Abstract
Gene-associated simple sequence repeat (SSR) markers were developed for Puccinia triticina through the data mining of existing EST libraries. Analysis of 7134 expressed sequence tags (ESTs) from cDNA libraries of P. triticina detected 204 EST-SSRs with a minimum of 12 repeating nucleotides. The majority of EST-SSRs contained short di- or tri-nucleotide repeats. These EST-SSRs were evaluated on 35 P. triticina isolates collected in Canada and 21 EST-SSRs were polymorphic and informative in determining intraspecific genetic diversity. A comparison of virulence and EST-SSR genotypes showed a strong correlation between virulence to Lr2a, Lr2c and Lr17a and EST-SSRs genotypes. The differentiation of the P. triticina population based on EST-SSR genotypes was comparable to that obtained with genomic SSRs, despite differences between two types of SSR markers. Eight of the 21 EST-SSRs produced the cross amplification in Puccinia coronata and Puccinia graminis, suggesting that EST-SSRs are more applicable than genomic SSRs for interspecific analysis. In summary, our study suggests that the data mining of EST databases is a feasible way to generate informative molecular markers for genetic studies of P. triticina.
Résumé: Des marqueurs microsatellites (SSR) associés à des gènes ont été conçus pour Puccinia triticina à partir l'exploration de données contenues dans des banques d'étiquettes de séquences exprimées (EST). L'analyse de 7134 EST issues de banques d'ADNc de P. triticina ont permis de détecter 204 EST-SSR avec un minimum de 12 répétitions de nucléotides. La majorité des EST-SSR contenaient de courtes répétitions di- ou tri-nucléotidiques. Ces EST-SSR ont été évaluées sur 35 isolats de P. triticina collectés au Canada. Parmi celles-ci, 21 étaient polymorphiques et ont fourni de l'information servant à établir la diversité génétique intraspécifique. Une comparaison de la virulence et des génotypes EST-SSR a montré une forte corrélation entre la virulence à l'égard de Lr2a, Lr2c et Lr17a ainsi qu'à l'égard des génotypes EST-SSR. La différenciation de la population de P. triticina, basée sur les génotypes EST-SSR, était comparable à celle obtenue avec les SSR génomiques, malgré les différences entre les deux types de marqueurs. Huit des 21 EST-SSR ont engendré la transférabilité chez Puccinia coronata et Puccinia graminis, ce qui suggère que les EST-SSR sont plus adaptables à l'analyse interspécifique que les SSR génomiques. En résumé, notre étude suggère que l'exploration de données des banques d'EST permet de générer des marqueurs moléculaires informatifs pour les études génétiques portant sur P. triticina.
Introduction
Wheat leaf rust, caused by Puccinia triticina Eriks, is a serious problem in the production of common wheat (Triticum aestivum L.) and durum wheat (Triticum turgidum L. var. durum) in Canada and worldwide (Samborski, Citation1985). Depending on the level of resistance in wheat cultivars or the developmental stage of crops during the initial infection, P. triticina can cause up to 25% yield loss (Kolmer, Citation2001).
Populations of P. triticina in North American exhibit a high degree of virulence and genetic variation. Over 40 P. triticina virulence phenotypes have been identified on an annual basis in Canada since the 1980s (Kolmer et al., Citation2005) and various molecular markers, such as amplified fragment length polymorphism (AFLP) (Kolmer, Citation2001; Mebrate et al., Citation2006) and random amplified polymorphic DNA (RAPD) (Kolmer & Liu, Citation2000; Kosman et al., Citation2004), have been used to characterize the population structure of P. triticina. Such information is fundamental to understand the genetic mechanism(s) underlying variations in P. triticina, and is crucial for the development of an effective control strategy for this disease.
Both RAPD and AFLP are dominant molecular markers, which cannot differentiate homozygous from heterozygous genotypes, whereas simple sequence repeat markers (SSRs) are codominant markers, which are more user-friendly and easier to adapt for automation (Zane et al., Citation2002). Unfortunately, the development of genomic SSRs can be very expensive and labour intensive (Zane et al., Citation2002; Dutech et al., Citation2007). The increasing availability of expressed sequence tags (ESTs) derived from expression libraries has provided an alternative resource for the generation of new codominant markers (Squirrell et al., Citation2003). These EST-SSR markers are also more transferable to related taxa than genomic SSRs due to the conserved nature of gene-associated sequences (Keiper et al., Citation2006). Although few genomic SSRs have been generated from enriched genomic DNA libraries of P. triticina (Duan et al., Citation2003; Szabo & Kolmer, Citation2007), EST-SSRs were not yet available in P. triticina. More SSRs are needed to better understand the population structure of P. triticina.
Herein, we report the development of EST-SSRs by the data mining of existing P. triticina EST databases. These markers were evaluated for polymorphism on 35 P. triticina isolates collected in Canada. The correlation between virulence and genetic variability was analyzed within and between groups of P. triticina isolates. Two clustering methods were applied to virulence and SSR datasets and results were then compared. Furthermore, we examined the transferability of P. triticina EST-SSRs in taxa related to P. triticina, such as P. coronata and P. graminis.
Materials and methods
Puccinia triticina isolates and determination of virulence phenotypes
Thirty-five genetically pure isolates of P. triticina collected in Canada during the 2007 annual field survey were used (). Most virulence phenotypes were represented by a single isolate. Virulence of P. triticina isolates was determined using the method described previously (McCallum & Seto-Goh, Citation2004, Citation2005). The infection types produced on a set of 16 standardized differentials [Set 1: Lr1, Lr2a, Lr2c, Lr3; Set 2: Lr9, Lr16, Lr24, Lr26; Set 3: Lr3ka, Lr11, Lr17a, Lr30; Set 4: LrB, Lr10, Lr14a, Lr18] were used to determine a four-letter code according to the virulence phenotype nomenclature proposed by Long & Kolmer (Citation1989).
Table 1. Virulence phenotypes and group designations of Puccinia triticina isolates
SSR characterization and primer design
A total of 7134 ESTs generated from five different life stages of P. triticina (Hu et al., Citation2007) were analyzed for the presence of SSR motifs using MICAS (http://210.212.212.7/MIC) and RepeatMasker (http://www.repeatmasker.org). Sequences that contained at least 12 repeating nucleotides were chosen and PCR primers were designed using PRIMER3 (Rozen & Skaletsky, Citation2000) to amplify flanking regions. Primers were synthesized by Invitrogen™. For the isolation of total genomic DNA, fresh urediniospores were germinated on the surface of distilled water and mats of germinated urediniospores were collected. Spore mats were then vacuum-dried and ground with glass beads. Genomic DNA was extracted from spore mats using the method described by Kolmer et al. (Citation1995). DNA concentration was determined using spectrophotometer readings at A260 and A280 and a working solution of 50 ng μL–1 was made. PCR was carried out in a 25 μL reaction volume containing 1 × PCR buffer (Applied Biosystem), 1.5 mM MgCl2, 0.2 mM dNTPs, 2.5 mM forward and reverse primers, 0.5 U Taq polymerase (Invitrogen) and 30 ng sample DNA. Amplifications were performed in a thermal cycler (GeneAmp PCR 9700) using the following temperature profile: initial denaturation step at 95 °C for 2 minutes, then 35 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 1 minute, followed by a final extension step at 72 °C 5 minutes after. PCR products were analyzed in a 6% polyacrylamide gel and visualized by silver staining (Chalhoub et al., Citation2007). Alleles were scored by length in base pairs using a 25 bp DNA ladder (Invitrogen) as size standard.
P. triticina EST-SSR genotypic analysis
The M13-tailed primer method (Zhou et al., Citation2001) was used to label amplicons for the visualization of PCR products on a capillary DNA analyzer. Forward primers were 5′-tailed with 23-base pair M13 (unit-43) sequence (AGGGTTTTCCCAGTCACGACGTT). The entire forward primer is similar to 5′- AGGGTTTTCCCAGTCACGACGTTXXXXX, whereas Xs denote SSR specific primer sequence. PCR was carried out in a 12.5 μL reaction volume containing 1 × PCR buffer (Applied Biosystem), 1.5 mM MgCl2, 0.2 mM dNTPs, 1.25 μM of each SSR specific primers, 0.5 μM of 5′-fluorescently labelled M13 (unit-43) primer, 0.5 U Taq polymerase and 30 ng sample DNA. M13 primer was 5′-fluorescently tagged with HEX, 6-FAM or NED to facilitate multiplexing. Amplifications were performed in a thermal cycler (GeneAmp PCR 9700) using the following temperature profile: initial denaturation step at 95 °C for 2 minutes, then 35 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 1 minutes, followed by a final extension step at 72 °C 5 mintues after. Amplification products were analyzed with a ABI3730 DNA analyzer. A fluorescent DNA size standard ranging from 50–500 was included in each run.
Data analysis
Virulence dissimilarity of P. triticina was calculated using the complement of simple match coefficient (Kolmer et al., Citation1995). For genetic diversity study, the alleles of each EST-SSR locus were scored based on the absence (0) or presence (1) of the allele. The dissimilarity of EST-SSR genotypes was determined using the complement of Dice-coefficient (Kolmer & Ordonez, Citation2007). The number of clusters within the datasets were analyzed with MODECLUS procedure (SAS Institute Inc., 1997) using the method described previously (St-Laurent et al., Citation2000; Harder et al., Citation2001). A smoothing parameter (K-value) was used to determine the region of stability for clusters. The clusters taken from the region of stability were then evaluated by subjecting the original data matrix to an ordination through canonical discriminant analysis (CDA) using SAS CANDICS procedure (SAS Institute Inc., 1997). The unweighted pair group arithmetic mean method (UPGMA) (SHAN program, NTSYS version 2.1) was used to construct dendrograms representing virulence and genetic dissimilarity. The COPH module in NYSYS-pc was used to derive a cophenetic value matrix from the dendrogram to confirm the correlation between the dendrogram and original matrix. The matrices derived from virulence and SSR genotypes were transformed with Dcenter module in NTSYS and the degree of correlation between matrices derived from virulence and EST-SSRs were determined with MXCOMP module in NTSYS-pc.
Results and discussion
Two hundred and four EST-SSRs containing a minimum of 12 repeating nucleotides were identified in 7134 Puccinia triticina EST unigene sequences and the number of repeats in EST-SSRs ranged from 3 to 32 with an average of 5. The incidence of EST-SSRs in P. triticina (2.9%) is not only lower than incidences of EST-SSRs in wheat (5.6%) (Holton et al., Citation2002) and in oat (3.2%) (Becher, Citation2007), but is also lower than those reported in fungal grass entophytes, Neptyphodium coenophialum (9.7%) and N. lolii (6.3%) (Jong et al., Citation2003) and in the other rust fungi, such as in P. coronata (5%) (Dracatos et al., Citation2007) and in P. graminis f. sp. tritici (6%) (Zhong et al., Citation2009). The rarity of SSR motifs in P. triticina ESTs indicates that SSRs are less abundant in P. triticina than in other fungal or plant species. It is also possible that this difference is merely caused by the different selection criteria of SSRs used in those studies. In addition, the number and length of ESTs can also affect the frequency of EST-SSRs since many ESTs in the databases do not represent the full length of the transcripts. The average length of EST-SSRs in P. triticina was shorter than those reported in wheat and oat (Holton et al., Citation2002; Becher, Citation2007) and EST-SSRs with long motifs were rare in P. triticina. Similarly, Lim et al. (Citation2008) reported that short SSRs with five to seven repeating motifs were predominant in genomes of 14 fungal species. The low density and shortness of SSRs in fungal genomes have rendered the isolation of fungal SSRs through enriched genomic DNA libraries very difficult and inefficient (Dutech et al., Citation2007). In this regard, the identification of SSRs from fungal ESTs provides a new alternative because large sets of ESTs can be screened for SSRs with short motifs through computation.
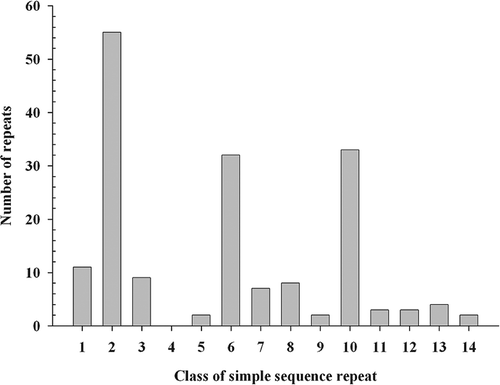
A variety of motifs were found in P. triticina EST-SSRs. Tri-nucleotide (45%) and di-nucleotide repeats (37%) were the most abundant, whereas other repeating structures occurred at much lower frequencies (tetra-nucleotide repeats: 13% and penta-nucleotide repeats: 5%). Tri-nucleotide motifs and low repeat numbers have been found to be common features of EST-derived SSRs, which differentiate them from genomic SSRs derived from enriched genomic libraries where di-nucleotide motifs predominate. The predominance of tri-nucleotide repeats in EST-SSRs is likely due to the suppression of non-trinucleotide SSRs in coding regions, which reduces frame-shift mutations within transcribed genes (Metzgar et al., Citation2008). Morgante et al. (Citation2002) suggested that high frequency of tri-nucleotide repeats in coding regions could have resulted from the mutation pressure and putative selection for specific amino acid stretches. Among 14 classes of di- and tri-nucleotide SSR motifs described in eukaryotes (Jurka & Pethiyagoda, Citation1995; Katti et al., Citation2001), ‘AG/GA/CT/TC’ (class 2) were the most predominant di-nucleotide motifs, whereas AAG/AGA/GAA/CTT/TTC/TCT (class 6) and AGG/GGA/GAG/CCT/CTC/TCC (class 10) were the most common tri-nucleotide motifs in P. triticina (). Similar di- and tri-nucleotide motifs were also found in EST-SSRs characterized from P. graminis f. sp. tritici ESTs (Zhong et al., Citation2009). Since P. triticina and P. graminis f. sp. tritici are closely related, the similarity between EST-SSR motifs from two cereal rust species is not surprising.
Fig. 1. Distributions of 14 classes of di- and tri-nucleotide simple sequence repeat motifs in the EST database of P. triticina. Classes of simple sequence repeats described by Jurka & Pethiyagoda (Citation1995) and Katti et al. (Citation2001) were listed as following: 1 = AT/AT; 2 = AG/GA/CT/TC; 3 = AC/CA/TG/GT; 4 = GC/CG; 5 = AAT/ATA/ TAA/ATT/TTA/TAT; 6 = AAG/AGA/GAA/CTT/TTC/TCT; 7 = A AC/ACA/CAA/GTT/TTG/TGT; 8 = ATG/TGA/GAT/CAT/A TC/TCA; 9 = AGT/GTA/TAG/ACT/CTA/TAC; 10 = AGG/GGA/GAG/ CCT/CTC/TCC; 11 = AGC/GCA/CAG/GCT/CTG/TGC; 12 = ACG/ CGA/GAC/CGT/GTC/TCG; 13 = ACC/CCA/CAC/GGT/GTG/TGG; 14 = GGC/GCG/CGG/GCC/CCG/CGC.
Two hundred and four primer pairs were designed to amplify flanking regions of P. triticina EST-SSRs. Seventy-six primer pairs produced amplification products with sizes consistent with initial primer designs. The failure in amplifications could be caused by the presence of introns, mutations, insertion or deletion in target regions which generate PCR products with excessive length or disrupt binding sites of PCR primers. In addition, the same annealing temperature was used in PCR during the initial screening process which might be only suitable for some primer pairs but not for others. Polymorphisms of these EST-SSRs were evaluated using 35 P. triticina isolates and 21 EST-SSRs were polymorphic among these isolates. The percentage of polymorphic EST-SSRs (10.2%) is lower than that reported in Puccinia genomic SSRs, which was in the range of 25–50% (Enjalbert et al., Citation2002; Szabo & Kolmer, Citation2007; Zhong et al., Citation2008). Two to four alleles were detected by EST-SSRs in P. triticina. The number of alleles detected by EST-SSRs is similar to those detected by genomic SSRs from Duan et al. (Citation2003) but lower than the number reported by Szabo & Kolmer (Citation2007), in which two to nine alleles per locus were detected by genomic SSRs from enriched P. triticina genomic libraries. Differences in the percentage of polymorphic loci and number of alleles per locus between EST-derived SSRs and genomic SSRs from enriched genomic libraries are likely due to the origin of these sequences, since sequences in coding regions are more conserved then sequences in non-coding regions in the fungal genome (Temnykh et al., Citation2000).
The majority of polymorphic di-nucleotide EST-SSRs contained ‘AG/GA/CT/TC’ motifs (class 2). Similar motifs were also found in genomic SSRs of P. triticina (Duan et al., Citation2003; Szabo & Kolmer, Citation2007). Most polymorphic tri-nucleotide EST-SSRs contained motifs from class 8, which were less common then class 6 and 10 motifs in P. triticina ESTs. Similar tri-nucleotide motifs were also common in polymorphic EST-SSRs found in P. graminis f. sp. tritici ESTs (Zhong et al., Citation2009). Studies on fungal SSRs have shown that most fungal genomes are dominated by AT rich motifs and highly iterated repeats often contain A and T bases (Lim et al., Citation2008). It is possible that the polymorphism of EST-SSRs in Puccinia species is not necessarily related to the abundance of certain motifs but more likely related to the nucleotide composition. Although SSRs with long motifs are usually ignored during the generation of genomic SSRs due to their rarity in the genome, one EST-SSR with tetra-nucleotide repeat motif was also polymorphic in our study, which suggested that they could also be useful for generating informative markers.
Since EST-SSRs are located within genes and tend to have lower variability values, such as the number of alleles, allele size and expected genetic diversity, than genomic SSRs (Cho et al., Citation2000), there is an obvious concern that they could be less polymorphic than genomic SSRs. Nevertheless, a recent study by Woodhead et al. (Citation2005) showed that the differentiation of population based on EST-SSRs were comparable to those based on genomic SSRs and AFLPs. In addition, a number of comparative studies also suggest that only a small percentage of genes are under the positional selection (Clark et al., Citation2003). In our study, the level of heterozygosity and deviation from Hardy–Weinberg equilibrium for EST-SSRs were analyzed on 35 P. triticina isolates using GENALEX6 (Peakall & Smouse, Citation2006) (). Seventeen EST-SSRs (80.9%) had significantly higher levels of observed heterozygosity than expected and 10 loci exhibited significant deviations from Hardy–Weinberg equilibrium corrected for multiple comparisons (P < 0.001). Given the fact that P. triticina isolates used in this study were generated from the asexual uredinial stage, these results were not surprising. Similar deviations from Hardy–Weinberg equilibrium were also found in asexual populations of P. triticina (Szabo & Kolmer, Citation2007) and P. psidii (Zhong et al., Citation2008) using genomic SSRs.
Table 2. Repeat motif, primer sequence, amplification conditions and characteristics
The structures in datasets derived from EST-SSRs and virulence were analyzed using MODECLUS () and UPGMA methods (, ). Both methods have been used previously in the population genetic study of several Puccinia species (Kolmer & Liu, Citation2000; Harder et al., Citation2001; Kolmer, Citation2001). When the dissimilarity matrix derived from EST-SSRs was analyzed with MODECLUS, the plot of K-value versus the number of clusters showed a large stable region (K = 7–17) at two clusters ( a). Subsequently, two clusters defined at the region of stability were analyzed in a 2-D PCA plot. P. triticina isolates that were avirulent on Lr2a and Lr2c, but virulent on Lr17a (group I) were separated from isolates that were avirulent on Lr17a (group II and group III) ( b). Similarly, two major groups of P. triticina isolates separated at 41% dissimilarity were found when the dissimilarity matrix derived from EST-SSRs was analyzed using the UPGMA method (). The first group contained P. triticina isolates that were avirulent on Lr2a and Lr2c but virulent on Lr17a (group I) and the second group included P. triticina isolates that were avirulent on Lr17a (group II and III). Similarly, Kolmer et al. (Citation1995; Kolmer, Citation2001) reported that the group of P. triticina isolates that was avirulent on Lr2a and Lr2c but virulent on Lr17a had molecular characteristics that were different from other P. triticina isolates in North America based on RAPD and AFLP analysis. It was proposed that this group of isolates could be recently introduced into North America, most likely from Mexico.
Fig. 2. The clustering structure in the data sets derived from virulence and EST-SSRs. a, Functions of K with the number of clusters for Puccinia triticina isolates based on virulence (□) and EST-SSR genotypes (▪) and b, two-dimensional depiction of clusters at K = 12 differentiated by virulence to 16 differential lines (□, ▵, ∘) and 21 EST-SSR markers (▪, ▴, •). Isolates were grouped based on the virulence to resistance gene Lr2a, Lr2c and Lr17a. Group I (□,▪) are avirulent to Lr2a and Lr2c but virulent to Lr17a. Group II (▵,▴) are avirulent to Lr2a, Lr2c and Lr17a. Group III (∘, •) are virulent to Lr2a and Lr2c but avirulent to Lr17a.
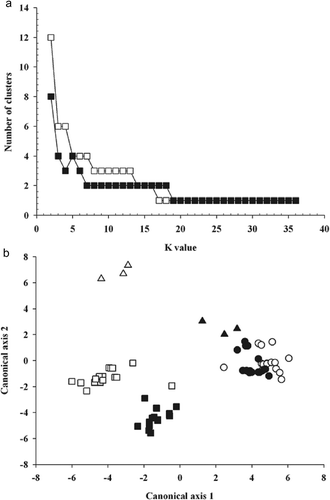
Fig. 3. Genetic dissimilarity dendrogram of 35 Puccinia triticina isolates collected in Canada based on the unweighted pair group method with arithmetic means clustering method using 1-Dice coefficient calculated from 21 EST-SSR markers. Numbers along the nodes are bootstrap values > 70% in 1000 replicas. The first four letters of isolate designation represent virulence phenotypes of P. triticina isolates based on 16 differential lines and the last letter represents the location where the isolate was collected. O, isolates collected in Ontario and Quebec; M, isolates collected in Manitoba and Saskatchewan; B, isolates collected in British Columbia and Alberta; P, isolates collected in Prince of Edward Island. Group I, II and III represent three groups of isolates clustered based on virulence to Lr2a, Lr2c and Lr17a.
Fig. 4. Virulence dissimilarity dendrogram of 35 Puccinia triticina isolates collected in Canada in 2007 based on the unweighted pair group method with arithmetic means clustering method using simple mismatch coefficient (1-simple match coefficient) calculated based on virulence on 16 leaf rust resistance genes. Numbers along the nodes are bootstrap values > 70% in 1000 replicas. The first four letters of isolate designation represent virulence phenotypes of P. triticina isolates based on 16 differential lines and the last letter represents the location where the isolate was collected. O, isolates collected in Ontario and Quebec; M, isolates collected in Manitoba and Saskatchewan; B, isolates collected in British Columbia and Alberta; P, isolates collected in Prince of Edward Island. Group I, II and III represent three groups of isolates clustered based on virulence to Lr2a, Lr2c and Lr17a.
In comparison, the dissimilarity matrix derived from virulence was subjected to MODECLUS analysis. A large region of stability were found at three clusters (K = 8–13) (). In the 2D-PCA plot, P. triticina isolates from group I (avirulent on Lr2a and Lr2c, virulent on Lr17a) remained separated from group II and group III, which were both avirulent on Lr17a (). However, the analysis of virulence dissimilarity matrix with the UPGMA method revealed two major groups of P. triticina isolates which were separated at 29% dissimilarity. Isolates in group I and group II were grouped together and were separated from isolates in group III (). Although this discrepancy in virulence and EST-SSR analysis could indicate that P. triticina virulence phenotypes with different genetic backgrounds were all under the selection of resistance genes deployed in the host population, we could not rule out the possibility that such discrepancy was merely caused by the limited P. triticina virulence phenotypes used in this study. More P. triticina virulence phenotypes, such as those avirulent on Lr2a but virulent on Lr2c and those avirulent on Lr2a, Lr2c and Lr17a, should be included to study the clustering of P. triticina virulence phenotypes in Canada.
To investigate the correlation between virulence and EST-SSR genotypes, the dissimilarity matrices from virulence and EST-SSRs were analyzed with MXCOMP module in NTSYS. A significant correlation was found (r = 0.685, P < 0.001) between the dissimilarity matrix derived from virulence and that from EST-SSRs. Dissimilarity matrices based on virulence to the single resistance gene were also compared with that from EST-SSRs. Virulence to Lr2a, 2c and Lr17a had the highest correlations with EST-SSR genotypes (0.636, 0.636 and 0.893, respectively; P < 0.001). A strong correlation between virulence and molecular phenotypes in P. triticina has been reported using RAPDs (Kolmer & Liu, Citation2000), AFLPs (Kolmer, Citation2001) and genomic SSRs (Kolmer & Ordonez, Citation2007). For example, Kolmer & Ordenez (Citation2007) found that virulence to Lr2a and Lr2c was associated with molecular phenotypes based on genomic SSR analysis. Since P. triticina population in North America is asexual in nature and comprised of a few genetic lineages which likely evolve over time, reflecting the selection of host resistance genes used. The non-random associations between virulence and molecular phenotypes in P. triticina could result from the asexual clonal reproduction and selection for virulence against resistance genes deployed in the hosts in North America (Kolmer, Citation2001). It is interesting that a strong correlation was found between EST-SSR genotypes and virulence to Lr17a in this study, while such correlation was not observed when genomic SSRs were used to analyze the population of P. triticina in central Asia and Caucasus (Kolmer & Ordonez, Citation2007). It is possible that the difference in the correlation between EST/genomic-derived SSR genotypes and virulence to Lr17a in P. triticina populations from two regions reflects the difference in the selection pressure from resistance genes deployed in those regions.
It has been suggested that EST-SSRs are more efficient than genomic SSRs in terms of transferability among related species (Dracatos et al., Citation2007). In our study, eight out of 21 pairs of primers produced amplification products from genomic DNA of P. graminis and P. coronata (). Three primer sets produced polymorphic products from single-pustule-derived genomic DNA samples of P. graminis and P. coronata (data not shown). These results indicate that EST-SSRs are capable of producing a high proportion of cross-species markers with distinct allele patterns in related species which give them one clear advantage over genomic SSRs.
In summary, EST-derived SSRs developed in this study are effective in differentiating P. triticina population despite the smaller number of alleles detected by EST-SSRs. While EST-SSRs are not without their drawbacks, EST-SSRs offer some advantages over genomic SSRs including rapid and inexpensive development and higher levels of cross-taxon portability.
Acknowledgements
The work described in this study was supported by Agriculture and Agri-Food Canada ‘Canadian Crop Genomics Initiative’ fund. The author is the recipient of a NSERC Visiting Fellowship in Government Laboratories. The author thanks Dr. Sylvie Cloutier for scientific advice and Travis Bank for the assistance with computer software.
References
- Becher , R. 2007 . EST-derived microsatellite as a rich source of molecular markers for oats . Plant Breed. , 126 : 274 – 278 .
- Chalhoub , B.A. , Thibault , S. , Laucou , V. , Rameau , C. , Hofte , H. and Cousin , R. 2007 . Silver staining and recovery of AFLP amplification products on large denaturing polyacrylamide gels . Biotechniques , 22 : 216 – 220 .
- Cho , Y.G. , Ishii , T. , Temnykh , S. , Chen , X. , Lipovich , L. , McCouch , S.R. , Park , W.D. , Ayres , N. and Cartinhour , S. 2000 . Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) . Theor. Appl. Genet. , 100 : 713 – 722 .
- Clark , A.G. , Glanowski , S. , Nielsen , R. , Thomas , P.D. , Kejariwal , A. and Todd , M.A. 2003 . Inferring non-neutral evolution from human-chimp-mouse orthologous gene trios . Science , 302 : 1960 – 1963 .
- Dracatos , P.M. , Dumsday , J.L. , Olle , R.S. , Cogan , N.O.I. , Dobrowolski , M.P. , Fujimori , M. , Roderick , H. , Stewart , A.V. , Smith , K.F. and Forster , J.W. 2007 . Development and characterization of EST-SSR markers for the crown rust pathogen of ryegrass (Puccinia coronata f.sp. lolii) . Genome , 49 : 572 – 583 .
- Duan , X. , Enjalbert , J. , Vautrin , S. , Solignac , M. and Giraud , T. 2003 . Isolation of 12 microsatellite loci, using an enrichment protocol, in the phytopathogenic fungus . Puccinia triticina. Mol. Ecol. Notes , 3 : 65 – 67 .
- Dutech , C. , Enjalbert , J. , Fournier , E. , Delmotte , F. , Barrcs , B. , Carlier , J. , Tharreau , D. and Giraud , T. 2007 . Challenges of microsatellite isolation in fungi . Fungal Genet. Biol. , 44 : 933 – 949 .
- Enjalbert , J. , Duan , T. , Giraud , D. , Vautrin , S. , Vallavieille-Pope De , C. and Solignac , M. 2002 . Isolation of twelve microsatellite loci, using an enrichment protocol, in the phytopathogenic fungus . Puccinia striiformis f.sp. tritici. Mol. Ecol. Notes , 2 : 563 – 565 .
- Harder , D.E. , Baum , B.R. and McCallum , B.D. 2001 . Virulence in populations of Puccinia graminis f.sp. tritici in Canada from 1952 to 1998: A non-parametric analysis . Can. J. Bot. , 79 : 556 – 569 .
- Holton , T.A. , Christopher , J.T. , McClure , L. , Harker , N. and Henry , R.J. 2002 . Identification and mapping of polymorphic SSR markers from expressed gene sequences of barley and wheat . Mol. Breed. , 9 : 63 – 71 .
- Hu , G.G. , Linning , R. , McCallum , B. , Banks , T. , Cloutier , S. , Butterfield , Y. , Liu , J. , Kirkpatrick , R. , Stott , J. , Yang , G. , Smailus , D. , Jones , S. , Marra , M. , Schein , J. and Bakkeren , G. 2007 . Generation of a wheat leaf rust, Puccinia triticina, EST database from stage-specific cDNA libraries . Mol. Plant Pathol. , 8 : 451 – 467 .
- Jong , E.v.Z.d , Spangenberg , G.C. and Forster , J.W. 2003 . Development and characterization of EST-derived simple sequence repeat (SSR) markers for pasture grass endophytes . Genome , 46 : 277 – 290 .
- Jurka , J. and Pethiyagoda , C. 1995 . Simple repetitive DNA sequences from primates: Complication and analysis . J. Mol. Evol. , 40 : 120 – 126 .
- Katti , M.V. , Ranjekar , P.K. and Gupta , V.S. 2001 . Differential distribution of simple sequence repeats in eukaryotic genome sequences . Mol. Biol. Evol. , 18 : 1161 – 1167 .
- Keiper , F.J. , Haque , M.S. , Hayden , M.J. and Park , R.F. 2006 . Genetic diversity in Australian populations of Puccinia graminis f. sp. avenae . Phytopathology , 96 : 96 – 104 .
- Kolmer , J.A. 2001 . Molecular polymorphism and virulence phenotypes of the wheat leaf rust fungus Puccinia triticina in Canada . Can. J. Bot. , 79 : 917 – 926 .
- Kolmer , J.A. and Liu , J.Q. 2000 . Virulence and molecular polymorphism in international collections of the wheat leaf rust fungus Puccinia triticina . Phytopathology , 90 : 427 – 436 .
- Kolmer , J.A. , Liu , J.Q. and Sies , M. 1995 . Virulence and molecular polymorphism in Puccinia recondita f. sp. tritici in Canada . Phytopathology , 85 : 276 – 285 .
- Kolmer , J.A. , Long , D.L. and Hughes , M.E. 2005 . Physiological specialization of Puccinia triticina on wheat in the United States in 2003 . Plant Dis. , 89 : 1201 – 1206 .
- Kolmer , J.A. and Ordonez , M.E. 2007 . Genetic differentiation of Puccinia triticina populations in Central Asia and the Caucasus . Phytopathology , 97 : 1141 – 1149 .
- Kosman , E. , Pardes , E. , Anikster , Y. , Manisterski , J. , Yehuda , P.B. , Szabo , L.J. and Sharon , A. 2004 . Genetic variation and virulence on Lr26 in Puccinia triticina . Phytopathology , 94 : 632 – 640 .
- Lim , S. , Notley-McRobb , L. , Lim , M. and Carter , D.A. 2008 . A comparison of the nature and abundance of microsatellites in 14 fungal genomes . Fungal Genet. Biol. , 41 : 1025 – 1036 .
- Long , D.L. and Kolmer , A.J. 1989 . A North American system of nomenclature for Puccinia recondita f. sp. tritici . Phytopathology , 79 : 525 – 529 .
- McCallum , B. and Seto-Goh , P. 2004 . Physiological specialization of wheat leaf rust (Puccinia triticna) in Canada in 2001 . Can. J. Plant Pathol. , 26 : 109 – 129 .
- McCallum , B. and Seto-Goh , P. 2005 . Physiologic specialization of wheat leaf rust (Puccinia triticina) in Canada in 2002 . Can. J. Plant Pathol. , 27 : 90 – 99 .
- Mebrate , S.A. , Dehne , H.W. , Pillen , K. and Oerke , E.C. 2006 . Molecular diversity in Puccinia triticina isolates from Ethiopia and Germany . J. Phytopathol. , 154 : 701 – 710 .
- Metzgar , D. , Bytof , J. and Wills , C. 2008 . Selection against frame shift mutations limits microsatellite expansion in coding DNA . Genome Res. , 18 : 72 – 80 .
- Morgante , M. , Hanafey , M. and Powell , W. 2002 . Microsatellites are preferentially associated with non-repetitive DNA in plant genomes . Nat. Genet. , 30 : 194 – 200 .
- Peakall , R. and Smouse , P.E. 2006 . GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research . Mol. Ecol. Notes , 6 : 288 – 295 .
- Rozen , S. and Skaletsky , H.J. 2000 . “ Primer3 on the WWW for the general users, biologists and programmers ” . In Bioinformatics methods and protocols: Methods in molecular biology , Edited by: Krawetz , S. and Misener , S. 365 – 386 . Totow, NJ : Humana Press .
- Samborski , D.J. 1985 . “ Wheat leaf rust ” . In The cereal rusts , Edited by: Roelfs , A.P. and Bushnell , W.R. Vol. 2 , 39 – 59 . Orlando, FL : Academic Press .
- Squirrell , J. , Hollingsworth , P.M. , Woodhead , M. , Russell , J. , Lowe , A.J. , Gibby , M. and Powell , W. 2003 . How much effort is required to isolate nuclear microsatellites from plants? . Mol. Ecol. , 12 : 1339 – 1348 .
- St-Laurent , L. , Baum , B.R , Akpagana , K. and Arnason , J.T. 2000 . A numerical taxonomic study of Trema (Ulmaceae) from Togo, West Africa . Syst. Bot. , 25 : 399 – 413 .
- Szabo , L.J. and Kolmer , J.A. 2007 . Development of simple sequence repeat markers for the plant pathogenic rust fungus Puccinia triticina . Mol. Ecol. Notes , 7 : 708 – 710 .
- Temnykh , S. , Park , W.D. , Ayres , N. , Cartinhour , S. , Hauck , N. , Lipovich , L. , Cho , Y.G. , Ishii , T. and McCouch , S.R. 2000 . Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.) . Theor. Appl. Genet. , 100 : 697 – 712 .
- Woodhead , M. , Russell , J. , Squirrell , J. , Hollingsworth , P.M. and Gibby , M. 2005 . Comparative analysis of population genetic structure in Athyrium distentifolium (Pterdophyta) using AFLPs and SSRs from anonymous and transcribed gene regions . Mol. Ecol. , 14 : 1681 – 1695 .
- Zane , L. , Bargelloni , L. and Patarnello , T. 2002 . Strategies for microsatellite isolation: A review . Mol. Ecol. , 11 : 1 – 16 .
- Zhong , S.B. , Leng , T.L. , Friesen , J.D. , Faris , J.D. and Szabo , L.J. 2009 . Development and characterization of expressed sequence tag-derived microsatellite markers for the wheat stem rust fungus Puccinia graminis f.sp. tritici . Phytopathology , 99 : 282 – 289 .
- Zhong , S.B. , Yang , B.J. and Alfenas , A.C. 2008 . Development of microsatellite markers for the guava rust fungus, Puccinia psidii . Mol. Ecol. Notes , 8 : 348 – 350 .
- Zhou , Y. , Bui , T. , Auckland , L.D. and Illiams , C.G. 2001 . Direct fluorescent primers are superior to M13-tailed primers for Pinus taeda microsatellites . Biotechniques , 31 : 24 – 28 .