Abstract
Ergot fungi produce toxic sclerotia that have significant impacts on the agricultural, food and pharmaceutical industries. These fungi were classified in various genera (such as Sclerotium clavus, or Spermoedia clavus) before Tulasne, in 1853, erected Claviceps and described three species based on variations in morphology and host ranges, viz. C. purpurea, C. microcephala, and C. nigricans. Since then, knowledge regarding the biological, clinical, and pharmaceutical perspectives of ergot has accumulated rapidly. However, a serious taxonomic examination was lacking until Langdon’s revision accepted 25 species in 1952. That was followed by intensive regional studies by Loveless in the 1960s for Rhodesia (now Zimbabwe, Africa), and Tanda in Japan (1970s–1990s). More species names were reported and currently over 90 named taxa (species, varieties) are recorded in fungal name repositories (Mycobank and Index Fungorum). Most species were described based on morphological characteristics (sclerotia, ascomata and conidia) and host ranges. Some were characterized by their alkaloid profiles. Recently, DNA multi-locus sequence analyses (MLSA) were applied to resolve species complexes. For instance, the C. purpurea complex was separated into four species, and additional new species were recognized from South Africa and Canada. Infra- and supra-specific level genetic variations were identified via multi-locus and genomic studies. Based on five-locus phylogenies, Píchová and colleagues separated Claviceps into four sections: C. sect. Claviceps, C. sect. Citrinae, C. sect. Paspalorum and C. sect. Pusillae, for 60 species. Among these sections, several doubtful species names require clarification. Careful research on type specimens combined with molecular analyses is essential for clarifying these names.
Résumé
Les champignons de l’ergot produisent des sclérotes toxiques qui ont de sérieuses répercussions sur les industries agricole, alimentaire et pharmaceutique. Ces champignons avaient été classifiés dans différents genres (comme Sclerotium clavus ou Spermoedia clavus) avant que Tuslane (1853) élève Claviceps au rang d’espèce et en décrive trois en se basant sur les variations morphologiques et les gammes d’hôtes, à savoir C. purpurea, C. microcephala et C. nigricans. Depuis, les connaissances relatives aux perspectives biologiques, cliniques et pharmaceutiques de l’ergot se sont accumulées rapidement. Toutefois, un examen taxinomique approfondi faisait défaut jusqu’à ce que la révision de Langdon (1952) reconnaisse 25 espèces. Cela a été suivi par d’intenses études régionales menées par Loveless dans les années 1960 en Rhodésie (maintenant le Zimbabwe, en Afrique) et à Tanda au Japon (1970-1990). Plus de noms d’espèces ont été rapportés et, actuellement, plus de 90 taxons nommés (espèces, variétés) sont enregistrés dans des bases de données mycologiques (Mycobank et Index Fungorum). La plupart des espèces ont été décrites en fonction de leurs caractéristiques morphologiques (sclérotes, ascomes et conidies) et de leurs gammes d’hôtes. Certaines ont été caractérisées selon les profils de leurs alcaloïdes. Récemment, des analyses de séquences multilocus de l’ADN (MLSA) ont été utilisées pour examiner soigneusement les complexes d’espèces. Par exemple, le complexe C. purpurea a été divisé en quatre espèces et de nouvelles espèces supplémentaires sud-africaines et canadiennes ont été reconnues. Des variations génétiques aux niveaux infra et supra-spécifiques ont été constatées dans le cadre d’études multilocus et génomiques. En se basant sur des phylogénies à cinq locus, Píchová et coll. ont séparé Claviceps en quatre sections: C. sect. Claviceps, C. sect. Citrinae, C. sect. Paspalorum et C. sect. Pusillae, et ce, pour 60 espèces. Parmi ces sections, plusieurs noms controversés d’espèces requièrent d’être débrouillés. Une recherche minutieuse sur les spécimens types, combinée aux analyses moléculaires, est essentielle pour débrouiller ces noms.
Introduction
Ergot fungi belong to the genus Claviceps and typically infect the florets of cereal crops, grasses (Poaceae), rushes (Juncaceae) and sedges (Cyperaceae), replacing the seeds with toxic sclerotia. Some ergot species, such as C. africana Freder., Mantle & De Milliano, C. sorghi B.G.P. Kulk., Seshadri & Hegde and C. purpurea (Fr.) Tul. can reduce the grain production in agricultural populations (Menzies and Turkington Citation2015; Miedaner and Geiger Citation2015). Often, they were considered pathogens also because of their detrimental effects on humans and animals. However, in natural populations, their aposematic function proves to be beneficial to their host plants, therefore, to a certain extent, they should be considered mutualistic parasites instead of antagonistic plant pathogens (Lev-Yadun and Halpern Citation2007; Wäli et al. Citation2013). The mycotoxins contained in ergots (sclerotia) attracted a wide scope of research, viz biochemistry, biology, food safety, pharmacology and plant pathology (Kren and Cvak Citation1999; Menzies and Turkington Citation2015; Arcella et al. Citation2017). Identifying and naming ergot fungi can be traced back to ancient folklores (Barger Citation1931). The first formal binomial name was given by de Candolle (Citation1815) as Sclerotium clavus, but Tulasne (Citation1853) had the first comprehensive study on ergot, discovered the sexual stage, erected the genus Claviceps and recognized three species. This is often considered the starting point of the taxonomy of ergot. Throughout history, ergot species were recognized based on the morphology of sclerotia, ascostromata (if applicable), honeydew conidia, colony features of axenic cultures, host ranges, chemistry features of the sclerotium, honeydew and culture, as well as molecular markers. The phylogenetic relationships of ergot fungi and the major characteristics of some accepted species were first reviewed by Langdon (Citation1952) and later by Pažoutová and Parberry (Citation1999). More recently, a polyphasic approach was applied to a large panel of species and resulted in a supra-specific classification of the genus into four sections (Píchová et al. Citation2018). In this short review, we highlight some major milestones in the history of ergot taxonomy and summarize recent progress in recognizing the species and genetic diversity through a molecular polyphasic approach.
Pre-history period
The 168 year history of ergot taxonomic study commences with the initial naming of Claviceps purpurea based on its sexual stage by Tulasne (Citation1853), and includes up to the year 2021, when this paper was presented at a Canadian Phytopathological Society symposium entitled ‘The increasing issue of ergot in cereal production and the biological questions being explored’. Prior to 1853, ergot fungi had various names in the folklores of various ethnic groups. A list of prescriptions in German from 1474 and its medicinal use seems to be the oldest documentation of ergots (Píchová et al. Citation2018), whereas the earliest reference in literature can be traced back to Adam Lonicer’s description of the ‘long black nails between rye grains’ in a German book, Kreuterbuch, in 1582, according to Barger’s (Citation1931) historical account. The name ‘ergot’ was used by the peasants of Sologne (France), referring to the shape of sclerotia resembling cock’s-spurs (Barger Citation1931). At that time, the fungus was referred to mostly as abnormal grains, i.e., sun-baked grains, excessive sap formation, or mutant grains. Claude Joseph Geoffroy, a French apothecary and chemist, was the first to recognize them as separate organisms from their host plants in the 1700s (Oberstockstall Citation2013). About 40–50 years later, Augustin Pyramus de Candolle (1778–1841, resourced from Index Fungorum http://www.indexfungorum.org/Names/AuthorsOfFungalNames.asp) confirmed this observation, included the fungus in his iconic treatise Famille des champignons in Flore française and named it Sclerotium clavus (de Candolle Citation1815). Elias Magnus Fries (1794–1878) erected the genus Spermoedia for the sclerotium state, and made the new combination Sp. clavus (DC) Fr., meanwhile also included Sp. paspali (Schwein.) Fr. in the genus (Fries Citation1822). Joseph-Henri Léveillé (1796–1870) described the asexual stage (conidia in honeydew) of the fungus on rye and other cereal crops and named it Sphacelia segetum (Léveillé Citation1827).
Louis Rene Tulasne (1815–1885) and the early twentieth century
Tulasne (Citation1853) discovered the sexual stage during his detailed studies on l’ergot des glumacées and considered it conspecific to Sphaeria purpurea, a name coined by Fries (Citation1823) in sanctioning Sphaeria entomorrhiza Schumach. (Schumacher Citation1803) based on a figure of stroma in Dickson, Fasc. pl. crypt. brit., 1, 1785 (resourced from Index Fungorum, accessed in December 2021). The genus Claviceps was erected to include both sclerotial and the sexual stages, the combination C. purpurea was made, and Sclerotium clavus DC was noted as the sclerotial state of the fungus. Besides C. purpurea, Tulasne also recognized two other species based on different host ranges and morphological and biological differences. Claviceps purpurea was on cereal crops (rye, wheat, oats) and various grasses, C. microcephala (Wallr.) Tul. on Phragmites communis Trin. and Molinia caerulea (L.) Moench, and C. nigricans Tul. on Scirporum (Scirpus, Cyperaceae). Morphologically, the ascomata of C. microcephala were characterized by producing long flexuous stipes, and very small soft heads (capitula). Those of C. nigricans had the perithecia located more distantly than the other two species. Biologically, C. microcephala differed from the other two species by the immediate germination of sclerotia on plants (Tulasne Citation1853). Between 1853 and the 1940s, over30 ergot species were described as Claviceps spp. by mycologists here and there (), some of which were reclassified to other genera by contemporary studies (more discussion in the section on the molecular era). A few species were described as Spermoedia spp. such as Sp. zizaniae Fyles, Sp. stevensii Seaver, Sp. rolfsii (F. Steven & J. G. Hall) Seaver, Sp. tripsaci (F. Steven & J. G. Hall) Seaver.
Table 1. Claviceps spp. described between 1853 and 1940s.
R.F.N. Langdon (1916 – 2014) – the first comprehensive revision
The first comprehensive revision was conducted by Langdon during the 1940s and the 1950s. He studied Australian ergot species and also did a systematic account on various aspects of twenty-five non-Australian Claviceps spp. including the informative characteristics for differentiating species, host ranges, distributions, and synonyms (Langdon Citation1952). His dissertation is still a relevant reference for current ergot taxonomy. Based on his observation, the characteristics that were useful for differentiating species include the shape and colour of sclerotia, the colour of ascostromata, the placement of perithecia (imbedded or elevated) etc. He considered other characteristics, such as the length of stipes and the dimension and shape of perithecia, as insignificant for differentiating species. The shapes and sizes of conidia varied between species, but are also variable within species (Langdon Citation1952), thus this might be useful to a certain extent.
A. R. Loveless – characteristics of honeydew conidia
Loveless put more emphasis on the morphology of conidia as useful features for distinguishing species. He studied the conidia from honeydew from 34 grass species collected around Rhodesia (a British colony in territory equivalent to present-day Zimbabwe). Using a piece of special graphic equipment, he outlined the shapes and sizes of conidia randomly selected from honeydew samples from 34 grass species, and categorized the conidial shapes and sizes into 13 groups, six of which were associated with a certain species, i.e. group 2, 5, 6, 8, 9 and 12 corresponding to C. paspali F. Stevens & J.G. Hall, C. digitariae Hansf., C. sulcata Langdon, C. maximensis T. Theis, C. pusilla (Ces.) Ces. and C. cynodontis Langdon. Consequently, he predicted seven more species to be described in Rhodesia (Loveless Citation1964). Later, several species described in the area indeed corresponded to the size ranges of conidial groups, such as C. africana to group 10 (Frederickson et al. Citation1991), C. fusiformis Loveless to group 7 (Loveless Citation1967), C. rhynchelytri Herd & Loveless to group 1 (Herd and Loveless Citation1965), Claviceps loudetiae Pažoutová & Freder. to group 13, and C. truncatispora Pažoutová & Freder. to group 11 (Pažoutová et al. Citation2011).
S. Tanda – ergot fungi in Japan between 1970s and 1990s
Another frequently encountered name in the ergot taxonomy literature is Seinosuke Tanda. Tanda had a series of publications on ergot fungi in Japan between the 1970s to 1990s. He described not only some species, C. imperatae Tanda & Kawat. (Tanda and Kawatani Citation1976), C. microspora Tanda (Tanda Citation1985), C. panicoidearum Tanda & Y. Harada (Tanda and Harada Citation1989), C. bothriochloae Tanda & Y. Muray (Tanda Citation1991), C. amamiensis Tanda (Tanda Citation1992), but also characterized a lot of varieties (Tanda Citation1977, Citation1978a, Citation1978b, Citation1978c, Citation1979a, Citation1979b, Citation1979c, Citation1980a, Citation1980b, Citation1981) based on very careful examinations, measurements, and comparisons using many aspects of features including sclerotia, sexual stage, asexual stage, culture, alkaloid tests, and sugar consumption in culture. Even though he noted the ergot on Phalaris arundinacea L. had significantly larger conidia, he was not convinced it was a new species. He called it a new variety, viz C. purpurea var. phalaridis (Tanda Citation1979b), which turned out to be a new species based on molecular evidence (see later discussion about C. humidiphila Pažoutová & M. Kolařík). In the same way, the real identities of those varieties he described need to be validated.
Chemistry features applied for Claviceps taxonomy
The sugar composition in honeydew and its significance for taxonomy was investigated by Mower and Hancock (Citation1975). Through analyzing the sugar composition of nine determined and several undetermined Claviceps spp., they found significant variation in sugar profiles associated with several taxonomic sub-groups. For example, C. gigantea S.F. Fuentes, C. cinerea Griffiths and C. grohii J.W. Groves were rich in arabinitol or/and mannitol, whereas C. purpurea, C. fusiformis, and C. nigricans were rich in glucose and fructose. In contrast, there was a lot more research on the alkaloid content in sclerotia or in vitro cultures as a result of their toxic effects and pharmaceutical potentials. As three groups of ergot alkaloids (EAs) were first isolated from the sclerotia of C. purpurea (Stoll Citation1950), a wide spectrum of active compounds have been continuously reported from various Claviceps spp. through many analytical technics (Tanaka and Sugawa Citation1952; Mantle Citation1968; Porter et al. Citation1974), also reviewed by Flieger et al. (Citation1997). Consequently, the characteristics of EA production were incorporated in species descriptions. Notable examples are the studies of ergot in Japan by Tanda in that the presence/absence of EA production was almost always featured in the descriptions of species or/and varieties (Tanda Citation1979b, Citation1991). Lately, the more advanced technologies, i.e. high performance liquid chromatography (HPLC) and ultra high performance liquid chromatography (UPLC) enabled the highly sensitive and reliable separation of compounds and profiling of EA production. Varied species can be characterized by a unique combination of EA components in different quantities (Negård et al. Citation2015; Liu et al. Citation2020). Furthermore, indole diterpenes or paspaline-like compounds were detected in several more species in addition to C. paspali and C. cynodontis (Uhlig et al. Citation2014, Citation2021; Negård et al. Citation2015). In addition to alkaloids, various pigments present in the sclerotia are often in equal or greater amounts (Buchta and Cvak Citation1999). Among them, ergochromes showing toxicity to animals can contribute to overall ergot toxicity, and their spectrum represents another distinguishing feature between species (Flieger et al. Citation2019).
Ergot taxonomy in the molecular era – a polyphasic approach
The Pažoutová group in the Czech Republic should be considered the pioneer in the application of molecular markers for differentiating Claviceps species with their publication of characterizing C. citrina Pažoutová, Fučík., Leyva-Mir & Flieger, a species on Distichlis spicata (L.) Greene from Mexico. They demonstrated the different fingerprints of the random amplified polymorphism DNA (RAPD) of C. citrina from eight other species and variations in the rDNA internal transcribed spacer (ITS) (Pažoutová et al. Citation1998). Combined with its specific host range and other unique morphological features, especially the lemon-yellow colour of capitula, C. citrina was so named. During their study, it was found that on a high-sugar content media (T2), most Claviceps species grow faster, with less aerial hyphae showing more compact colonies that differed in texture and colour (Pažoutová et al. Citation1998, Citation2008a). Conidia formation on T2 medium appeared more abundant, which allowed more opportunity to observe the features of conidia and conidiogenesis. Building on previous observations of the phialidic conidiogenesis in Claviceps (Luttrell Citation1980; Rykard et al. Citation1984) and the occasional formation of the secondary conidia, Pažoutová and colleagues observed pleomorphic conidiation, i.e. primary enteroblastic (Sphacelia-like), holoblastic formation of secondary conidia (such as in C. africana) and sympodial holoblastic in C. citrina and C. zizaniae producing conidia with truncate ends (Pažoutová et al. Citation2004). The presence/absence of micro-conidia and the secondary conidia proved to be species-specific characteristics.
A phylogenetic study of C. purpurea and close relatives using RAPD fingerprinting and ITS rDNA sequences showed the presence of three genetic groups, G1 – G3, distinguishable by conidia morphology and alkaloid spectrum (Pažoutová et al. Citation2008b). This was later verified by multi-locus sequence analyses, and the three genetic groups were proven to be associated with different ecological niches, suggesting ecological cryptic speciation (Douhan et al. Citation2008). Until 2015, a formal taxonomic treatment was applied, and species names were designated to the three groups, i.e. G1 = C. purpurea s. str, G2 = C. humidiphila and G3 = C. spartinae (R.A. Duncan & J.F. White) Pažoutová & M. Kolařík. In addition, the new lineage G2a was named C. arundinis (Pažoutová et al. Citation2015). In terms of the host range and morphology, C. arundinis corresponds well with Tulasne’s concept of C. microcephala (Wallr.) Tul. However, the type specimen connected to C. microcephala (basionym: Kentrosporium microcephalum Wallr. 1846) was on an insect larva, which conflicts with the host of C. arundinis, therefore a new name was proposed with a holotype on Phragmites australis (Cav.) Trin. Ex Steud. (PRM922710). Claviceps humidiphila was characterized by often producing larger conidia than C. purpurea, and commonly found on Calamagrostis, Deschampsia cespitosa (L.) P. Beauv. and Phalaris arundinacea, corresponding well to C. purpurea var. phalaridis Tanda, therefore Pažoutová et al. (Citation2015) considered these two conspecific and designated a specimen collected by Tanda from Japan as holotype and a specimen from Germany as an epitype. A follow-up study proved the loss of the holotype specimen, and another specimen collected from the type location indicated C. purpurea var. phalaridis Tanda belonged to a different species from the epitype of C. humidiphila from Germany. A taxonomic rectification was conducted in that the specimen from Japan was designated as a neotype of C. humidiphila, whereas the German species was given a new name C. bavariensis (Liu et al. Citation2022). The delimitation of these species was confirmed by a study of Norweigian samples and also supported by the different indole alkaloid profiles produced by these species (Negård et al. Citation2015).
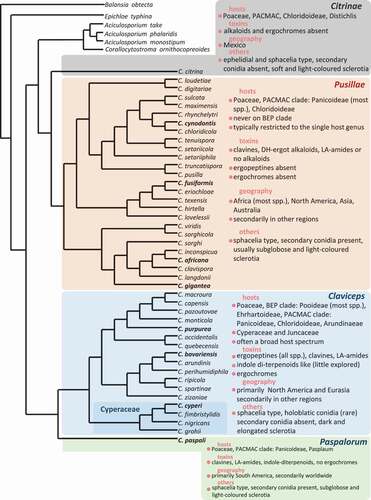
Host specificity of species and/or varieties was a long-standing topic of ergot study in that some species have a restricted host range, while others are more generalists (Campbell Citation1957; Alderman et al. Citation2004). The four phylogenetic species mentioned earlier showed a complex pattern of host preference. Some plant species (universal host) can host more ergot species (e.g. Poa, Dactylis) than others, whereas in particular areas (Central Europe vs Scandinavia) host specificity is more evident at a certain level, such that cultivated cereals in Europe are exclusively infected by C. purpurea s.str. (Negård et al. Citation2015; Pažoutová et al. Citation2015). Pažoutová et al. (Citation2008b) observed that C. purpurea s.str. prefers a dryer and open localities, whereas C. humidiphila is more often in wet and shady places. This pattern was challenged by later studies (Negård et al. Citation2015; Liu et al. Citation2020). Further evaluation with proper sampling on universal hosts could bring more insights. The DNA sequence-based polyphasic approach was also applied to genus-wide phylogenetic study and classification. Píchová et al. (Citation2018) conducted comprehensive phylogenetic analyses based on multiple protein-coding genes, resulting in the supra-specific classification of the genus into four sections, i.e. Claviceps sect. Claviceps, C. sect. Citrinae, C. sect. Paspalorum, and C. sect. Pusillae, associated with host and geographic ranges, and ergochrome and ergot alkaloid profiles. This comprehensive phylogenetic tree was further supplemented with a few more species from Canada (). In a supplementary Table, Píchová et al. (Citation2018) also listed a tentative assessment for ninety published names of Claviceps spp. building a foundation for a taxonomic revision.
Fig. 1 (Colour online) Phylogenetic relationships of ergot species with available DNA sequences. The classification into four sections (Citrinae, Claviceps, Paspalorum, Pusillae) and their characterization (toxicity, host range, geography and others), were based on.Píchová et al. (Citation2018) and Liu et al. (Citation2020). Ergot species in bold are of agricultural importance: C. africana on sorghum, C. cynodontis on Bermuda grass, C. fusiformis on pearl millet, C. gigantea on maize, C. paspali on paspalum and C. purpurea on cereals and forage grasses are important food and feed contaminants causing toxicity to human and livestock. Claviceps bavariensis infecting common pasture grasses may impact livestock. The maximum likelihood tree generated in PhyML 3.0. was constructed using ITS-LSU rDNA, TUB2, TEF1-α and RPB2 sequences from Píchová et al. (Citation2018) and Liu et al. (Citation2020).
Genetic diversity in Canada and the western USA
In the Canadian National Mycological Herbarium (DAOM), 90% of over 700 specimens of ergot fungi were identified as C. purpurea partly because ergot has never been systematically studied by taxonomists in Canada. In addition to C. purpurea, five other species were in the collection, including C. grohii, C. junci J. Adams, C. microcephala (possibly C. arundinis, see more discussion earlier), C. nigricans and C. zizaniae (Fyles) Pantidou ex Redhead, M.E. Corlett & M.N.L. Lefebvre.
Multi-gene DNA sequence analyses of the samples collected in Canadian agricultural and surrounding areas recovered seven lineages within the pre-molecular concept of C. purpurea (Shoukouhi et al. Citation2019). Three of them corresponded to C. humidiphila (now C. bavariensis), C. spartinae, and C. purpurea s.str., whereas the other four new lineages represented four new species (Liu et al. Citation2020), i.e. C. ripicola M. Liu, J.G. Menzies & S.A. Wyka on Ammophila, Phalaris and Calamagrostis from semi-wet habitat, river/lake banks, and sand dunes (British Columbia, Manitoba, Ontario, Quebec; USA: Colorado), C. perihumidiphila M. Liu on Agrostis capillaris L. and Elymus albicans (Scribn. & J. G. Sm.) Á. Löve from marshlands or terrestrial (Alberta, Newfoundland), C. quebecensis M. Liu & J. Cayouetteon Ammophila breviligulata Fernald (Quebec) distinguishable by ovoid shaped conidia, C. occidentalis M.Liu as axenic culture from Canadian Collection of Fungal Culture, and originally on Bromus inermis Leyss., and Phleum pratense L. from Alberta. The genetic diversity and distribution of C. purpurea s.str. in Canada and the Western USA were investigated through phylogenetic network analyses and population demographic parameters, resulting in the discovery of three subdivided genetic clusters, however the subdivision is not correlated with geographic regions or host groups. It was speculated that the intrinsic mechanisms that led to the cessation of gene flow among these three generic clusters might be mating and/or vegetative incompatibility, genomic adaptation etc. (Liu et al. Citation2021).
Challenges and future perspectives
The internal transcribed spacer (ITS) region was widely accepted as a marker in species identifications and phylogenetic analyses for many fungal groups and was proposed by the Consortium for the Barcode of Life as the primary universal DNA barcoding locus for fungi (Schoch et al. Citation2012). However, for some fungal groups, the ITS appeared to be a poor marker as a result of the presence of non-orthologous copies (O’Donnell and Cigelnik Citation1997), or low resolution at the species level (Geiser et al. Citation2007). In Claviceps, non-orthologous ITS copies were found in C. purpurea s.str. (G1) and C. occidentalis (G4) through examination of the ITS tree generated for Canadian samples (Shoukouhi et al. Citation2019). The low resolution at the species level is another shortcoming from which the ITS region in Claviceps spp. suffers; for instance, it could neither separate C. ripicola (G6, 7) from C. spartinae (G3), nor C. arundinis (G2a), C. humidiphila (G2) and C. perihumidiphila (G2b) (Negård et al. Citation2015; Pažoutová et al. Citation2015; Shoukouhi et al. Citation2019). In contrast, the protein-coding genes provide better resolutions, although, for certain species, a fragment of the second largest subunit of RNA polymerase II (RPB2) and the translation elongation factor 1-α (EF1-α) displayed a varied pattern of affinities among species. For example, RPB2 inferred C. humidiphila (G2) and C. arundinis (G2a) are more closely related than each of them to C. perihumidiphila (G2b), whereas TEF1 indicated C. humidiphila is more closely related with C. perihumidiphila (Shoukouhi et al. Citation2019). A follow-up study using the neutrality test indicated that these two genes are likely linked with genes under positive selection, and have undergone ‘genetic hitchhiking’ or ‘selective sweeping’ (Liu et al. Citation2021). Given these limitations of individual genes, it is recommended to apply a holistic MLSA approach for species recognition in Claviceps.
The drastically increased number of whole genome sequences of Claviceps confirmed the species recognition based on MLSA, and also allowed the genome level understanding of the speciation mechanism associated with genome landscape and host expansion (Wyka et al. Citation2021). This could also provide opportunities for developing more gene markers for species recognitions and species-level phylogenetic studies. The question then arise is what would be the best strategy to apply genome data in fungal taxonomy. Similar to the selection of standard DNA barcoding loci, there is a need to select a standard panel of genes for species recognition in various fungal groups. Target enrichment approaches increased the efficiency of obtaining pre-selected genome regions that are suitable for specific phylogenetic levels (Albert et al. Citation2007; Gnirke et al. Citation2009; Jones and Good Citation2016). The technology has been used for certain fungi and fungus-like groups for exploring phylogenetic informative markers (Grewe et al. Citation2020; Nguyen et al. Citation2021). In the course of selecting the target genome regions, the paralogous gene copies caused by ancient lineage sorting, hybridization, recombination, horizontal gene transfer etc. were still among the biggest concerns (Andermann et al. Citation2020; Dettman and Eggertson Citation2021). Some strategies were proposed for distinguishing them, including identifying the unusually high levels of variations or checking the presence of extra haplotypes. However, none of the tactics can ensure the elimination of paralogous genes (Andermann et al. Citation2020). A thorough understanding of specific genome structures through careful genome wide examination is deemed to be an essential step. The concept and recognition of species have been a centre of debates since the birth of biological species concepts (Mayr Citation1942; Dobzhansky Citation1970) that criticized the shortcomings of the traditional typological species concepts or morphological species concepts (Mayr Citation1996). The debates became more heated with the emergence of diverse species concepts coming along with the molecular markers applied for classification, for example, the genealogical species concept (Baum and Shaw Citation1995), Hennigian species concept (Meier and Willmann Citation2000), monophyletic species concept (Donoghue Citation1985), phylogenetic species concepts (Nixon and Wheeler Citation1990) etc, see review by Luckow (Citation1995). In fungal molecular systematic, the Genealogical Concordance Phylogenetic Species Recognition (GCPSR, Taylor et al. (Citation2000)) was widely applied in fungal species recognition although many researches demonstrated that paraphyletic groups could be considered as species as well (Hörandl and Stuessy Citation2010). It is foreseeable that the debates over how to recognize fungal species will continue in the genomic era (Matute and Sepúlveda Citation2019; Xu Citation2020). We argue that despite the rapid advances in DNA-based technology may reveal more genotypic diversity, the recognition of species should still be based on the combination of genotypic and phenotypic features (polyphasic approach) because it is the phenotypes that are tangible features directly impacting humans and eco-systems (Lücking et al. Citation2020).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 4(11):903–905. doi:10.1038/nmeth1111
- Alderman SC, Halse RR, White JF. 2004. A reevaluation of the host range and geographical distribution of Claviceps species in the United States. Plant Dis. 88(1):63–81. doi:10.1094/PDIS.2004.88.1.63.
- Andermann T, Torres Jiménez MF, Matos-Maraví P, Batista R, Blanco-Pastor JL, Gustafsson ALS, Kistler L, Liberal IM, Oxelman B, Bacon CD, et al. 2020. A guide to carrying out a phylogenomic target sequence capture project [Review]. Front Genet. 10(1407). doi:10.3389/fgene.2019.01407.
- Arcella D, Gómez Ruiz JÁ, Innocenti ML, Roldán R, European Food Safety Authority. 2017. Human and animal dietary exposure to ergot alkaloids. EFSA J. 15(7):4902.
- Barger G. 1931. Chapter I. Ergot: historical. In: Barger G, editor. Ergot and Ergotism — a Monograph. London: Gurney and Jackson; p. 1–13.
- Baum DA, Shaw KL. 1995. Genealogical perspectives on the species problem. In: Hoch PC, Stephenson AG, editors. Experimental and molecular approaches to plant biosystematics. St. Louis: Missouri Botanical Garden; p. 289–303.
- Buchta M, Cvak L. 1999. Ergot alkaloids and other metabolites of the genus Claviceps. In: Kren V, Cvak L, editors. Ergot: the genus Claviceps. Amsterdam (The Netherlands etc): Harwood Academic Publishers; p. 25–56. (republished 2006 by Taylor and Francis e-Library).
- Campbell WP. 1957. Studies on ergot infection in gramineous hosts. Can J Bot. 35(3):315–320. doi:10.1139/b57-028.
- de Candolle AP. 1815. Famille des champignons. In: Candolle AP, editor. Flore française. Paris: Desray; p. 115.
- Dettman JR, Eggertson Q. 2021. Phylogenomic analyses of Alternaria section Alternaria: a high-resolution, genome-wide study of lineage sorting and gene tree discordance. Mycologia. 113(6):1218–1232. doi:10.1080/00275514.2021.1950456.
- Dobzhansky T. 1970. Genetics of the Evolutionary Process. New York: Columbia University Press.
- Donoghue MJ. 1985. A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist. 88(3):172–181. doi:10.2307/3243026.
- Douhan GW, Smith ME, Huyrn KL, Westbrook A, Beerli P, Fisher AJ. 2008. Multigene analysis suggests ecological speciation in the fungal pathogen Claviceps purpurea. Mol Ecol. 17(9):2276–2286. doi:10.1111/j.1365-294X.2008.03753.x.
- Flieger M, Wurst M, Shelby R. 1997. Ergot alkaloids–sources, structures and analytical methods. Folia Microbiol (Praha). 42(1):3–29. doi:10.1007/BF02898641.
- Flieger M, Stodůlková E, Wyka SA, Černý J, Grobárová V, Píchová K, Novák P, Man P, Kuzma M, Cvak L, et al. 2019. Ergochromes: heretofore neglected side of ergot toxicity. Toxins. 11(8):439. doi:10.3390/toxins11080439
- Frederickson DE, Mantle PG, Wajd M. 1991. Claviceps africana sp. nov.; the distinctive ergot pathogen of sorghum in Africa. Mycol Res. 95(9):1101–1107. doi:10.1016/S0953-7562(09)80555-8.
- Fries EM. 1822. Systema mycologicum: sistens fungorum ordines, genera et species, huc usque cognitas, quas ad normam methodi naturalis determinavit. Lundae: Ex Officina Berlingiana; p. 268–269.
- Fries EM. 1823. Systema mycologicum: sistens fungorum ordines, genera et species, huc usque cognitas, quas ad normam methodi naturalis determinavit. Lundae: Ex Officina Berlingiana; p. 325.
- Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA. 2007. The current status of species recognition and identification in Aspergillus. Stud Mycol. 59:1–10. doi:10.3114/sim.2007.59.01.
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 27(2):182–189. doi:10.1038/nbt.1523
- Grewe F, Ametrano C, Widhelm TJ, Leavitt S, Distefano I, Polyiam W, Pizarro D, Wedin M, Crespo A, Divakar PK, et al. 2020. Using target enrichment sequencing to study the higher-level phylogeny of the largest lichen-forming fungi family: parmeliaceae (Ascomycota). IMA Fungus. 11(1):27. doi:10.1186/s43008-020-00051-x
- Herd GW, Loveless AR. 1965. Studies of Rhodesian ergots III. Claviceps rhynchelytri sp. nov. and Claviceps digitariae Hansford. Kirkia. 5(1):21–24.
- Hörandl E, Stuessy TF. 2010. Paraphyletic groups as natural units of biological classification. Taxon. 59(6):1641–1653. doi:10.1002/tax.596001.
- Jones MR, Good JM. 2016. Targeted capture in evolutionary and ecological genomics. Mol Ecol. 25(1):185–202. doi:10.1111/mec.13304.
- Kren V, Cvak L. 1999. Ergot: the Genus Claviceps. The Netherlands: Harwood academic publishers. (republished 2006 by Taylor and Francis e-Library).
- Langdon RFN. 1952. Studies on ergot (Thesis). University of Queensland.
- Lev-Yadun S, Halpern M. 2007. Ergot (Claviceps purpurea)-An aposematic fungus. Symbiosis. 43(2):105–108.
- Léveillé MJ-H. 1827. Sur l’ergot, ou nouvelles recherches sur la cause et les effects de l’ergot considéré sous le triple rapport botanique, agricole et médical. Mem Soc Linn Paris. 5:578.
- Liu M, Overy DP, Cayouette J, Shoukouhi P, Hicks C, Bisson K, Sproule A, Wyka SA, Broders K, Popovic Z, et al. 2020. Four phylogenetic species of ergot from Canada and their characteristics in morphology, alkaloid production, and pathogenicity. Mycologia. 112(5):974–988. doi:10.1080/00275514.2020.1797372
- Liu M, Shoukouhi P, Bisson KR, Wyka SA, Broders KD, Menzies JG. 2021. Sympatric divergence of the ergot fungus, Claviceps purpurea, populations infecting agricultural and nonagricultural grasses in North America. Ecol Evol. 11(1):273–293. doi:10.1002/ece3.7028.
- Liu M, Tanaka E, Kolařík M. 2022. Neotypification of Claviceps humidiphila and recognition of C. bavariensis sp. nov. Mycotaxon. 137(1):73–87. doi:10.5248/137.73.
- Loveless AR. 1964. Use of the honeydew state in the identification of ergot species. Trans Brit Mycol Soc. 47(2):205–213. doi:10.1016/S0007-1536(64)80054-1.
- Loveless AR. 1967. Claviceps fusiformis sp.nov., the causal agent of an agalactia of sows. Trans Brit Mycol Soc. 50(1):15–18. doi:10.1016/S0007-1536(67)80058-5.
- Lücking R, Aime MC, Robbertse B, Miller AN, Ariyawansa HA, Aoki T, Cardinali G, Crous PW, Druzhinina IS, Geiser DM, et al. 2020. Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 11(1):14.
- Luckow M. 1995. Species concepts: assumptions, methods, and applications. Syst Bot. 20(4):589–605. doi:10.2307/2419812.
- Luttrell ES. 1980. Host–parasite relationships and development of the ergot sclerotium in Claviceps purpurea. Can J Bot. 58(8):942–958. doi:10.1139/b80-118.
- Mantle PG. 1968. Studies on Sphacelia sorghi McRae, an ergot of Sorghum vulgare. Pers. 62(3):443–449.
- Matute DR, Sepúlveda VE. 2019. Fungal species boundaries in the genomics era. Fungal Genet Biol. 131:103249. doi:10.1016/j.fgb.2019.103249.
- Mayr E. 1942. Systematics and the origin of species from the viewpoint of a zoologist. New York: Columbia University Press.
- Mayr E. 1996. What is a species and what is not? Philos Sci. 63:262–277. doi:10.1086/289912.
- Meier R, Willmann R. 2000. The Hennigian species concept. In: Wheeler QD, Meier R, editors. Species concepts and phylogenetic theory. New York: Columbia University Press; p. 30–43.
- Menzies JG, Turkington TK. 2015. An overview of the ergot (Claviceps purpurea) issue in western Canada: challenges and solutions. Can J Plant Pathol. 37(1):40–51. doi:10.1080/07060661.2014.986527.
- Miedaner T, Geiger HH. 2015. Biology, genetics, and management of ergot (Claviceps spp.) in rye, sorghum, and pearl millet. Toxins. 7(3):659–678. doi:10.3390/toxins7030659.
- Mower RL, Hancock JG. 1975. Sugar composition of ergot honeydews. Can J Bot. 53(23):2813–2825. doi:10.1139/b75-309.
- Negård M, Uhlig S, Kauserud H, Andersen T, Høiland K, Vrålstad T. 2015. Links between genetic groups, indole alkaloid profiles and ecology within the grass-parasitic Claviceps purpurea species complex. Toxins. 7(5):1431–1456. doi:10.3390/toxins7051431.
- Nguyen HDT, McCormick W, Eyres J, Eggertson Q, Hambleton S, Dettman JR. 2021. Development and evaluation of a target enrichment bait set for phylogenetic analysis of oomycetes. Mycologia. 113(4):856–867. doi:10.1080/00275514.2021.1889276.
- Nixon KC, Wheeler QD. 1990. An amplification of the phylogenetic species concept. Cladistics. 6:211–223. doi:10.1111/j.1096-0031.1990.tb00541.x.
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 7(1):103–116. doi:10.1006/mpev.1996.0376.
- Oberstockstall T. 2013. Ergot. The fungi that ate medieval Europe. RAMS digital.
- Pažoutová S, Fucikovsky L, Leyva-Mir SG, Flieger M. 1998. Claviceps citrina sp. nov., a parasite of the halophytic grass Distichlis spicata from Mexico. Mycol Res. 102:850–854. doi:10.1017/S095375629700573X.
- Pažoutová S, Parbery DP. 1999. The taxonomy and phylogeny of Claviceps. In: Kren V, Cvak L, editors. Ergot: the Genus Claviceps. Amsterdam (The Netherlands): Harwood Academic Publishers; p. 25–56. (republished 2006 by Taylor and Francis e-Library).
- Pažoutová S, Kolařík M, Kolínská R. 2004. Pleomorphic conidiation in Claviceps. Mycol Res. 108(2):126–135. doi:10.1017/S0953756203009067.
- Pažoutová S, Kolařík M, Odvody G, Frederickson D, Olšovská J, Man P. 2008a. A new species complex including Claviceps fusiformis and Claviceps hirtella. Fungal Divers. 2008:95–111.
- Pažoutová S, Olšovská J, Šulc M, Chudíčková M, Flieger M. 2008b. Claviceps nigricans and Claviceps grohii: their alkaloids and phylogenetic placement. J Nat Prod. 71(6):1085–1088. doi:10.1021/np8001173.
- Pažoutová S, Odvody GN, Frederickson DE, Chudíčková M, Olšovská J, Kolařík M. 2011. New Claviceps species from warm-season grasses. Fungal Divers. 49(1):145–165. doi:10.1007/s13225-011-0102-4.
- Pažoutová S, Pešicová K, Chudíčková M, Šrůtka P, Kolařík M. 2015. Delimitation of cryptic species inside Claviceps purpurea. Fungal Biol. 119(1):7–26. doi:10.1016/j.funbio.2014.10.003.
- Píchová K, Pažoutová S, Kostovčík M, Chudíčková M, Stodůlková E, Novák P, Flieger M, Van der linde E, Kolařík M. 2018. Evolutionary history of ergot with a new infrageneric classification (Hypocreales: clavicipitaceae: claviceps). Mol Phylogenet Evol. 123:73–87. doi:10.1016/j.ympev.2018.02.013.
- Porter JK, Bacon CW, Robbins JD. 1974. Major alkaloids of a Claviceps isolated from toxic Bermuda grass. J Agric Food Chem. 22(5):838–841. doi:10.1021/jf60195a007.
- Rykard DM, Luttrell ES, Bacon CW. 1984. Conidiogenesis and Conidiomata in the Clavicipitoideae. Mycologia. 76(6):1095–1103.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Barcoding Consortium F. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 109(16):6241. doi:10.1073/pnas.1117018109.
- Schumacher HCF. 1803. Enumeratio plantarum in partibus Saellandiae septentrionalis et orientalis. Hafnia: Apud Fridericum Brummer. Typis Zach. Breum.
- Shoukouhi P, Hicks C, Menzies JG, Popovic Z, Chen W, Seifert KA, Assabgui R, Liu M. 2019. Phylogeny of Canadian ergot fungi and a detection assay by real-time polymerase chain reaction. Mycologia. 111(3):493–505. doi:10.1080/00275514.2019.1581018.
- Stoll A. 1950. Recent investigations on ergot alkaloids. Chem Rev. 47(2):197–218. doi:10.1021/cr60147a002.
- Tanaka K, Sugawa T. 1952. Studies on ergot alkaloids by paper partition chromatography. I YAKUGAKU ZASSHI = J Pharma Soc Jpn. 72(5):616–620.
- Tanda S, Kawatani T. 1976. A new species of Claviceps parasitic on Imperata cylindrica Beauv. var. koenigii Durand et Schinz. Trans Mycol Soc Japan. 17:289–294.
- Tanda S. 1977. Mycological studies on ergot in Japan (Part 3): a new variety of Claviceps purpurea Tul. parasitic on Alopecurus aequalis Sobol. var. amurensis Ohwi. J Agric Sci, Tokyo Nogyo Daigaku. 22:293–299.
- Tanda S. 1978a. Mycological studies on ergot in Japan (Part 4): two varieties of Claviceps purpurea Tul. parasitic on timothy. Phleum pretense L. J Agric Sci, Tokyo Nogyo Daigaku. 23:141–150.
- Tanda S. 1978b. Mycological studies on ergot in Japan (Part 6): a physiologic race of Claviceps purpurea Tul. var. alopecuri Tanda collected from Trisetum bifidum Ohwi. J Agric Sci, Tokyo Nogyo Daigaku. 23:207–214.
- Tanda S. 1978c. Mycological studies on ergot in Japan (Part 7): two varieties of Claviceps purpurea Tul. parasitic on Agrostis spp. J Agric Sci, Tokyo Nogyo Daigaku. 23:215–221.
- Tanda S. 1979a. Mycological studies on ergot in Japan (Part 8): ergots on Calamagrostis spp. J Agric Sci, Tokyo Nogyo Daigaku. 23:67–78.
- Tanda S. 1979b. Mycological studies on ergot in Japan (Part 9): distinct variety of Claviceps purpurea Tul. on Phalaris arundinacea L. and P. arundinacea var. picta L. J Agric Sci, Tokyo Nogyo Daigaku. 24:84.
- Tanda S. 1979c. Mycological studies on ergot in Japan (Part 10): ergots on Velvet grass, Holcus lanatus L. J Agric Sci, Tokyo Nogyo Daigaku. 24:145–148.
- Tanda S. 1980a. Mycological studies on ergot in Japan (Part 11): ergots on Polypogon fugax. Trans Mycol Soc Japan. 21:97–101.
- Tanda S. 1980b. Mycological studies on ergot in Japan (Part 16): ergots on orchard grass, Dactylis glomerata L. J Agric Sci, Tokyo Nogyo Daigaku. 25:263–271.
- Tanda S. 1981. Mycological studies on ergot in Japan (Part 19): ergots on bluegrass (Poa spp.). J Agric Sci, Tokyo Nogyo Daigaku. 26(1):179–192.
- Tanda S. 1985. Mycological studies on the ergot in Japan (XXI): a new species of Claviceps parasitic on Arundinella hirta (Thunb.) C. Tanaka. J Agric Sci, Tokyo Nogyo Daigaku. 30(2):94–99.
- Tanda S, Harada Y. 1989. Mycological studies on the ergot in Japan (XXII) A new ergot parasitic on Isachne globosa. Trans Mycol Soc Japan. 30:105–109.
- Tanda S. 1991. Mycological studies on the ergot in Japan, 26: a new ergot, Claviceps bothriochloae parasitic on Bothriochloa parviflora. J Agric Sci, Tokyo Nogyo Daigaku. 36(1):36–42.
- Tanda S. 1992. Mycological studies on the ergot in Japan, 27: an undescribed species Clavieps on Digitaria. J Agric Sci, Tokyo Nogyo Daigaku. 36(3):182–187.
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 31:21–32. doi:10.1006/fgbi.2000.1228.
- Tulasne LR. 1853. Mémoire sur l’ergot des Glumacées. Ann Sci Nat Bot. 20:5–56.
- Uhlig S, Egge-Jacobsen W, Vrålstad T, Miles CO. 2014. Indole-diterpenoid profiles of Claviceps paspali and Claviceps purpurea from high-resolution Fourier transform orbitrap mass spectrometry. Rapid Commun Mass Spectrom. 28(14):1621–1634. doi:10.1002/rcm.6938.
- Uhlig S, Rangel-Huerta OD, Divon HH, Rolén E, Pauchon K, Sumarah MW, Vrålstad T, Renaud JB. 2021. Unraveling the ergot alkaloid and indole diterpenoid metabolome in the Claviceps purpurea species complex using lc–hrms/ms diagnostic fragmentation filtering. J Agric Food Chem. 69(25):7137–7148. doi:10.1021/acs.jafc.1c01973.
- Wäli PP, Wäli PR, Saikkonen K, Tuomi J. 2013. Is the pathogenic ergot fungus a conditional defensive mutualist for its host grass? PLOS ONE. 8(7):e69249. doi:10.1371/journal.pone.0069249.
- Wyka SA, Mondo SJ, Liu M, Dettman J, Nalam V, Broders KD. 2021. Whole-genome comparisons of ergot fungi reveals the divergence and evolution of species within the genus claviceps are the result of varying mechanisms driving genome evolution and host range expansion. Genome Biol Evol. 13(2):evaa267. doi:10.1093/gbe/evaa267.
- Xu J. 2020. Fungal species concepts in the genomics era. Genome. 63(9):459–468. doi:10.1139/gen-2020-0022.
