS. 2017. Characterization of seed rot resistance to Pythium aphanidermatum in soybean. Crop Sci. 57:1394–1403. doi:https://doi.org/10.2135/cropsci2016.08.0669.
- Urrea K, Rupe JC, Rothrock CS. 2013. Effect of fungicide seed treatments, cultivars, and soils on soybean stand establishment. Plant Dis. 97:807–812. doi:https://doi.org/10.1094/PDIS-08-12-0772-RE.
- Voldeng HD, Guillemette RJD, Leonard DA, Cober ER. 1996. Maple Glen soybean. Can J Plant Sci. 76:475–476. doi:https://doi.org/10.4141/cjps96-085.
- Voldeng HD, Seitzer JF, Hamilton RI. 1985. Maple Isle soybean. Can J Plant Sci. 65:777–779. doi:https://doi.org/10.4141/cjps85-099.
- Warton DI, Hui FKC. 2011. The arcsine is asinine: the analysis of proportions in ecology. Ecology. 92:3–10. doi:https://doi.org/10.1890/10-0340.1.
- Wei L, Xue AG, Cober ER, Babcock C, Zhang J, Zhang S, Li W, Wu J, and Liu L. 2011. Pathogenicity of Pythium species causing seed rot and damping-off in soybean under controlled conditions. Phytoprotection. 91:3–10. doi:https://doi.org/10.7202/1008539ar.
- Zhang BQ, Yang XB. 2000. Pathogenicity of Pythium populations from corn-soybean rotation fields. Plant Dis. 84:94–99. doi:https://doi.org/10.1094/PDIS.2000.84.1.94.
- Zitnick-Anderson KK, Nelson BD Jr. 2015. Identification and pathogenicity of Pythium on soybean in North Dakota. Plant Dis. 99:31–38. doi:https://doi.org/10.1094/PDIS-02-14-0161-RE.