Abstract
Economically, legumes (Fabaceae) represent the second most important family of crop plants after the grass family, Poaceae. Grain legumes account for 27% of world crop production and provide 33% of the dietary protein consumed by humans, while pasture and forage legumes provide vital part of animal feed. Fabaceae, the third largest family of flowering plants, has traditionally been divided into the following three subfamilies: Caesalpinioideae, Mimosoideae, and Papilionoideae, all together with 800 genera and 20,000 species. The latter subfamily contains most of the major cultivated food and feed crops. Among the grain legumes are some of mankind's earliest crop plants, whose domestication parallelled that of cereals: Soybean in China; faba bean, lentil, chickpea and pea in the Fertile Crescent of the Near East; cowpeas and bambara groundnut in Africa; soybean and mungbeans in East Asia; pigeonpea and the grams in South Asia; and common bean, lima bean, scarlet runner bean, tepary bean and lupin in Central and South America. The importance of legumes is evidenced by their high representation in ex situ germplasm collections, with more than 1,000,000 accessions worldwide. A detailed knowledge of the phylogenetic relationships of the Fabaceae is essential for understanding the origin and diversification of this economically and ecologically important family of angiosperms. This review aims to combine the phylogenetic and genetic diversity approaches to better illustrate the origin, domestication history and preserved germplasm of major legume crops from 13 genera of six tribes and to indicate further potential both for science and agriculture.
I. INTRODUCTION
Fabaceae (Leguminosae), with 800 genera and 20,000 species (Lewis et al., Citation2005), is the third largest family of flowering plants, after the Orchidaceae and Asteraceae. It is an extremely diverse family with worldwide distribution, encompassing a broad range of life forms, from arctic alpine herbs and temperate or tropical perennial shrubs to annual xerophytes and equatorial giant trees. Some legumes are weeds of cereal agriculture, while others are major grain crops in their own right. These latter species are known as grain legumes, or pulses, and together with two pasture and forage legumes are the focus of this review. Members of the Fabaceae are characterized by the distinct fruit, termed a legume, which gives the family its original name. Flower structure is highly variable; however, the butterfly-like (papilionoid) flower is almost universal in the Papilionoideae subfamily (∼14,000 species). Fabaceae includes many economically important and versatile species, with the majority providing grains and pulses. Among the grain legumes are some of humanity's earliest crop plants, including soybean and mungbean in East Asia; faba bean, lentil, chickpea and pea in the Fertile Crescent of the Near East; and common bean or lupin in Central and South America. Legumes’ symbiosis with nitrogen-fixing bacteria provides not only added value in agriculture, but also plays an important role in natural ecosystems. Moreover, the legume species Pisum sativum L., pea, was the key experimental organism for Mendel's pioneering work (1866) in establishing the underlying basis of heredity (Smýkal, 2014).
Reconstructing the phylogenetic relationship of the Fabaceae is essential to understanding the origin and diversification of this family. Phylogenetic analyses of Fabaceae began with the plastid rbcL gene (Doyle, Citation1995; Kass and Wink, Citation1997), followed by analyses including the more variable matK gene (Wojciechowski et al., Citation2004; reviewed in Lewis et al., Citation2005). Both are now accepted as barcoding regions for plants (CBOL, Citation2009). The picture is far from complete, however, as many species have not yet been sequenced or are represented by just one or two accessions. Nonetheless, for chickpea, common bean, cowpea and soybean, as well as for the model legumes Medicago truncatula Gaertn. and Lotus japonica (Regel) K. Larson, rapid progress has been made. The monophyly of the family has been repeatedly demonstrated through molecular systematics (Doyle et al., 1995; Kass and Wink, Citation1997; Wojciechowski, Citation2003). Currently, based on morphological characters, the following three major groups are recognized and regarded as subfamilies: The mimosoid legumes, Mimosoideae (sometimes regarded as family Mimosaceae with four tribes and 3,270 species); the papilionoid legumes, Papilionoideae (or family Fabaceae/Papilionaceae with 28 tribes and 13,800 species); and the caesalpinioid legumes, Caesalpinoideae (or family Caesalpiniaceae with four tribes and 2,250 species) (Lewis et al., Citation2005). Estimates for the date of origin and early evolution of the legumes vary, but a rich Eocene macrofossil record shows that some lineages of the family existed by around 50 million years ago (Mya). The earliest known legume pollen remains date back to about 60-75 Mya (Lavin et al., Citation2005; Wojciechowski, Citation2003), predating the macrofossils.
Papilionoideae is a monophyletic group, according to all recent phylogenetic analyses, making it by far the largest subfamily, with 476 genera and about 14,000 species. It is estimated that all papilionoids shared a common ancestor, which experienced a 50 kb inversion in its chloroplast genome (Doyle et al., 1995; Lavin et al., Citation2005), around 50 Mya. The largest group of papilionoids is Hologalegina, with nearly 4,000 species in 75 genera, including the large galegoid tribes (Galegeae, Fabeae, Trifolieae, Genisteae, etc.), united by the loss of one copy of the chloroplast inverted repeat (IR), often referred as the IR-loss clade. Of great economic and scientific interest are the Fabeae and Trifolieae, which together comprise 11 genera and nearly 800 species. The tribe Genisteae belongs to the basal Genistoid clade, which diverged early in the evolution of the Papilionoid legumes (Lavin et al., Citation2005). Like other Genistoid legumes, Genisteae species synthesize quinolizidine alkaloids, bitter compounds that provide a defense against pathogens and predators (Bunsupa et al., Citation2013; Wink and Mohamed, Citation2003).
When cultivated grain legumes, or pulses, are considered, the Papilionoideae can be divided into the following four clades (): (1) Phaseoloids (Glycine Willd., Phaseolus L., Cajanus L. and Vigna Savi), (2) Galegoids (Pisum L., Lens Mill., Lathyrus L., Vicia L., Medicago L. and Cicer L.), (3) Genistoids (Lupinus L.) and (4) Dalbergoids (Arachis L. and Stylosanthes Sw.) (Lewis et al., Citation2005). Phaseoloids are pan-tropical and often referred to as “warm season,” “tropical” or “millettioid” clade. By contrast, Galegoids are often referred to as “cool-season,” “temperate” or “Hologalegina” legume crops, since they are mainly distributed in temperate regions of the world, such as Europe and the Mediterranean.
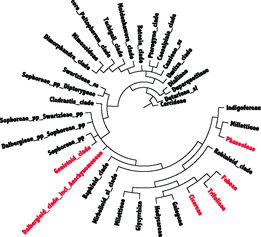
The Mimosoideae and Caesalpinoideae are mostly woody trees and shrubs. Many are valuable for timber (Acacia spp. Mill., Albizia Benth., Dalbergia L.f.), dyes (Indigofera tinctoria L., Haematoxylon campechianum L., Caesalpina brasiliensis L.), tannins (Acacia dealbata Link., A. decurrens Desv.), resins (Trachylobium verrucosum (Gaertn.) Oliv), gums (Senegalia senegal Britton), insecticides (Derris elliptica (Wall.) Benth), medicines (Cassia alata (L.) Roxb., Senna occidentalis (L.) Link.), food (Tamarindus indicus L., Ceratonia siliqua L., Leucaena esculenta Sesse & Moc. Ex DC.) Benth.) and animal fodder (Bauhinia spp. L.).
This review aims to combine the phylogenetic and genetic diversity approaches to better illustrate the origin, domestication history and preserved germplasm of major legume crops from 13 genera of six tribes () and to indicate further potential in both science and agriculture.
TABLE 1 List of taxonomical species, and current number of germplasm stored accessions of major legume crops and their gene pools
A. Genetic Diversity Conserved in Ex Situ Germplasm Collections and Its Characterization
Ex situ conservation was pioneered by N.I. Vavilov in 1926. Currently, 1,750 germplasm collections hold over 7 million crop plant accessions worldwide (FAO, Citation2010). Legumes (grain and forage) constitute the second largest group (1,041,345 accessions, 15% of all [FAO, Citation2010]) after cereals. The study of genetic diversity for both germplasm management and breeding has received much attention, especially following the introduction of the core collection concept by Frankel and Brown (Citation1984). However, in practice, even the core collection approach did not help to fully characterize genetic diversity or use it in breeding.
The conserved germplasm is characterized for distinct morpho-agronomic traits, using sets of crop-specific descriptors. Approximately 78% of the 146,837 grain legume germplasm accessions held in CGIAR centers have been characterized for morphological traits, including for resistance to biotic and abiotic stresses. However, only a small percentage of these collections have been characterized for biochemical traits. More emphasis and funding are needed in order to generate data on biochemical characteristics associated with biotic and abiotic stresses (Upadhyaya et al., Citation2011). The Global Crop Diversity Trust (GCDT) supports the development of global crops (including legumes) and regional strategies for ex situ conservation and utilization of crop diversity. These strategies represent a major investment in the field of plant genetic resources (PGR), mobilizing experts to collaboratively plan for the more efficient and effective conservation and the use of crop diversity. The themes viewed under these strategies include regeneration, crop wild relatives (CWR), collecting, crop descriptors, information systems, user priorities, new technologies and research, and challenges to building a strategy for rational conservation (Khoury et al., Citation2010). There is an urgency to ensure that the diversity in landraces is sampled and conserved in ex situ genebanks, especially as farming evolves from subsistence to a market-orientated endeavor dominated by modern cultivars and resulting in the erosion of crop genetic diversity. A similar caution also applies to the in situ populations of wild relatives, as land use intensifies with urban expansion and climate change threatens ecology (Snook et al., Citation2011). A further development in targeting germplasm for key traits is to use a GPS map of landrace collection sites, which can be overlaid with climate data corresponding to vegetative and reproductive growth stages, and to identify landraces corresponding to sites with severe abiotic stresses during the previous 25 years. The landraces encompass much of the original diversity, plus accumulated mutations and genetic recombinations since domestication. These landraces are now disappearing, replaced by improved, higher yielding modern cultivars from modern breeding programs, as exemplified on recent collecting missions in China (Bao et al., Citation2008; He at al., 2008). On the premise that such landraces may have had natural selection for tolerances to these stresses, Li Ling et al. (Citation2013) identified pea landraces from China as priority candidates to evaluate for tolerance of reproductive frost stress, reproductive heat stress, and drought tolerance.
Adoption of the Germplasm Resource Information Network (GRIN-Global) database by international centers and national genebanks will allow for online queries across multiple genebanks for client-selected accessions, including multi-trait queries.
With a wide range of approaches now available for genotyping, and with the declining cost of whole genome sequencing, the greatest limitation for gene banks is precise phenotyping, not only for descriptive traits, but also for agriculturally relevant quantitative traits relating to the expression of yield, crop growth and disease resistance. To increase precision, a single seed should be used for self-pollination to provide genetically uniform progeny for genotypic and phenotypic analysis. This level of precision is desirable if the key alleles of genes for important agronomic traits are to be identified, but broad characterization of diversity in germplasm can be based on a pooled DNA sample and phenotyping done on the bulked landrace mixture. Multi-environment analysis of quantitative variation involving multi-trait evaluation is far more informative than a single-environment trial and potentially provides some prediction for performance in other environments (Redden et al., 2012). The challenge for gene bank curators is to strategically sample collections and maximize information from costly evaluation trials. One approach is to use core collections, geographically sub-sampled or sampled using molecular marker diversity to characterize species diversity, or to sample based on priority traits. This has led to the use of climatic site descriptors for characterization of natural selection, focusing on abiotic stress response, and has therefore provided lists of prospective germplasm with potential tolerances to heat, frost, and drought stresses (Li et al., Citation2012, Citation2013). With current advances in genotyping and phenotyping methods, it is possible to effectively mine and explore the diversity stored in germplasm collections (McCouch, Citation2013).
B. Botanical Gardens and Herbariums as Sources of Information and Complementary Conservations
In addition to gene banks, botanical gardens offer an ex situ alternative to seed conservation. Botanical gardens face both challenges and opportunities in responding to global trends and, in particular, to climate change. The increased number of species at risk as a result of the changing climatic conditions will force many botanical gardens to refocus, to strengthen their conservation policies and to increase their participation in recovery programs for critically endangered species, including crop wild relatives (CWR). In addition, botanical gardens face an unprecedented opportunity to develop their role as introduction centers and play a major role in the assessment of new germplasm, both of ornamentals as well as other economically important plants (Heywood, Citation2011). Historically, botanical gardens have played a major role in plant introduction with far-reaching impacts and have been major drivers in human population growth and economic development of crops. Many botanical gardens manage seed banks of horticultural and wild species (such as the Millennium Seed Bank managed by Royal Botanic Garden at Kew, UK), have well-curated herbarium collections, are involved in re-introduction programs, and contain DNA storage facilities (DNA banks). Although herbarium and DNA banks are of relatively little practical use to conserve diversity, both provide valuable resources for studying CWR genetic diversity and information that can be used in gap analysis, as in the case of Phaseolus (Ramírez-Villegas et al., Citation2010) and Lathyrus (Shehadeh et al., Citation2013). Moreover, digitalization of and public access to herbarium vouchers allows for the remote study of morphological traits. These institutions often have the most direct knowledge and access to existing genetic diversity preserved in situ. Unfortunately, there is often an information gap between gene banks, botanical gardens and universities, which needs to be overcome in the near future by means of workshops, conferences and informal meetings.
C. In Situ Conservation of Crop Wild Relatives
In situ genetic reserve conservation may be defined as “the location, designation, management and monitoring of genetic diversity in natural wild populations within defined areas designated for active, long-term conservation” (Maxted et al., Citation1997). A genetic reserve is actively managed, even if the management involves only regular monitoring of the target CWR taxa; as long as the target population levels are maintained above the minimum viable population of approximately 5,000 individuals no further conservation action may be required (Dulloo et al., Citation2008). Importantly, in situ conservation action is a long-term commitment because significant resources have to be invested in order to establish a genetic reserve. Although the conservation goal is to always implement complementary conservation involving the parallel application of in situ and ex situ conservation techniques, there exists a preference for in situ conservation, primarily due to the overall need to maintain ecosystem health, but also because it has the advantage of maintaining the dynamic evolution of the CWR diversity itself in relation to parallel biotic and abiotic changes. Furthermore, due to the sheer number of CWR involved, the need to maintain effective genetic representation and the difficulty in precisely identifying which CWR or traits are required by plant breeders currently and in the future, in situ conservation is highly recommended, even if the main access route for breeders to diversity is via backup in situ samples deposited in ex situ genebanks.
All species in protected areas are passively conserved if the entire ecosystem or habitat is stable; however, without monitoring and active management, the genetic diversity within and between individual CWR populations could be eroded, and entire populations could even go extinct. Nonetheless, Stolten et al. (2006) emphasize that many protected areas already play an important role in the conservation of CWR species, even though many managers may be unaware that the land under their stewardship contains important crop genetic diversity. However, if our goal is to conserve the maximum genetic diversity within CWR taxa, then we need to study and monitor the genetic diversity and natural dynamics of CWR populations; otherwise, our efforts in establishing protected areas for these taxa may be wasted. It should also be noted that the in situ management of CWR may differ significantly from that required for more traditional protected areas whose objective is to sustain climax communities. For example, CWR of major crop plants are often located in pre-climax communities (Lathyrus ervoides Grande, Lens orientalis Popow, Cicer bijugum Rech. f.) where the site management is comparatively intense, or the CWR may be closely associated with traditional farming practices (Vicia johannis Tamamschjan, Lathyrus cicera L., Pisum sativum subsp. elatius Asch. & Graebn.), in which case, genetic reserve management would need to be associated with maintenance of the traditional farming/ranching system (Lawn, 2014). Detailed guidelines on how to undertake in situ CWR conservation are provided by Iriondo et al. (Citation2008); minimum standards for managing CWR genetic reserves are provided by Iriondo et al. (Citation2012).
Specifically, there has been very limited effort to conserve legume CWR diversity in situ. This has in part been due to two related disconnects: (a) the disconnect between academic studies identifying where genetic reserves or less formal in situ management activities should be established and their actual implementation, and (b) lack of collaboration between the plant genetic resource and protected area communities (Meilleur and Hodgkin, Citation2004; Maxted and Kell, Citation2009). Consequently, native legume populations are susceptible to genetic erosion or even extinction (Maxted and Bennett, Citation2001). What was potentially the first recomendation for the establishment of in situ genetic reserves for legume CWR diversity was made by Maxted (Citation1995), who proposed four locations to conserve Fabeae species in Syria and Turkey. Subsequently, three reserves were established within the Global Environment Facility project in Turkey, one of which, Ceylanpinar (Tan, Citation1998; Tan and Tan, Citation2002), emphasizes legume (and cereal) CWR in situ conservation as a priority. Within Syria, one of the sites recommended by Maxted (Citation1995) has been established for in situ legume conservation in Suweida province (Amri et al., Citation2008a, b). Further genetic reserves to conserve legume CWR have been established for Lathyrus grimesii Barneby in Nevada, USA (Hannan and Hellier, in Pavek and Garvey, 1999); for Vavilovia formosa (Stev.) Fed. at Akna Lich, on the Geghama mountain ridge, Yerevan province, Armenia and other legumes within the Erebuni Reserve near Yerevan, Armenia (Avagyan, Citation2008); and for wild bean populations (Phaseolus spp.) in Costa Rica (Baudoin et al., Citation2008). However, admittedly none of these genetic reserves to date meets the minimum standards for managing CWR genetic reserves proposed by Iriondo et al. (Citation2012), though the in situ conservation now in place is an important step forward.
Wild soybean (Glycine soja Willd.) is presumed to share a common ancestor with cultivated soybean (Hymowitz, Citation1970). Apart from ex situ conservation, in situ strategy is also used to conserve wild soybean, since populations of G. soja typically show high levels of genetic heterogeneity. In China, more than 40 in situ conservation sites located in 15 provinces and regions have been established (Zhao et al., Citation2009). Their genetic diversity is identified by genotyping 40 individuals at 20 SSR marker loci for each population, and the results showed that at least 90% of the total genetic diversity was present (Guan et al., Citation2006; Zhao et al., Citation2006).
There have also been a number of gap analysis studies that have proposed where in situ genetic reserves might be sited. Gap analysis (Maxted et al., Citation2008) involves four steps: (a) identify priority taxa; (b) identify genetic (or ecogeographic as a proxy for genetic) diversity and complementary hotspots using distribution and environmental data; (c) match current in situ and ex situ conservation actions with the identified genetic (or ecogeographic) diversity and complementary hotspots to identify the so-called ‘gaps;’ and (d) formulate revised in situ and ex situ conservation actions derived from identification of the gaps. This methodology has been applied for several legume CWR groups, including vetch Vicia subgenus Vicia (Maxted, Citation1995), lentils Lens (Ferguson et al., Citation1998), Asiatic Vigna (Tomooka et al., Citation2002), African Vigna species (Maxted et al., Citation2004), perennial Medicago (Bennettet al., 2006), 14 (including garden pea, faba bean and cowpea) globally important food crop gene pools (Maxted and Kell, Citation2009), Medicago of the Mediterranean Basin (Al-Atawneh et al., Citation2009), Phaseolus species (Ramírez-Villegas et al., Citation2010), Medicago species in the Former Soviet Union (Greene et al., Citation2012), wild Glycine in Australia (Gonzalez-Orozco et al., 2012) and Lathyrus species (Shehadeh et al., Citation2013). However, in terms of establishing in situ conservation priorities, it is of greater practical value and is more cost efficient to establish multi-genepool conservation targets irrespective of individual genepool results. This multi-genepool approach has recently been used by Maxted et al. (Citation2012) for the temperate legume genera Cicer, Lathyrus, Lens, Medicago, Pisum and Vicia species. This involved the collation of 200,281 unique geo-referenced records (Cicer - 452, Lathyrus - 61,081, Lens - 672, Medicago - 42,248, Pisum - 728 and Vicia - 95,100) collected between 1884 and 2008. The analysis identified the western Fertile Crescent (South-Central Turkey, western Syria and northeast Lebanon) as the area in which to focus in situ conservation efforts. The highest concentration of all priority species, and therefore the most species-rich hotspot, is in the north of the Bekaa valley in Lebanon and the adjoining Tel Kalakh region in Homs province, Syria, but there is currently no in situ conservation in this area, even though it has been shown to be suffering extensive genetic erosion (Keiša et al., 2007). Undertaking similar multi-crop genepool analysis based perhaps on the legume species found in each of the Vavilov Centers should be a globally important priority. Once in situ locations are identified, they should be implemented to help improve global food security. New initiatives led by the Global Crop Diversity Trust (GCDT) (together with the Millenium Seed Bank, Royal Botanic Gardens, Kew) (Guarino and Lobell, Citation2011 and www.cwrdiversity.com) and the Food and Agriculture Organisation of the UN (FAO, Citation2013) are attempting to systematically plan and implement effective conservation of global CWR diversity, with the GCDT project focusing on ex situ conservation and the Food and Agriculture Organisation focusing on in situ conservation, with both projects promoting the use of conserved CWR diversity. The foundation of both projects is an annotated inventory of global priority CWR taxa for 173 priority crops, the Harlan and de Wet inventory (www.cwrdiversity.org/checklist/). Within the inventory, the family with the most CWR is the Fabaceae, with 253 global priority CWR from the genera Arachis, Cajanus, Cicer, Glycine, Lablab Adans., Lathyrus, Lens, Lupinus, Medicago, Phaseolus, Pisum, Vicia and Vigna. The GCDT ex situ project has collated over 8 million unique geo-referenced records for the ex situ gap analysis. There is a now an urgent priority to undertake the complementary in situ gap analysis for the legume taxa in order to identify globally where in situ conservation is required.
II. DOMESTICATION OF LEGUMES
Members of the Fabaceae family were domesticated as grain legumes in conjunction with the domestication of grasses for cereals (De Candolle, Citation1884; Vavilov, Citation1951; Smartt, Citation1990; Zohary and Hopf, Citation2000; Abbo et al., Citation2012). However, more legumes were domesticated overall, resulting in Fabaceae becoming the family to contain the largest number of domesticates. Pea, faba bean, lentil, grass pea and chickpea are some of the world's oldest domesticated crops and arose in the Fertile Crescent of Mesopotamian agriculture. These legumes accompanied cereal production and formed important dietary components of early civilizations in the Middle East and the Mediterranean.
Archaeological evidence dates the existence of pea back to 10,000 BC in the Near East (Baldev, Citation1988; Zohary and Hopf, Citation2000) and Central Asia (Riehl et al., Citation2013). In Europe, pea has been cultivated since the Stone and Bronze Ages and in India from 200 BC (De Candolle, Citation1884). Cultivation of pea spread from the Fertile Crescent into today's Russia, and westwards along the Danube valley into Europe and/or to ancient Greece and Rome, which further facilitated its spread to northern and western Europe. In parallel, pea cultivation moved eastward to Persia, India and China (Makesheva, 1979; Chimwamurombe and Khulbe, Citation2011).
Like pea, faba bean is an historically important crop. Faba bean remains have been found in archeological sites at Tell-el-Kerkh in northwest Syria, indicating that faba bean originated during the 10th millenium BC (Tanno and Wilcox, Citation2006). More recent, large-seeded, major type faba bean remains from the Mediterranean basin have been dated to the 2nd to 3rd millennia BC (Cubero, Citation1973) and likely represent a secondary center of domestication (Muratova, Citation1931), which was followed by their further spread into Europe. From their primary center in southwestern Asia, faba bean probably spread to Ethiopia. Introduction to South America in the 15th century has resulted in Peruvian and Bolivian faba bean landraces displaying a wide range of seed trait variability (Duc et al., Citation2010).
Lentil is closely associated with wheat and barley cultivation in the Near East and is regarded as a founder crop of Old World Neolithic agriculture (Zohary and Hopf, Citation1973). Carbonized lentil seeds were retrieved from pre-farming (9,200-7,500 BC) Mureybit and Tell Abu Hureyra in Syria and from Netic Hagdud in Israel (cited in Zohary and Hopf, Citation2000). Charred lentil seeds dating to the 8th and 7th millennia BC were found in most of the Pre-Pottery Neolithic B early farming villages in the Near East. In later Neolithic settlements, lentil seeds were larger than 4 mm in diameter, indicating advanced domestication. In the 6th and 5th millennia, lentil spread into southeast Europe and later into Central Europe. Lentil accompanied wheat and barley in their spread southwards to Egypt and eastwards along the Caspian Sea to India. Charred lentil seeds were found in Afghanistan and dated to 2000 BC. However, archeological remains of lentils do not provide conclusive evidence of lentil's domestication, as the only indicative trait is the increase in seed size, which was slow and gradual (Zohary and Hopf, Citation2000).
The earliest archaeological evidence of grasspea (Lathyrus sativus) comes from Jarmo in Iraqi Kurdistan and is dated to 8000 BC. Remains of Lathyrus species have also been found at Ali Kosh (9500-7600 BC) and Tepe Sadz (7500-5700 BC) in Iran and are among the most common foods recorded at these sites (Jackson and Yunus, Citation1984). At Azmaska Moghila, in Bulgaria, remains dated at ca. 7000 BC have been tentatively identified as L. cicera L. (Renfrew, Citation1969). The species L. sativus is probably a derivative from the genetically closest species, L. cicera (Hopf, Citation1986). This somewhat smaller-seeded grasspea grows in countries from Greece to Iran and Transcaucasia. Remains of L. sativus have also been reported in India and have been dated back to 2000-1500 BC by Saraswat (Citation1980) who indicated the possibility of diffusion of the crop from West Asia. Vicia faba L. and V. ervilia (L.) Willd. were already used by Neolithic and Bronze Age cultures in the eastern Mediterranean and in Asia Minor (Zohary and Hopf, 1973).
In contrast to the other crops domesticated during the Neolithic period, chickpea has followed a distinct evolutionary path, a series of bottlenecks from its narrow origin as a southeast Anatolian winter annual (Cicer reticulatum Ladiz.) to its current status as a South Asian and spring-sown Mediterranean crop (van Maesen, 1987; Abbo et al., 2003). The earliest archeological remains of chickpea (10th millennium BP) were discovered within (Pasternak, Citation1998; van Zeist and de Roller, Citation1991, Citation1992) or close (Tanno and Willcox, Citation2006) to the current distribution of C. reticulatum in south-east Anatolia (Berger et al., Citation2003). Thereafter, chickpea spread throughout the Eastern Mediterranean, presumably as a winter annual, like its wild progenitor, and was spread throughout the Mediterranean basin by the Greeks, Romans and Phoenicians. More recent chickpea remains are scarce, re-emerging only in Bronze Age sites in South Asia and in a much reduced, more southern Mediterranean distribution (Berger, Citation2013; Redden and Berger, Citation2007). Chickpea appeared in Ethiopia during the Iron Age (Dombrowski, Citation1970). The Spanish and Portuguese brought chickpea to the New World in the 16th century, while kabuli types were brought to India through Central Asia via the Silk Route in the 18th century (see references in Redden and Berger, Citation2007). Following the early Mediterranean change from autumn- to spring-sowing, and concomitant movement to warmer climates to the south and southeast (Africa and South Asia), chickpea escaped low winter temperatures both in time and space (Berger, Citation2013). This evolutionary trajectory had important ramifications on chickpea lineage's capacity to deal with biotic and abiotic stresses.
The warm-season legumes of the Phaseolid group have been domesticated somewhat later than the cool-season legumes. Common bean in the Americas probably has the longest history as a domesticate, originating in parallel in two separate centers of domestication, one in the Andean mountains of South America, giving rise to the Andean genepool, and one in the Central American highlands and lowlands, giving rise to the Mesoamerican (Middle American) genepool (Blair et al., Citation2009). Early archeological remains in caves of the Ayacucho and Guerrero regions of Peru and Mexico, respectively, suggest that domestication could have occurred as early as 10,000 years ago in the Andes and around 7,500 years ago in Central America. Four other related cultivated species in the genus Phaseolus were probably domesticated at a later date, as indicated by the lack of archeological records. Among these Phaseolus species, tepary bean (P. acutifolius A. Gray) was probably domesticated once or twice near the Mexico-USA border from wild populations of the same species, including P. acutifolius var. tenuifolius A. Gray, suffering a large bottleneck in the process (Blair et al., Citation2012a). Some studies suggest that lima bean domestication may be similarly as old as common bean and occurred in parallel but over a broader region, including Central America and the Caribbean, all the way to the Amazon, Andes and Peruvian coast. This wide geographic span led to the creation of at least two genepools, again classified as Andean and Mesoamerican, but with four subgroups based on grain type. A closer relative to common bean and of more recent origin, the scarlet runner bean (P. coccineus L.) was domesticated exclusively in Central America and may have crossed naturally with common bean, resulting in the intermediate year-long bean (P. dumosus Macfad.).
The domestication of various Vigna species occurred over a wide range of Old World centers of domestication and additional regions not widely considered in crop history. The most important of these species is cowpea (V. unguiculata (L.) Walp., which was domesticated in the Sahel region of West Africa with influences from a large group of wild relatives found from West to East and Southern Africa, all the way to current Botswana. The oldest evidence that cowpea existed in West Africa was obtained from carbon dating specimens from the Kimtampo rock shelter in central Ghana (Flight, Citation1976). A minor relative of the cowpea was domesticated for its underground pods and is commonly known as Bambara groundnut (Vigna subterranea (L.) Verdc). This species was also domesticated in Africa, but its exact origin is unclear.
Meanwhile, in Asia, a range of important Vigna grain legumes was domesticated. These include mungbean (V. radiata (L.) R. Wilczek) and the grams (V. mungo (L.) Hepper), from South and East Asia, respectively. Vigna have been domesticated in an arc from the Indian subcontinent to the Far East (Smartt, Citation1990). Remains of Asian Vigna dating to 3500 to 3000 BC were found in archeological sites at Navdatoli in Central India (Jain and Mehra, Citation1980). However, the domestication dates of other Vigna crops, especially those from Africa, are largely unknown due to a lack of research and the tropical climates, which create poor conditions for preservation of archeological remains.
For pigeonpea (Cajanus cajan L.), historical evidence suggests a relatively short cultivation history, starting in 400 BC to 300 AD. Until recently, the origin of pigeonpea was unclear, with some researchers suggesting an African origin, others India. However, a number of archaeological, taxonomic and modern DNA-based studies now suggest India as single center of origin (Vavilov, 1928; van der Maesen, Citation1990; Kass et al., Citation2012). From India, it traveled to East Africa and continued to the American continent with the misfortunes of the African slave trade.
The Phaseolid group contains a legume tree species domesticated for grain rather than fruit. This unique tree is Erythrina edulis Triana ex Micheli, which produces a large bean seed called Chachafruto, and which was domesticated along with a suite of Solanaceae shrubs and small trees for agroforestry systems in the Andes. Other legume trees produce edible pulp around their seeds, including species of Inga Mill., from South America and the Caribbean, and carob (Ceratonia siliqua L.), from the Mediterranean region. Other examples are the Mimosoid legume tree Leucaena Benth., which is used as a food crop throughout south-central Mexico, as well as tamarind (Tamarindus indica L.), a tree from India. Many sub-tropical and tropical legumes also produce valuable wood, resins, decorative beads, and medicinal products or toxins used for hunting and fishing. This shows the multi-functional nature of legumes, one of the reasons for their success and presence around the world.
The Dalbergoid clade contains the smallest number of domesticated legumes, with just one of worldwide importance, the cultivated peanut (groundnut) (Arachis hypogaea L.), and a few forage species of local importance, such as the genus Stylosanthes Sw., which has only recently been developed as a crop. The history of domestication of peanut dates back approximately 7,600 years in the Pantanal across the whole tropical world since the sixteenth century, mainly by Spanish and Portuguese traders (Krapovickas and Gregory, 1994; Valls and Simpson, 2005). The cultivated A. hypogaea is probably derived from the spontaneous inter-specific hybridization of two wild sympatric Arachis species; their genome combine as an allotetraploid, an event which makes all cultivars of peanuts highly monomorphic.
Based on the distribution of Glycine soja Willd. in China, Japan, Korea, and the far eastern Russia in East Asia, it has been suggested that domestication occurred simultaneously at multiple sites (Xu et al., Citation2002; Lee et al., Citation2010). However, most recent studies indicate that cultivated soybean was domesticated only in China, which also has the earliest written historical records of soybean cultivation (Qiu et al., Citation2010). Soybean is mentioned in many Chinese books dating back 4,500 years. Based on 14C radiocarbon dating, these soybean remains are more than 2,590 years old. However, the exact site of domestication has not been identified until now, although several candidate locations have been proposed, including the Huanghuai Region (Yellow River valley) (Li et al., Citation2010a; Li et al., Citation2013a; Guo et al., Citation2010).
Finally, for the Genisteae, two lupin species, Lupinus albus L. and L. mutabilis Sweet, were introduced into agriculture 3,000–4,000 years ago in Egypt and the Andes, respectively. The timing of domestication of L. mutabilis in the New World closely mirrors that of L. albus in the Old World. As a cultigen with no known wild counterparts, it is cultivated from Venezuela to northern Argentina (Wolko et al., Citation2011). Eastwood and Hughes (Citation2008a) identified L. piurensis C.P. Sm. as the likely progenitor of L. mutabilis, which would place the origin of L. mutabilis in northern Peru and southern Ecuador. Lupinus angustifolius L. and L. luteus L. were introduced into agriculture more recently in Northern Europe in the nineteenth century. Three Old World lupin species (L. angustifolius L. luteus and L. albus) and one New World lupin species (L. mutabilis) have been domesticated and bred as grain crops. It is thought that cultivation of L. albus first occurred in Egypt around 2000–1000 BC and that its use spread around the Mediterranean as a fodder crop, a green manure crop, and a grain crop (Gladstone, Citation1970). Even in ancient times, soft-seeded and indehiscent types were available, but up until the twentieth century, all cultivated cultivars were bitter, and seeds had to be soaked and boiled to remove the alkaloids. In Germany, in the 1930s, von Sengbusch identified natural sweet-seeded mutants, which heralded the beginning of modern L. albus breeding (Gladstone, Citation1970). The first successful use of L. angustifolius in modern agriculture was as a fodder and green manure crop in France, Germany and the UK in the early nineteenth century (Wolko et al., Citation2011). Domestication began in earnest at the start of the twentieth century with the development of sweet (low-alkaloid) cultivars in Germany, Poland and Russia (Wolko et al., Citation2011). The process of domestication was completed in Australia through the incorporation of pod indehiscence, soft-seededness and early flowering (removal of vernalization responsiveness) genes (Berger et al., Citation2012a). Australian sweet cultivars contain a recessive gene for white flower and seed color that distinguishes them from bitter, blue-flowered wild types. Since the final domestication of L. angustifolius, cultivars have been developed across Europe and in Australia in particular.
Unlike grain legumes, domesticated forage legumes retain many of the characteristics of their wild relatives, such as seed shattering and small seed size, since breeding emphasis has focused on fodder production and persistence under grazing and hay production. Also, unlike many domesticated crops, wild forms of forage legume species continue to occur naturally, and there are areas of the world where domesticated and wild forms hybridize readily. The wild nature of forage legumes makes it difficult to trace when domestication actually occurred (Small, Citation2011). However, there are records that early cultivation of clover (Trifolium L.) most likely started in southern Spain around 1000 AD. From there it spread to the Netherlands and Italy. Here the rotational cultivation of clover was recommended for the improvement of poor soils (Camillo Tarello in Ricordo d’agricultura, 1567). By the end of the seventeenth century, clover cultivation had spread over most of Europe, reaching the northern areas by the end of the eighteenth century (Kjaergaard, Citation2003). Cultivated clovers generally have larger leaves and flower earlier and more prolifically than their wild ancestors or relatives (Ravagnani et al., Citation2012).
Fourteen annual Medicago species are cultivated (Wiersema and León, Citation2013), the most significant being Medicago sativa L. (alfalfa or lucerne) and annual medics (Medicago spp.), of which, Medicago truncatula Gaertn. (barrel medic), serves also as important model for legume genomics (Cook, Citation1999). Annual medics are even less domesticated than perennial alfalfa. In alfalfa (Medicago L.), the crop progenitor is thought to be Medicago sativa subsp. caerulea (Less. ex Ledeb.) Schmalh., which is a purple-flowered diploid that continues to have a sympatric range with the wild purple-flowered tetraploid, Medicago sativa subsp. sativa, whose domesticated form is alfalfa. The general consensus is that the crop originated in Vavilov's “Near Eastern Center” (Vavilov, Citation1951), which includes Asia Minor, Transcaucasia, Iran and Turkistan (Bolton et al., Citation1962; Small, Citation2011). There is some suggestion that domestication may have occurred more than once and likely occurred in areas where horses were raised (Small, Citation2011). Lesins (Citation1976) suggested the irrigated valleys of eastern Turkey and oases along the Central Iranian plateau may have been the first places alfalfa was cultivated between 5,500 and 4,000 BC. The earliest recorded use of alfalfa was found on brick tablets from central Turkey dated between 1,400 and 1,200 BC (Bolton et al., 1962). Small (Citation2011) suggests that alfalfa spread from north central Asia eastward into China and India and westward into the Middle East and northern Africa. By the fifth century BC, it had been spread to Europe by the Roman Empire, transported as the primary fodder for horses and other livestock. In the sixteenth century, alfalfa was introduced into South America, and by the eighteenth century, it had been introduced into New Zealand, Australia, South Africa and the Eastern United States. Alfalfa was introduced into the Western United States in the mid nineteenth century when South American cultivars were brought to California. Up until the sixteenth century, cultivated alfalfa was predominantly purple-flowered M. sativa subsp. sativa. Michaud et al. (Citation1988) suggested that when purple-flowered alfalfa was introduced into Germany and Northern France around the sixteenth century, it hybridized with the yellow-flowered subspecies, M. sativa subsp. falcata (L.) Arcang., to form variegated alfalfa (M. sativa nothosubsp. varia (Martyn) Arcang.). Variegated alfalfa had greater winter hardiness, greater disease resistance, and was more tolerant to acidic soils; these hybrid forms were domesticated as well (Small, Citation2011). Although the yellow-flowered alfalfa contributed important agronomic characteristics that expanded the production of alfalfa, it has been minimally domesticated and continues to be a poor seed producer. Relatively few yellow-flowered cultivars have been developed compared to purple-flowered and variegated alfalfa.
A. Genetic Aspects of Legume Domestication
Despite legumes’ crucial role in providing much of the protein in the human diet and animal feed, comparably little is known about their domestication. The “domestication syndrome” for legumes includes changes in plant architecture, seed gigantism, transition from outcrossing to selfing, reduced seed dispersal and loss of seed dormancy (Hammer, Citation1984).
An increase in the seed size of domesticates compared to their wild relatives is suggested to be related to greater planting depth in agricultural systems, with larger seeds producing more vigorous seedlings (Abbo et al., Citation2011). At the same time early farmers may have selected for a higher proportion of starch, oil and protein. Seed shattering was avoided during the selection process in order to reduce the occurrence of the natural explosive seed pod opening mechanism of wild legumes. Experiments growing wild peas and lentil have demonstrated that both seed dormancy and pod dehiscence cause poor crop establishment via reduced germination, as well as dramatic yield losses via seed shattering (Abbo et al., Citation2011, 2013).
The loss of seed shattering has been a fundamental characteristic under selection in most legume grain crops in order to facilitate seed harvesting, while in wild plants, shattering is a fundamental trait for assuring seed dispersal. The evolution of the non-shattering trait would have occurred automatically as a result of harvesting that favored non-shattering mutants in harvested populations that were subsequebtly sown. Central to the ballistic mechanisms of seed dispersal in pea is the dehiscent pod (single carpel fused along its edges), where the central pod suture undergoes an explosive rupturing along a dehiscence zone (Ambrose and Ellis, Citation2008). In domesticated species, this is removed or delayed. Breeding experiments have shown single-locus control of pod dehiscence in lentil (Erskine, Citation1985), two in mungbean (Isemura et al., Citation2012), yardlong bean (Kongjaimun et al., Citation2012) and pea (Weeden, Citation2007). One locus controls the number of twists along the length of the shattered pod, while the second locus controls the percentage of shattered pods (Weeden and Wolko, Citation2001; Ambrose and Ellis, Citation2008).
Probably the second most important domestication trait in grain legumes relates to seed dormancy, often called hard-seededness due to the physical barrier of testa water permeability. As greater seed size was selected and seeds were stored from one season to another, the potential for absorption of water and germination during storage made it necessary to select for seed dormancy. Moreover, seed imbibition plays a crucial role in reducing the cooking time of most grain legumes. Hence, reducing seed coat thickness led to a concurrent reduction of seed coat impermeability during domestication. This was largely overcome in all domesticated grain legumes (Werker et al., Citation1979; Smartt, Citation1990; Weeden, Citation2007). A single recessive locus has been reported in lentil (Ladizinsky, Citation1985), while Weeden (Citation2007) has identified two to three loci involved in pea seed dormancy, mediated by testa thickness and the structure of the testa surface. Among the legumes, unlike most cereal families (except for the millets), related species within a single genus have been domesticated at different stages, periods and places but with similar results in terms of cultivated crop characteristics. As well as suggesting that the domestication of plant crops was a directed, intentional and transmitable process, this makes several genera of legumes valuable and interesting for further study. For example, Lupinus provides a useful system for exploring plant domestication, as there are four related species whose domestication spans ancient (L. albus and L. mutabilis) and recent (L. angustifolius and L. luteus) times. All four species share the same domestication traits (reduced alkaloids, seed indehiscence, soft-seededness, and removal of the vernalisation requirement). Identification of the causal genes would allow for the development of diagnostic markers to improve the efficiency of introgression of genetic and phenological diversity from wild into domesticated germplasm. Similarly, in the genera Phaseolus and Vigna, there are clusters of cultivated species that allow for comparisons of the domestication traits, genes and mechanisms. This is in contrast to Arachis, Cicer, Cajanus, Erythrina and Glycine, which have only single domesticates. However, the last three of these genera can be compared to the other tropical legumes, which is why common bean and cowpeas have been suggested as models for soybean and pigeonpea.
In order to identify genetic basis of domestication traits, genetic analysis has been performed in wild to cultivated mungbean and pigeonpea crosses (Isemura et al., Citation2012; Kongjaimun et al., Citation2012; Kassa et al., Citation2012), resulting in the identification of quantitative trait loci (QTLs) for 38 domestication-related traits or single-nucleotide polymorphisms derived from 670 genes. Comparably smaller numbers and mostly anonymous markers were used for mapping pea domestication traits, resulting in the identification of around 20 loci (Weeden, Citation2007). In common bean, seed size appears to be under multi-genic control, with 10 QTL found in wild x cultivated advanced backcross population analysis (Blair et al., Citation2006; Blair and Izquierdo, Citation2012). Seed coat color genes are divided into those that control pattern and those that control tone of color (Caldas and Blair, Citation2009). These genes are underlayed by pro-anthocyanidin pathway genes, resulting in the accumulation of tannins and anthocyanins (Díaz et al., Citation2010). Determinacy is also believed to be an important trait in the domestication of common bean from a viny wild phenotype to a short, rapidly maturing phenotype in some regions of early agriculture in the Americas.
III. TRIBE FABEAE RCHB.
The tribe Fabeae contains the following five accepted genera: Lathyrus L. (grasspea/vetchling; about 160 species); Lens Mill. (lentil; 4 species); Pisum L. (pea; 2-3 species); Vicia L. (vetch; about 160-200 species) and the monotypic genus Vavilovia Fed. (Smýkal et al., Citation2011; Schaefer et al., Citation2012). The tribe Fabeae is sister to the Trifolieae tribe (Steele and Wojciechowski, Citation2003; Schaefer et al., Citation2012). The tribe is morphologically characterized by mostly paripinnate, often tendrillous leaves and a pubescent style or a pollen brush (Lavin and Delgado, 1990). Stylar shapes and hair patterns are one of the principal diagnostic characteristics within the genera of Fabeae (Gunn and Kluve, Citation1976; Kupicha, Citation1981; Schaefer et al., Citation2012). The clade is considered one of the youngest tribes among legumes (Kupicha, Citation1981; Steele and Wojciechowski, Citation2003; Wojciechowski et al., Citation2004; Lock and Maxted, Citation2005), and estimates based on rates of evolution in the maturase K chloroplast gene place the age of the crown node at 17.5 Mya, in the mid-Miocene (Lavin et al., Citation2005). A recent Bayesian molecular clock analysis of combined plastid and nuclear sequences also suggests a crown age of 23 – 16 Mya (Schaefer et al., Citation2012).
Ancestral range reconstructions (Kenicer et al., Citation2005; Schaefer et al., Citation2012) place the area of origin in the Eastern Mediterranean, which is also the current center of diversity of the tribe (Kupicha, Citation1981). From there, a minimum of three dispersal events to the middle-Atlantic islands and seven to the Americas are required to explain the current distribution pattern (Schaefer et al., Citation2012). South America was probably colonized twice from the Mediterranean and once via range expansion into North America. In Africa, Fabeae species, all descendants of Mediterranean lineages, occur only in the northern regions with extension into tropical mountains (D. R. Congo, Tanzania, Uganda) and into Ethiopia (Schaefer et al., Citation2012).
A. Genus Lathyrus L.
The greatest diversity of Lathyrus species is found in Europe, Asia and North America (Kupicha, Citation1981; Kupicha, Citation1983; Kenicer et al., Citation2005; Schaefer et al., Citation2012), but its distribution extends to South America and East Africa. Most species are adapted to temperate regions, but some can be found at high altitudes in tropical Africa. The genus contains many restricted endemic species. Lathyrus underwent several long-distance dispersal events, from Asia via Beringia to North America and into South America (Schaefer et al., Citation2012). Most species of Lathyrus are mesophytes of open woodlands, forest margins, and roadsides, with several-drought tolerant and halophytic species.
The generic boundaries between Lathyrus and the other Fabeae genera have been much debated (Kupicha, Citation1981; Schaefer et al., Citation2012). This taxonomic confusion has led to an abundant and complex synonymy. Molecular study of Lathyrus phylogeny, based on nuclear (ITS) and chloroplast (trnL-F, trnS-G) markers using a large set of geographical and taxonomic samples, was done by Kenicer et al. (Citation2005). Based on a very limited sampling, Steele and Wojciechowski (Citation2003) suggested that Lathyrus, Lens and Pisum might all be nested in Vicia. This was recently confirmed in a comprehensive analysis by Schaefer et al. (Citation2012), who demonstrated that nuclear ITS and chloroplast DNA regions lead to phylogeny estimates with Pisum and Vavilovia nested in Lathyrus, Lens nested in Vicia s. str. and all four genera (Pisum, Vavilovia, Lathyrus, Lens) nested in Vicia s. l. Consequently, a monophyletic Lathyrus will have to include both Pisum and Vavilovia. To maintain most of the species names in Lathyrus, a recircumscription of Vicia has been proposed, which would mean that the clade containing Vicia tetrasperma (L.) Schreb. and another clade containing Vicia hirsuta (L.) Gray and Vicia sylvatica L., among others, will be split from Vicia s. str. and become two additional genera (Schaefer et al., Citation2012).
Several methods have been used to study the phylogenic relationships among different Lathyrus species, including morphological traits, crossability, karyotype analysis, chromosome banding and in situ hybridization and molecular markers (reviewed in Kumar et al., Citation2013). Lathyrus is predominantly a true diploid with a chromosome number of 2n = 2x = 14 (also reconstructed as the ancestral number), with a few exceptions having 2n = 28 or 42 (Schaefer et al., Citation2012 and references therein). There are a few polyploid species among the perennials, including hexaploid (L. palustris L., 2n = 6x = 42) and tetraploid (L. venosus Muhl ex Willd., 2n = 4x = 28). Natural and induced autopolyploids have also been reported in L. sativus, L. odoratus L., L. pratensis L. and L. venosus. Genome size of 52 measured species ranges from 1C = 3.43 pg (L. miniatus) to 18.2 pg (L. sativus) (RBG Kew DNA C-values database).
1. Crop grasspea (Lathyrus sativus L.)
The most widely cultivated Lathyrus species for human consumption is the grasspea (L. sativus) which serves as a key famine food for rural populations in countries like Kenya, Ethiopia, India, and Bangladesh. Other species, which are grown for forage and/or grain purposes, are L. cicera, L. ochrus (L.) DC., L. clymenum L., L. tingitanus L., L. latifolius L. and L. sylvestris L. (IPGRI, 2000, ), which are important animal fodders. Lathyrus cicera is cultivated in Greece, Cyprus, Iran, Iraq, Jordan, Spain and Syria, and L. ochrus in Cyprus, Greece, Syria and Turkey (Saxena et al., Citation1993). Other species, like L. hirsutus and L. clymenum, are cultivated as minor forage or fodder crops in the southern United States and Greece (Sarker et al., Citation2001). The primary gene pool of cultivated Lathyrus sativus and L. cicera is limited to cultivars, landraces and escapes from cultivation, while the secondary gene pool includes L. chrysanthus Boiss., L. gorgonii Parl., L. marmoratus Boiss. & Blanche, L. pseudocicera Pamp., L. amphicarpos L., L. blepharicarpos Boiss., L. chloranthus Boiss. & Balansa, L. cicera, L. hierosolymitanus Boiss. and L. hirsutus L. The remaining species are included in the tertiary gene pool (). Although the progenitor of L. sativus remains unknown, L. cicera is the most probable candidate, as it is morphologically and cytogenetically closest to the cultivated species (Jackson and Yunus, Citation1984; Hopf, Citation1986).
Ex situ Lathyrus sativus germplasm collections total 38,360 accessions, with the largest number held at the International Center for Agriculture in Dry Areas (ICARDA) (9,000), followed by China (5,200) and Australia (2,445) (Cambel, 1997; Kumar et al., Citation2013, ). Besides cultivated L. sativus, there are smaller collections of the remaining Lathyrus species, which may be better represented in botanical gardens; however, such displays often offer only a single accesion per species. Australian Grain Genebank holds 553 acc. of 37 Lathyrus species, EURISCO lists 1,645 acc. of 66 species, USDA holds 445 acc. of 49 species and VIR has 5,500 acc. of 58 species. Breeding efforts focused on three species―L. sativus, L. cicera and L. ochrus, and, to a lesser extent, L. clymenum―with an aim at improving grain yield, biomass, resistance to biotic and abiotic stresses, and, most importantly, to reduce the neurotoxin in its seeds. Unfortunately Lathyrus seeds, apart of being protein rich, contain the water soluble non-protein amino acid β-diaminopropionic acid (ODAP), which has been found to be a neurotoxin linked to an irreversible neurological disorder called lathyrism (Barrow et al., Citation1974). Several low β-diaminopropionic acid (with < 0.1% ODAP) cultivares were developed through intraspecific hybridization in Bangladesh, ICARDA, Ethiopia, Canada and Australia.
Evaluation of Lathyrus germplasm has been undertaken for different traits in order to identify useful donors for important parameters, including low ODAP content, phenology and yield-related traits. For example, Chowdhury and Slinkard (Citation2000) studied genetic diversity in 348 accessions of L. sativus from 10 geographical regions using polymorphism for 20 isozymes. They observed the closest genetic distance between populations from the Near East and North Africa. The most extensive study of 1,082 accessions belonging to 30 species evaluated for 21 descriptors and agronomic traits was performed at ICARDA (Cambell, 1997). A detailed cataloguing of grasspea germplasm comprising characterization and evaluation information on 63 traits for 1,963 accessions was perfromed in India (Kumar et al., Citation2013). ODAP content in seeds was found to vary from 0.02% to 2.59%. Interestingly, ODAP concentration in L. cicera is lower compared to that of L. sativus (reviewed in Kumar et al., Citation2013).
Genetic diversity of Lathyrus has experienced serious genetic erosion, largely as a result of intensification of agriculture, overgrazing, and the decline of permanent pastures (Maxted and Bisby, 1986, 1987). Despite L. sativus being among the hardiest crop species, able to withstand conditions from flooding to severe drought, the genetic diversity of grasspea has suffered a great deal from a ban on its sale, causing serious erosion of landraces. Development of less toxic cultivars that retain palatability remains a holy grail in arid land crop research. Grasspea is one of the hardiest but most underutilized crops for adaptation to fragile agro-ecosystems due to its ability to survive under extreme climatic conditions, such as drought, water stagnation and heat stress (Vaz Patto et al., Citation2006; Kumar et al., Citation2013). It is an annual, cool-season legume crop of economic and ecological significance in South Asia, sub-Saharan Africa, and, to a lesser extent, in Central and West Asia, North Africa, southern Europe and South America. It is grown mainly for food in India, Bangladesh, Nepal, Pakistan, Ethiopia and for feed and fodder purposes in other countries (Kumar et al., Citation2013). Grasspea grains provide a good protein supplement (24–31%) to the cereal-based diet of poor people in areas of its major production. Globally, the area under grass pea cultivation is estimated at 1.50 million ha, with annual production of 1.20 million tonnes (Kumar et al., Citation2011).
B. Genus Vicia L.
The genus Vicia, with 160-200 species, is rich in diversity. The uncertain number of species reflects the abundant use of synonymous names and similar species descriptions. The distribution and species numbers of Vicia parallel that of Lathyrus, although it might be argued that Vicia shows less morphological diversity. The best known species of the genus are the faba bean, V. faba, and V. sativa.
Kupicha (Citation1976) undertook a comprehensive revision of the genus, dividing the species into two subgenera, Vicilla and Vicia, based on flower arrangement and the presence/absence of nectariferous spots on the stipules. Kupicha's subgenus Vicilla is further divided into 17 sections, while the subgenus Vicia is divided into five sections with 38 species. Section Faba of Kupicha (Citation1976) includes V. faba, V. galilaea Plitmann & Zohary, V. johannis Tamamschjan, V. narbonensis L., V. hyaeniscyamus Mouterde and V. bithynica. Vicia faba stands out due to its karyological (2n = 12) crossing barrier (Maxted, Citation1995), and this different characteristic is also supported by a recent phylogenetic study based on chloroplast and ITS sequences (Schaefer et al., Citation2012). A biosystematic study of the genus was conducted by Hanelt and Mettin (Citation1989) using morphological characteristics and classical karyology, which largely agreed with Kupicha's results. The presence of pubescence on the adaxial side of the style is typical for the groups Lathyrus and Pisum (Kupicha, Citation1981). However, several Vicia species, including V. ervilia and V. koeieana Rech. f., also show this type of pubescence and in fact do not appear in the core Vicia clade (Schaefer et al., Citation2012). Species in the genus Lens also have this pubescence pattern, rendering it of little use for morphology-based classification. Problems over the taxonomic distinction within species can be attributed to large variation in morphology and karyotypes (Maxted et al., 1991). Maxted (1993, Citation1995) revised Vicia subgenus Vicia and proposed nine sections and nine series. Chromosome numbers in Vicia vary between 2n = 10, 12, 14, 28, and 42, with an ancestral number of 2n = 14 (Schaefer et al., Citation2012 and references cited therein). Notably, cultivated V. faba with a chromosome number of 12 and genome size of 1C = 13.3 pg is reproductively isolated from its closest relatives. Faba bean is partially allogamous, with the rate of out-crossing differing between environments and genotypes and ranging from 4 to 84% (Bond and Poulsen, Citation1983; Suso et al., Citation2001; Hu et al., Citation2011). The genome size of 88 analyzed Vicia species varies from 1C = 1.83 pg (V. lunata (Boiss. & Balansa) Boiss. & Balansa) to 27.4 pg in V. faba (RBG Kew DNA C-values database).
Faba bean stands as an exception among the cultivated legumes, as there is no known wild progenitor. Vicia faba subsp. paucijuga (Alef.) Murat. from Pakistan and Afghanistan has been suggested as the progenitor, because it showed primitive characteristics. Another proposed, close wild relative, V. pliniana (Trabut) Murat. from Algeria (Muratova, Citation1931), is currently considered to be only a morphological variety of V. faba subsp. faba var. minor. Morphological similarity led Hopf (1973) to propose V. narbonensis as the faba bean ancestor; however, its crossing barriers and phylogenetic results (Schaefer et al., Citation2012) do not support this hypothesis. In summary, we do not know the faba bean progenitor and cannot be sure it is not extinct. Once more genomic information has been gathered, more light may be shed on this question. No chloroplast DNA variation has been detected among V. faba genotypes (Shiran and Mashayek, Citation2004), whereas mitochondrial DNA displays variation in sequence and size (Flamand et al., Citation1993). The extreme genome size of V. faba (13,000 Mb) can be explained by a high number of retrotransposons (Pearce et al., 1996).
Except for V. faba and V. sativa, the species of the large Vicia genus are poorly systematically conserved ex situ. The VIR collection has the largest set of 5,500 acc. of 58 Vicia species; the USDA holds 942 acc. of 51 species; the Australian Grain Genebank has 1,924 acc. of 60 species, plus 1,013 acc. of V. sativa; and EURISCO lists 7,303 acc. of 117 species and 6,968 acc. of V. sativa, the majority of which are at the Gatersleben genebank in Germany.
1. Forage vetches (Vicia sp.)
Numerous species of the vetch genus (Vicia L.) represent a frequent component of local floras and bring an essential contribution to the quality of pasture and meadow communities and soil fertility. Among such are narrow-leafed vetch (V. sativa subsp. nigra (L.) Ehrh.), large-flowered vetch (V. grandiflora Scop.), hairy vetch (V. villosa Roth), Hungarian vetch (V. pannonica Crantz) and tiny vetch (V. hirsuta (L.) Gray). Of these, common vetch (V. sativa) is the most commonly used, providing palatable forage (fresh, hay and silage) and grain to livestock. V. sativa originated from southern Europe and is now widespread in the Mediterranean basin, in west and central Asia, China, eastern Asia, India and in the USA. It is moderately tolerant of cold and can grow in areas with mild winters. V. sativa is found in areas with annual rainfall ranging from 310 mm to 1630 mm and on a large variety of soils with a preference for well-drained, moderately fertile soils with soil pH ranging from 6.0 to 7.0. It is not tolerant of drought during the early stages of establishment and it is advisable to plant it in autumn (FAO, Citation2010). It can withstand short waterlogging periods but no extended flooding periods. Like other Vicia species, the seeds of V. sativa contain numerous antinutritional factors, notably cyanogenic amino acids and cyanogenic glycosides that are toxic to monogastric animals. The nutritive value of common vetch hay is higher than that of alfalfa and sainfoin at similar vegetation stage (Heuzé et al., Citation2013). These species have not been bred and cultivars are result of selection from wild populations. These species are of interest for drought prone regions in order to provide good quality animal feedstock with minimal input.
2. Crop faba bean (Vicia faba L.)
In the case of faba bean, there are 37 collections, holding approximately 38,000 accessions (Duc et al., Citation2010, ), which comprise around 17% of total grain legume accessions worldwide (Suso et al., Citation2006). Most of the accessions from European collections are listed in the EURISCO database with available passport data (Duc et al., Citation2010). Of these, 30% have cultivar names, and 52% are of European origin. Since faba bean is an open-pollinated species, it is important to prevent cross-pollination during regeneration in order to preserve the unique genetic identity of the landraces (Suso et al., Citation2006; Hu et al., Citation2011). In addition, a large proportion of the germplasm is not stored under long-term storage conditions and is subject to a wide variety of regeneration cycles ranging from 5 to 35 years. Interestingly, winter faba beans from China were found to be distinct from the winter gene pool in the rest of the world (Bao et al., 2006; Zong et al., 2009).
A global composite collection of 1,000 accessions was collaboratively developed under the Generation Challenge Program (GCP) using genomic microsatellites (Sadiki et al., Citation2006). Repetitive sequences and other DNA-based markers have been used to assess genetic diversity (Zeid et al., Citation2003; Gutierrez et al., Citation2006; Torres et al., Citation2006; Sanz et al., Citation2007; Terzopolous and Bebeli, 2008; Zong et al., 2009; Kwon et al., Citation2010) among cultivated faba genotypes. Phenotypic traits of agronomic interest were used to evaluate ex situ faba bean germplasm collections in Europe, at ICARDA and in China (reviewed in Duc et al., Citation2010). Particularly, flowering response and stem architecture (internode length and strength, branching, determinate growth) were investigated in relation to crop adaptation to diverse agronomic practices and climatic zones. Drought tolerance was detected in accessions from the Mediterranean region, while frost tolerance was found in German landraces (Arbaoui et al., 2008). Sources of resistance/tolerance to various fungal, viral and pest biotic stresses were identified and used in breeding (Sillero et al., Citation2010). Genetic variation was used to develop zero-tannin-content and low-vicine, convicine faba bean cultivars (reviewed in Duc et al., Citation2010). The low-vicine, convicine “Fevita” types remove the danger of favism, which has been linked to a genetic variant of glucose-6-phosphate dehydrogenase deficiency in humans (Arese and De Flora, Citation1990). Significant genetic variation for all these traits of interest exists within faba bean germplasm, providing an excellent resource for plant breeders (Duc et al., Citation2010). Fast and reliable screening methods have been adjusted to fulfil the needs of breeding programmes both for fungal diseases (Sillero et al., Citation2006), parasitic weeds (Rubiales et al., Citation2006) and abiotic stresses (Stoddard et al., Citation2006). Many of these traits of interest have already been incorporated into modern cultivars but several others, many of which are controlled quantitatively by multiple genes, have been more difficult to manipulate.
Faba bean is now cultivated over a latitudinal range from the equator to approximately 50 °N and 40 °S and an altitude range from sea level to above 3,000 m. The long period of cultivation across such diverse environments has resulted in the differentiation of germplasm into distinct groups based on seed size (paucijuga, minor, equina and major) and region of adaptation (winter, spring types) (Flores et al., Citation2013). Despite the complicated, large genome, significant efforts have been made to understand the genetics and genomics of faba bean. Linkage maps of faba bean have been constructed based on various marker types, and various QTLs (Quantitative Trait Loci) have been identified (reviewed in Gnanasambandam et al., Citation2012; Ma et al., Citation2013).
The long history of cultivation, wide distribution across various climate environments and the response to human selection have caused faba bean to become a most versatile crop for use as food, feed, forage, vegetable and as a cover crop. According to FAO, the annual harvested faba bean acreage averaged 2.5 million hectares from 2005 to 2010, with an annual production of approximately 4.2 million tonnes and mean yield of 1,666 kg/ha (FAOSTAT, Citation2012). The high-protein seeds of faba bean are a staple in the diets of many societies in the Middle East and North Africa. Snacks made from faba bean have been marketed in China and elsewhere. In China, substantial amounts of faba bean are used to produce pastes or doubangjiang, an indispensible condiment in Chinese cuisine. Although global faba bean acreage and production has experienced a steady reduction over the past four decades, demand for faba bean in the world market has driven the production upwards in Australia and France in recent years (FAOSTAT, Citation2012). Immature faba bean seeds (fresh or frozen) are a favored vegetable in many countries. Like other food legumes, faba bean contains numerous phytonutrients, such as vitamins, minerals and phenolics, which contribute to the overall antioxident activities of plant foods (Oomah et al., Citation2011; Baginsky et al., Citation2013). Faba bean provides an alternative to soybean meal for animal feed in temperate regions where soybean cannot be grown. Faba bean is one of the a few plant species capable of producing the medicinally important molecule, L-3,4-dihydroxyphenylalanine (L-DOPA), the major ingredient of several prescription drugs used to treat Parkinson's disease (Apaydin et al., Citation2000).
Faba bean is an ideal cover crop or for green manure, particularly for organic growers, since it has been documented as having the highest capacity for fixing atmospheric nitrogen among the major cool-season food legumes (Herridge et al., Citation1994). Faba bean has undergone significant improvement in yield and other agronomic traits during the past half century. However, similar to other temperate-region legume crops, faba bean faces major challenges from more profitable crops, such as wheat, corn and soybean, for a place in growers’ crop rotation plan. Despite the fact that the global acreage dedicated to faba bean production dropped significantly, from six million ha in the 1960s to 2.5 million ha in recent years (FAOSTAT, Citation2012), there remains potential for increased faba bean cultivation world-wide remains good because of the steady growth of the consumer population. New advances in genomics are expected to have enormous impact on the genetic improvement of the faba bean crop. As in the case of other grain legumes, yield stability is a major challenge for faba bean breeding. Many efforts have been made recently to improve abiotic tolerance and climate adaptation, though progress has been slow, since the partially allogamous nature of faba bean slows the process of developing pure lines. Nevertheless, useful genetic resources with good drought tolerance have been identified (Khan et al., Citation2007). Enhancing winter-hardiness of faba bean will mitigate the cold damage and ensure crop productivity (Arbaoui et al., 2008; Link et al., Citation2010; Hu et al., Citation2011; Flores et al., Citation2012). Other abiotic stresses, like heat tolerance and water logging, also require attention. Identifying and incorporating host resistance to biotic stresses is needed (Infantino et al., Citation2006; Tivoli et al., Citation2006; Pérez-de-Luque et al., Citation2010; Sillero et al., Citation2010).
Tannins and vicine-convicine are the two major antinutritional elements of concern for faba bean. Tannins impart a bitter flavor to the seed and reduce the digestibility of protein, while vicine-convicine aglycone derivatives may induce favism (Arese and De Flora, Citation1990). Useful natural mutants have been identified in faba bean germplasm. Tannins can be removed by incorporating one of the two independent, recessive genes named zt1 and zt2 (Picard, Citation1976), which also determine the white-flower phenotype. Vicine and convicine can be lowered by incorporating the spontaneous recessive mutant (Duc et al., Citation1989). Molecular markers linked to these genes have been developed (Gutierrez et al., Citation2006; 2007 and 2008), and these genes have been incorporated into a few European cultivars (Duc, personal communication).
In recent years, crosses were made using very diverse germplasm, and elite breeding lines adapted to various production regions were developed. The success of this breeding effort demonstrates the benefit of including diverse germplasm from different origins (Gnanasambandam et al., Citation2012).
C. Genus Pisum L.
The Pisum genus is distinguished morphologically from the related genera Lathyrus and Vavilovia by the presence of large, leafy stipules, which are semi-amplexicaul. The genus Pisum contains the flavonoid phytoalexin pisatin, which is shared with genus Lathyrus but not found in Vicia species (Bisby et al., Citation1994), which have wyerone instead. The P. sativum L. complex (P. sativum subsp. sativum and subsp. elatius Asch. & Graebn.) is native to the Europe-Mediterranean region and Middle and northwest Asia, whereas P. fulvum Sibht -Sm. is restricted to the Middle East. Pisum sativum subsp. abyssinicum A. Braun is found in cultivation (together with P. sativum subsp. sativum) in Eastern Africa. This taxon is native to Ethiopia and Yemen, has very low genetic diversity (see later section) and possesses a distinct set of phenotypic characteristics (early flowering, an adaptation useful for avoiding drought periods; unipinnate and strongly serrate leaflets), as well as unique alleles at particular loci. The classification of Pisum L. has changed over time from a genus with five species, to a monotypic genus, to a genus with two species (reviewed in Smýkal et al., Citation2011, 2013). In Yarnell's review (1962), P. humile Mill. (P. syriacum (A. Berger) C. O. Lehm., P. sativum var. pumilio Meikle), P. elatius M. Bieb., P. abyssinicum, and P. sativum were considered conspecific, even though they often differ by inversions and translocations. The most appropriate status for P. sativum subsp. abyssinicum is still under debate, as it has been resurrected as a third species by some authors (Maxted and Ambrose, Citation2001; Vershinin et al., Citation2003; Jing et al., Citation2007). The most recent studies place it between P. fulvum Sibth. & Sm. and a subset of P. elatius (Vershinin et al., Citation2003; Jing et al., Citation2010). While most authors agreed with the original suggestion of Linné, who described the genus Pisum as distinct from Lathyrus (Linné, 1753), the recent molecular phylogenetic analysis (Schaefer et al., Citation2012) finds it deeply nested in Lathyrus. Interestingly, Lamarck (Citation1778) described the garden pea as Lathyrus oleraceus Lam., disagreeing openly with Linné's suggestion. Depending on how that complex is treated, the genus Pisum may be incorporated into a larger genus Lathyrus to represent a natural (monophyletic) group.
The primary gene pool consists of the Pisum sativus/elatius complex, although it is difficult to specify concisely because of the fertility barriers, caused by nucleo-cytoplasmic conflict, which exist within the species P. sativum (Bogdanova et al., Citation2009). A secondary gene pool generally extends to the other species in the genus, P. fulvum and P. sativum subsp. abyssinicum (), but with new knowledge regarding relationships with sections of Lathyrus, especially the closely related L. ochrus, L. clymenum, L. articulatus L. and L. neurolobus Boiss. & Heldr. (Schaefer et al., Citation2012), it may be useful to examine these sections more thoroughly. The tertiary gene pool currently consists of Vavilovia formosa (Stev.) Fed. (= Pisum formosum (Steven) Alef., Pisum aucheri Jaub. & Spach.), which might be reconsidered to be within the secondary pool, as shown by Golubev (Citation1990), and after the tribe circumscription, it would consist of numerous Lathyrus and Vicia species ().
The beautiful Vavilovia (Vavilovia formosa) was first described in 1813 by Steven and assigned to the genus Orobus L. It was later associated with both Lathyrus and Pisum. Fedorov (Citation1939) revised the taxonomy based on morphological characteristics, such as flower and stipule shape, absence of tendrils, presence of creeping, and thread-like rhizomes, as well as characteristics of disjunctive distribution range, ecology and perennial habit. Fedorov (Citation1939) ultimately separated Orobus L. into the monotypic genus Vavilovia. Vavilovia combines several of the morphological traits of the genera Lathyrus and Pisum, and Makasheva (1979) proposed that Vavilovia be considered the ancestor of both genera. Recent molecular phylogenetic analyses have shown its sister group relationship to Pisum (Oskoueiyan et al., Citation2010; Schaefer et al., Citation2012; Mikič et al., Citation2013).
Pea genetic diversity spreads in the area of the Fertile Crescent through Turkey, Syria, Iraq, Israel and Lebanon, to Central Asia and the Mediterranean region (Smýkal et al., Citation2011). There are two wild P. sativum populations, which are morphologically, ecologically and genetically distinct, variously described as subspecies of P. sativum or as species, P. sativum subsp. elatius M. Bieb. and P. sativum subsp. sativum (formerly P. humile (syn. P. syriacum) (Ben-Ze'ev and Zohary, Citation1973; Smýkal et al., Citation2011). Recent phylogenetic studies based on retrotransposon insertion markers support the model of P. sativum subsp. elatius as a paraphyletic group, within which all P. sativum are nested (Vershinin et al., Citation2003; Jing et al., Citation2005, 2010; Nasiri et al., Citation2010). The phylogenetic relationships of pea have been reconstructed by Ellis et al. (Citation1998) and by Pearce et al. (Citation2000) using molecular, multiloci approaches, finding that P. fulvum and P. sativum subsp. abyssinicum formed sister clades. Pisum sativum subsp. elatius is positioned between P. fulvum - P. sativum subsp. abyssinicum and cultivated P. sativum. On the same set of pea accessions, Vershinin et al. (Citation2003) separated P. fulvum as an ancient lineage, while P. sativum subsp. elatius accessions formed a polytomy with P. humile and P. sativum subsp. abyssinicum accessions. Extremely low diversity of P. sativum subsp. abyssinicum was detected in several studies (Pearce et al., Citation2000; Weeden and Wolko, Citation2001; Vershinin et al., Citation2003; Jing et al., Citation2005, 2010), which could be explained by its passage through a bottleneck caused by a putative hybridization event between P. fulvum and P. sativum, as suggested by Kloz (Citation1971). A phylogenetic analysis based on the combination of mitochondrial, chloroplast and nuclear genome markers placed the P. sativum accessions in a distinct clade separated from all P. fulvum and P. sativum subsp. abyssinicum accessions (Kosterin and Bogdanova, Citation2008; Kosterin et al., Citation2010) and suggested that all wild forms of Pisum sativum should be included in a paraphyletic P. sativum subsp. elatius.
All Pisum species are diploid with 2n = 14. For cultivated pea, nuclear genome size estimates have been produced for several accessions using different methods and are estimated to be 1C = 4.4 to 4.8 pg DNA corresponding to the haploid genome size (1C) of 4.45 Gb, with a large part (75 to 97%) comprised of repetitive sequences (reviewed in Smýkal et al., Citation2012).
1. Crop pea (Pisum sativum L.)
There is no international genetic resource center for pea, following the relinquishing of this role by the International Center for Agricultural Research in Dry Areas (ICARDA) in the early 2000s. However, the ICARDA pea collection is still conserved, though not actively curated. There are substantial national pea collections: 98,947 accessions are distributed over 28 genebanks, comprised of landraces (38%), commercial cultivars (34%), mutant/genetic stocks (5%), wild relatives (2.6%) and breeding lines (13%) (Smýkal et al., Citation2013, ). The actual number of unique lines is substantially less due to duplication of stocks among genebanks. Of these, only 1,876 (2%) are wild pea relatives; approximately one quarter (24,000) are commercial cultivars and landraces; and 8,500, 600 and 6,000 represent breeding and recombinant inbred lines or mutant stocks, respectively. Moreover, there is a large bias (17%) towards Western and Central European accessions. Less represented are the Mediterranean (2.5%) and the Balkan (2%) regions, as well as the Caucasus (0.8%) and Central Asia (2%), the centers of pea crop domestication and diversity (Smýkal et al., Citation2013). The largest pea germplasm collections are held by INRA France (8,839 accessions with 4,818 acc. of TILLING mutants); the Australian Grains Genebank (AGG; formerly Australian Temperate Field Crops Collection, 7,432 acc.); the Vavilov Institute, Russia (6,790 acc.); the USDA (6,827 acc.); ICARDA (6,105 acc.); the Leibniz Institute of Plant Genetics and Crop Plant Research, Germany (5,343 acc.); Instituto Di Genetica Vegetale Italy (4,558 acc.); the Institute of Crop Sciences, China (3,837 acc.); the National Bureau of Plant Genetic Resources, India (3,609 acc.); and the John Innes Center, UK (3,567 acc.) (Smýkal et al., Citation2013). Some of these genebanks have also identified a core collection comprising around 5–10% of the collection size to represent a cross section of the diversity, usually based on geographic diversity of collection sites for landraces, to facilitate a search of the germplasm for alleles of key traits (Redden et al., Citation2005).
In addition to wild and cultivated accessions, there are also pea mutant stocks held at John Innes Collection, Norwich, UK (575 accessions); the Institute of Plant Genetics Resources collection, Plovdiv, Bulgaria (122 accessions); and a targeted-induced local lesions in genomes (TILLING) population of 4,818 lines (1,840 described by phenotype and 93 symbiotic mutants for 26 genes) held at INRA, Dijon, France. In addition, fast neutron generated deletion mutant resources (around 3,000 lines) are available for pea, which have been useful in identifying several developmental genes. These genetic stocks have well-identified phenotypic markers of high penetrance, ranging across seed and pod types to stem fasciation and internode length (Redden et al., Citation2005).
Several studies of pea germplasm using morphological descriptors and agronomical traits have been published (Ali et al., 2007; Sardana et al., 2007; Smýkal et al., Citation2008a; Sarikamis et al., 2010; Azmat et al., 2012). Different molecular techniques were applied over the last two decades to study pea genetic diversity. Using these markers, several major world pea germplasm collections have been analyzed and representative core collections formed (Baranger et al., Citation2004; Loridon et al., Citation2005; Jing et al., Citation2005, 2007, 2010, 2012; Smýkal et al., Citation2008a, 2011, 2013; Zong et al., 2009; Nasiri et al., Citation2010; Kwon et al., Citation2012; Majeed et al., Citation2012). All these studies give a consistent view: the Pisum genus is very diverse, and the diversity is structured, showing a range of degrees of relatedness that reflect taxonomic identifiers, eco-geography and breeding gene pools (Ellis, Citation2011; Jing et al., 2012; Smýkal et al., Citation2011, Citation2013).
Pea is an important food legume in the temperate and elevated sub-tropical cropping zones, grown as dry grain, as green unripe fresh grain for vegetable use and for canning, and as green leaves (Muelbauer and Tullu, 1998). The crop is also used for fodder. The total world grain production fluctuates 10 – 12 million tonnes, with Canada as the leading producer, followed by USA, India, Russia, France and China (see Smýkal et al., Citation2012 for review). However mean yield is relatively low 1,558 kg/ha, while European records are around 6,000 kg/ha, indicating a gap in yield stability and biological potential of the crop. Up to half of the area in which pea is sown may be used for the production of vegetables, green snap bean pods, green seed for vegetables (fresh, frozen or canned), green leaves and for direct livestock grazing. China, with 1.3 million ha sown, is a major producer of peas for green pods/seed (FAOSTAT Citation2012). Although pea is currently recognized as a protein crop (20-25%), its potential is also as a source of high-quality starch (up to 50%) and even putatively as an oil (1-5%) source (Khodapanahi et al., Citation2012). The key breeding objectives involve increasing yield potential by improving biotic and abiotic stress resistances and enhancing quality for diverse food markets. Quality includes improved appearance of the seeds, as well as improved nutritional value, cooking quality and flavor.
The energy and health benefits of peas derive mainly from the concentration and properties of starch, protein, fiber, vitamins, minerals and phytochemicals. Fiber from the seed coat and the cell walls of the cotyledon contributes to gastrointestinal function and health and reduces the digestibility of starch in peas. The intermediate amylose content of pea starch also contributes to its lower glycaemic index and reduced starch digestibility. Pea protein, when hydrolyzed, may yield peptides with bioactivities, including angiotensin I-converting enzyme inhibitor activity and antioxidant activity. The vitamin and mineral contents of peas may play important roles in the prevention of deficiency-related diseases, specifically those related to deficiencies of Se or folate (Dahl et al., Citation2012). Peas contain a variety of phytochemicals once thought to be only anti-nutritive factors. These include polyphenolics, in colored seed coat types in particular, which may have antioxidant and anticarcinogenic activity, saponins, which may exhibit hypocholesterolaemic and anticarcinogenic activity, and galactose oligosaccharides, which may exert beneficial prebiotic effects in the large intestine (Dahl et al., Citation2012). Development of low-phytate cultivars has become an important objective in several crops, including pea. This is because a major part of the total phosphorus in pea seeds is stored as phytate, an organic molecule that binds with some mineral cations and is excreted due to the lack of phytase enzymes in humans and non-ruminant animals. This causes nutrient deficiency as well as environmental pollution. Chemical mutagenesis led to identification of low-phytate pea lines (Rehman et al., Citation2012).
Genetic diversity available in wild Pisum species has been so far poorly exploited. Several large studies were published primarily for quantitative disease reactions (Infantino et al., Citation2006; Tivoli et al., Citation2006; Sillero et al., Citation2006). Many pea germplasm screens have been conducted for biotic and abiotic stresses, agronomic traits and seed quality (e.g., nutrition) but the studies are small (less than 20 accessions), the data is unavailable, published in difficult-to-access sources or unpublished. The most attention has been given to P. fulvum, as a donor of bruchid resistance and source of novel powdery mildew resistance (Clement et al., Citation2002, 2009; Fondevilla et al., Citation2007, 2008; Byrne et al., Citation2008). Incomplete levels of resistance to powdery mildew, rust (Uromyces pisi (Pers.) Wint.), crenate broomrape (Orobanche crenata) and Mycosphaerella pinodes are available in accessions of P. sativum subsp. sativum, subsp. abyssinicum, subsp. elatius and P. fulvum (Fondevilla et al., Citation2005, 2007, 2011; Rubiales et al., Citation2005, 2009; Barilli et al., Citation2009). Wild Pisum in its native range displays a typical winter habit in which plants germinate in autumn, overwinter in the vegetative state, and flower in response to increasing day-length in spring (Abbo et al., 2003; Weller et al., Citation2009, 2012). Wild Pisum was identified as a source of alleles of flowering locus Hr implicated to influence winter frost tolerance (Lejeune-Hénaut et al., Citation2008). Moreover, the flowering allele Hr enhances the capacity of pea photoperiodic lines to produce basal laterals, which is often found in primitive accessions of Pisum sativum subsp. sativum; P. sativum subsp. elatius and P. fulvum (Weller et al., Citation2009, 2012). The majority of cultivated pea accessions from higher latitudes have a quantitative long-day response and are grown as a spring crop, while the obligate or near-obligate requirement for long-days suits pea to a winter cropping cycle and has been retained in some forage cultivars (Weller et al., Citation2009, 2012). There is some interest to develop pea as winter crop in order to escape drought and heat stress during flowering and seed set periods.
D. Genus Lens Miller
The lentils (Lens Miller), a small genus of Mediterranean origin, are nested in the core clade of the genus Vicia (Schaefer et al., Citation2012). Different taxonomists have recognized different numbers of species within the genus. There were considered to be five lentil species: Lens culinaris Medik., L. orientalis Popow, L. ervoides Grande, L. nigricans (M. Bieb.) Godr. and L. montbretii (Fisch. & C. A. Mey.) P. H. Davis & Plitm. (Cubero, Citation1981).
Lens montbretii has been transferred from the genus Lens to the genus Vicia on the basis of its different morphology and cytology, with 2n = 12 chromosomes (Ladizinsky and Sakar, Citation1982). Lens lamottei was distinguished on the basis of an herbarium specimen of L. nigricans (Czefranova, Citation1971). An additional species L. odemensis was recognized by Ladizinsky et al. (Citation1984) as a new species due to the difference in stipules from L. nigricans (Ladizinsky, Citation1986). As the last discovered taxon, L. tomentosus was described as distinct from L. orientalis by tomentose pods, a minute satellite and one large, metacentric chromosome (Ladizinsky, Citation1997). Current classification recognizes one cultivated lentil (L. culinaris subsp. culinaris) and six related taxa: L. culinaris subsp. orientalis, L. culinaris subsp. tomentosus, L. culinaris subsp. odemensis, L. ervoides, L. nigricans and L. lamottei (Ferguson et al., Citation2000). The wild relatives of the cultivated lentil have a wide distribution. L. culinaris subsp. orientalis (Boiss.) Ponert, naturally distributed from Turkey to Uzbekistan, is considered the putative progenitor of the cultivated lentils (Ladizinsky, Citation1979a). Lens culinaris subsp. tomentosus is restricted to northern Syria, Iraq and eastern Turkey; L. ervoides occurs along the eastern Mediterranean coast to former Yugoslavia, often in shady habitats, such as pine plantations; L. lamottei is found in Morocco, Spain and Southern France; and L. nigricans occurs from southwest Turkey to the southwestern Mediterannean (Ferguson and Erskine, Citation2001).
The cultivated lentils were divided into two subspecies by Barulina (Citation1930) and two races by Cubero (Citation1981), the large-seeded macrosperma and small-seeded microsperma race. The small-seeded varietal group is traditionally grown in the Middle East, South Asia, North Africa, and mainly in Turkey, whereas the large-seeded varietal group is usually grown in Canada (FAOSTAT, Citation2013) and large seeded varietal group is grown in the Americas and Southern Europe (Sekhon et al., 2007).
Lentil is a self-pollinated species with cleistogamous flowers and consequently usually has <0.8% natural cross pollination (Wilson and Law, Citation1972). All species of Lens have a chromosome number of 2n = 2x = 14, which is also inferred as the ancestral number for the clade (Schaefer et al., Citation2012). The genome size is estimated to be 1C = 4.20 pg, corresponding to 4,063 Mb/C (Arumuganathan and Earle, Citation1991).
Phylogeny and genetic diversity of the genus Lens has been studied first by seed protein electrophoresis (Ladizinsky, Citation1979b; Sultana et al., Citation2006) and isozyme markers (Zamir and Ladizinsky, Citation1984; Pinkas et al., Citation1985; Hoffman et al., Citation1986; Muehlbauer et al., Citation1989; Erskine and Muehlbauer, Citation1991; Mayer and Soltis, Citation1994; Ferguson and Robertson, Citation1996). The DNA-based markers, such as RFLP (Havey and Muehlbauer, Citation1989; Muench et al., Citation1991; Rajora and Mahon, Citation1994), RAPD (Sharma et al., Citation1995; Ferguson et al., Citation2000; Sonnante and Pignone, Citation2001; Toklu et al., Citation2009; Tewari et al., Citation2012), AFLP (Sharma et al., Citation1996; Zavodna et al., Citation2000; Duran and Perez de le Vega, 2004; Kahraman et al., Citation2004; Rubeena et al., Citation2006; Fiocchetti et al., Citation2009; Toklu et al., Citation2009), microsatellite markers (Duran and Perez de le Vega, 2004; Hamwieh et al., Citation2009; Liu et al., Citation2008; Babayeva et al., Citation2009; Reddy et al., Citation2010; Gupta et al., Citation2012, Tewari et al., Citation2012; Zaccardelli et al., Citation2012), inter simple sequence repeat (ISSR) (Zavodna et al., Citation2000; Sonnante and Pignone, Citation2001; Toklu et al., Citation2009), internal transcribed spacers (ITS) (Fernandez et al., Citation2000; Mayer and Bagga, Citation2002; Sonnante et al., Citation2003), non-transcribed spacer (NTS) (Fernandez et al., 2005), sequenced tagged microsatellite site (STMS) (Inder et al., Citation2008; Datta et al., Citation2011), single-nucleotide polymorphisms (SNPs) (Alo et al., Citation2011), and resistance gene analogue (RGA) (Yaish et al., Citation2004; Sari et al., Citation2013) were used to analyze phylogenetic relationships among taxa in the genus Lens, as well as the genetic diversity of cultivated lentil.
1. Crop Lentil (Lens culinaris Medik.)
Ex situ collections of lentil number over 58,407 and are held in 12+ collections (Tullu et al., Citation2011, ). This world collection is shared among 40 genebanks, which have a large amount of cross-duplication. The world lentil collection is held by ICARDA and holds 10,000+ accessions, followed by India (7,712 acc.), Australia (5,254 acc.), USA (3,187 acc.) and the Vavilov Institute, Russia (2,556 acc.). Most other national collections hold some portion of the subsets of this collection (Coyne and McGee, Citation2013). The ICARDA has globally mandated research for lentil improvements; it holds roughly 600 wild accessions (Redden et al., Citation2007) and the largest collection of wild Lens accessions from 12 countries (Furman, Citation2006). The wild relative collections total 852 accessions (GENESYS Citation2013, Redden pers comm, for ATFCC) of 6 wild Lens taxa (L. culinaris subsp. orientalis, L. odemensis, L. tomentosus, L. ervoides, L. montbretii, and L. nigricans) (Ferguson et al., Citation2000; Sarker and Erskine, Citation2006) representing 24 countries. Ex situ collections give priority to the conservation of lentil landraces in order to maximize domestic diversity, as well as to conserve cultivars and landraces that have valuable combinations of traits and assembled linkage groups of valuable genes (Furman et al., Citation2009). Documentation of agronomic and descriptor traits in the lentil genepool exists across more than half of the world lentil collection at International Lentil Information System (ILIS). ILIS enabled the lentil germplasm to be explored for multiple traits a with search/query program, to target the preferred germplasm for breeders to assess, or for targeted acquisition of germplasm from other genebanks (Balachandra et al., Citation2006; Redden et al., 2007). With the integration of the ATFCC collections into the Australian Grains Genebank (AGG) in 2013, the ILIS database will be transferred into GRIN-Global.
Large-scale genotypic characterization of lentil genetic resources is lacking. The largest published study used sequences of 22 genes from 308 lentil accessions (133 cultivated and 175 wild) to determine the population structure (K = 8) and to propose theories on taxonomy and domestication origins (Alo et al., Citation2011). The next largest study was conducted on an ICARDA core collection of 57 cultivated and 52 wild lentil accessions (Hamwieh et al., Citation2009). Cluster analysis based on SSRs defined the two groups into two unsurprising clusters, cultivated and wild. A comparison of 30 landraces from South Asia to 130 from 13 other countries using RAPDs and isozymes was performed by Ferguson et al. (Citation1998). Numerous small-scale (less than 45 lentil lines) studies have been published using various DNA marker classes: RAPDs (Fikiru et al., Citation2010; Hoque and Hasan, Citation2012), ISSRs (Fikiru et al., Citation2011), AFLPs and SSRs (Sultana and Ghafoor Citation2008; Babayeva et al., Citation2009; Reddy et al., Citation2010; Datta et al., Citation2011; Zaccardelli et al., Citation2012). Seed proteins of 13 polymorphic peptides were investigated in 144 lentil landraces collected in Pakistan (Sultana et al., Citation2006). Several lentil core collections have been assembled (Erskine and Muehlbauer, Citation1991; Simon and Hannan, Citation1995; Furman, Citation2006; Hamwieh et al., Citation2009). 4,036 ICARDA accessions were evaluated for quantitative characteristics of time to flower, time to maturity, plant height, and lowest pod height (Erskine et al., Citation1989). Another study examined the relationship between the yield of seed and straw of 3,586 ICARDA accessions (Erskine, 1983). Response to temperature and photoperiod effect on time to flowering was investigated in 231 and 369 ICARDA accessions (Erskine et al., Citation1990, Citation1994, respectively). A study of 3,512 ICARDA accessions noted the presence of iron deficiency in a calcareous soil linked to geographical origin (Erskine et al., Citation1993). Similarly, within 495 ICARDA accessions grown in boron-deficient soil, yields revealed striking genetic differences associated with geographic origin (Srivastava et al., Citation2000). Next, a study screened 310 lines for growth in soil with a high concentration of boron and identified tolerance in accessions from Afghanistan and Ethiopia (Hobson et al., Citation2006). Characterization of the USDA lentil core collection of 287 lines identified useful trait variation for phenology, morphology, seed and straw yields for use in breeding (Tullu et al., Citation2001). Lentil is confounded by a number of important production constraints, particularly biotic stresses (Chen et al., Citation2009, 2011; Podder et al., Citation2012). The most important of these diseases on a global scale are ascochyta blight (Ascochyta lentis Bond. & Vassil.), rust (Uromyces viciae-fabae (Pers.) Schroet), botrytis grey mold (Botrytis cinera Pers. Ex Fr. and Botrytis fabae Sard), anthracnose (Colletotrichum truncatum (Schw.) Andrus & Moore), stemphylium blight (Stemphylium botryosum Wallr.), powdery mildew (Erysiphe pisi DC.), fusarium wilt (Fusarium oxysporum Schlecht. :Fr. f.sp. lentis Vasudeva and Srinivasan), sclerotinia stem rot (Sclerotinia sclerotiorum (Lib.) de Bary) and broomrape (Orobanche crenata Forssk) with several wild lentil species and accession found to be source of resistances (Buchwaldt et al., Citation2004; Fernández-Aparicio et al., Citation2008, 2009; Chen et al., Citation2009; Tullu et al., Citation2006; 2010; Vail et al., Citation2012; Rubiales et al., Citation2013; Shaikh et al., Citation2013).
Lentil is a highly nutritious food, high in protein, minerals and vitamins (Bhatty, Citation1988). It is consumed as a soup and forms the protein staple for a large portion of Asia. Consumption is particularly high in Sri Lanka and Nepal, followed by Syria and Turkey (Erskine, Citation2009). Lentils are combined with the carbohydrate staples rice and wheat, forming a complete protein diet (Erskine, Citation2009). Lentil has the potential to reduce micronutrient (iron and zinc) deficiency though the consumption of 100 g daily (Thavarajah et al., Citation2009). Additionally, lentil is a promising source of antioxidant phenolics and could serve as a dietary supplement (Zou et al., Citation2011). Lentil is typically grown in dryland cropping systems in rotation with grains, such as wheat and rice (Materne and Siddique, Citation2009). World production was 4.55 M tons in 2012 with mean yield of 1,070 kg/ha. Leading producers include Canada, with 1.49 M tons, followed by India (0.95 M tons), Australia (0.46 M tons), Turkey (0.44 M tons), USA (0.24 M tons) and Nepal (0.21 M tons) (FAOSTAT, Citation2012). While India has increased production over the last five years, Canada's production more than doubled in the same time period (Erskine, Citation2009; FAOSTAT Citation2012). Regionally, Asia produces close to half of the world's lentils (2.07 M tons).
IV. TRIBE TRIFOLIEAE (BRONN) ENDL.
A. Genus Medicago L.
The tribe Trifolieae, subtribe Trigonellinae, includes Medicago L. (alfalfa) and Trifolium L. (clover), along with Melilotus (L.) Mill. (sweet clover) and Trigonella L. (Maureira-Butler et al., Citation2008), with a total of c. 87 species in Medicago. Phylogenetic relationships within the genus have proved difficult to resolve (Maureira-Butler et al., Citation2008; Steel et al., Citation2010). Using morphological traits, Small and Jomphe (Citation1989) proposed 12 sections and 8 subsections. However, incongruence has been reported between morphological and molecular inferences (Bena, Citation1998). Incongruence has also been reported in an analysis using two nuclear-encoded genes and a mitochondrial gene (Maureira-Butler et al., Citation2008). Results of a phylogenetic analysis of plastid trnK/matK and nuclear GA3ox1 sequences supported the previously recognized groups, sect. Medicago and sect. Buceras, but suggested that M. arborea L., M. citrina (Font Quer) Greuter and M. strasseri Greuter, Matthäs & Risse, currently in sect. Dendrotelis, be moved to sect. Medicago. There was also little support for sect. Lupularia and sect. Platycarpa (Steele et al., Citation2010). Difficulty in resolving the Medicago phylogeny may be due to historic and ongoing hybridization within the genus (Maureira-Butler et al., Citation2008; Steele, et al., Citation2010). More recently, Yoder et al. (Citation2013) used whole-genome sequence data from 29 Medicago taxa to examine phylogenetic relationships. Using 87,596 polymorphic single-nucleotide sites, Yoder et al. (Citation2013) found that the consensus topography was consistent with previous classifications of major sections and subsections and was also able to resolve ambiguities among several species.
Alfalfa is part of the Medicago sativa complex that includes diploid (2n = 16) and tetraploid (2n = 32) forms that have either blue flowers and coiled pods or yellow-flowers and sickle-shaped pods. Some forms are characterized by the presence of glandular trichomes on pods. Small (Citation2011) proposed the most recent taxonomic classification based on ploidy level, hybridization, flower color, fruit coiling and the presence of glandular hairs on fruits. Using the classification of Small (Citation2011), taxa within the M. sativa complex fall into either the primary or secondary gene pool of alfalfa, depending on ploidy level. Crop wild relatives (CWR) in the primary alfalfa gene pool include the tetraploid forms of M. sativa subsp. falcata, M. sativa L. subsp. glomerata (Balb.) Rouy, M. sativa subsp. sativa, M. sativa L. nothosubsp. tunetana Murb., M. sativa L. nothosubsp. varia, and M. sativa subsp. falcata var. viscosa (Rchb.) Posp. (USDA GRIN 2013, ). References outlining the intraspecific and interspecific crossing studies used to designate the alfalfa gene pool can be found in Small (Citation2011).
Alfalfa is a highly diverse crop. It is a perennial, autotetraploid (2n = 4x = 32) (McCoy and Bingham, Citation1988), and is cross-pollinated by insects, predominately bees. The facts that taxa within the Medicago sativa complex occur sympatrically, that taxa at the same ploidy level freely intercross, and that the ploidy barrier is relatively weak due to the frequency of gamete reduction contribute substantially to the diversity seen in alfalfa (Kaljund and Leht, Citation2013). Taxa within the Medicago sativa complex extend across Eurasia, from the British Isles into Eastern Siberia, southward around the northern rim of the Mediterranean and the Black Sea, and south into Eastern Turkey, Northern Iraq and Iran, and East into Kazakhstan. M. sativa subsp. glomerata extends into Northern Algeria. Phylogeographic analysis provides ample evidence that germplasm within the Medicago sativa complex exhibits extensive adaptation to a broad array of environments (Sakiroglu et al., Citation2010; Sakiroglu and Brummer, Citation2013). Medicago sativa subsp. sativa is found growing in the steppe, in fertile meadows, and even on sand dunes. The species grows best in fertile, moist soil at a pH of 6.0-6.5. The diploid version of subsp. sativa, M. sativa subsp. caerulea is more drought tolerant, and some ecotypes are adapted to saline soils (Lubenets, Citation1953). Medicago sativa subsp. falcata is adapted to the dry cold steppe area and may be more broadly adapted than subsp. sativa (Oakley, Citation1917; Lesins and Lesins, Citation1979). M. sativa subsp. glomerata occurs in moist, montane areas (Small, Citation2011). Medicago sativa subsp. falcata var. viscosa and M. sativa subsp. varia possess the same broad adaptation as M. sativa subsp. falcata. Although these infraspecific taxa can be distinguished by flower color and pod shape, numerous ecotypes reflect extensive variation within taxa with regard to leaf size and shape, growth habit (upright to prostrate), leaf and pod pubescence, and degree of pod curl. This tremendous diversity is reflected in the numerous synonyms associated with the species. Small (Citation2011) provides a list of synonyms in his recent monograph.
1. Crop alfalfa (Medicago sativa L.)
There are over 91,000 accessions of Medicago held at major gene banks globally (FAO Citation2010, ). Almost half of the global collection is represented by wild species (47%), while landraces, breeding lines and advanced cultivars make up 6%, 7%, and 6% of the global collection, respectively. Thirty-four percent of the collection is made up of unknown accession types (FAO, Citation2010). Greene et al. (Citation2012) examined the representation of global ex situ collections for alfalfa CWR species native to the Russian Federation and neighboring countries. They found that representation of the Crimea, Mountain Central Asia (with the exception of subsp. sativa) and Eastern Siberia was weak, despite the fact that these are important areas of diversity and adaptation to extreme environments. Taxa with limited representation included M. sativa subsp. falcata var viscosa and M. sativa subsp. glomerata, whose tetraploid versions are in the primary gene pool and whose diploid versions are in the secondary gene pools (). Underrepresented CWR in the tertiary genepool included M. saxatilis, M. papillosa, M. rupestris, M. daghestanica, and M. marina (Greene et al., Citation2012). Parts of the world where valuable alfalfa germplasm occurs but has not been extensively sampled include Iran, Iraq, Afghanistan and Northern Pakistan (Bauchan and Greene, Citation2002). In a recent global gap analysis of 13 alfalfa CWR species, 70% of the species were ranked as high-priority species to collect, 15% were medium priority and 15% were low priority (CWR and Climate Change Citation2013a). Examining the online interactive map, collecting gaps for high-priority alfalfa CWR taxa include the southeastern part of the Crimea peninsula, southern Georgia, Armenia and parts of Turkey (CWR and Climate Change, 2013b). Although there are collection gaps, utilization of alfalfa germplasm is not hampered by a lack of diversity in ex situ collections, but rather by the challenges of evaluation and prebreeding. Substantial effort was made from the early 1980s to the mid 1990s to evaluate the USDA alfalfa collection. Recent efforts have provided chromosome counts on taxa within the Medicago sativa complex that have diploid and tetraploid forms (Brummer et al., Citation1999; Sakiroglu and Brummer, Citation2011; Sakiroglu and Kaya, Citation2012). Currently, about a third of the collection has been evaluated for resistance to 13 diseases, seven insects, seven feed quality traits, and five abiotic stress tolerance traits. These data are publically available in GRIN (Bauchan and Greene, Citation2002).
Alfalfa, also known as lucerne, is the most widely grown forage legume in the world. It is difficult to overstate the importance of alfalfa in the world economy. In the United States, alfalfa routinely places among the top five crops in the nation in terms of both farmgate value and total acreage. In terms of protein production, alfalfa placed third, behind soybeans and corn, in 2009. From a global perspective, alfalfa is among the top 10 crops for protein production (Sumner and Rosen-Molina, Citation2011). In 2009, the FAO estimated that alfalfa was grown on approximately 30 million hectares worldwide; 66% was produced in North America and Europe, 23% in South America, and the remainder in Asia, Africa and Oceania (FAO, Citation2013). Valuable characteristics of alfalfa include adaptation to a wide range of climates, ability to fix up to 560 kg/ha atmospheric nitrogen per year, production of large amounts of biomass that is highly nutritious and between 15 and 22% crude protein, production of sweet nectar that attracts bees, deep tap roots that improve soil tilth and a perennial growth habit that reduces soil erosion. Alfalfa is used primarily as animal feed in the form of forage and fodder. It is especially important for dairy cows but is also an important feed for horses, beef cattle, sheep, chickens, turkeys and other farm animals. Alfalfa is also used as a green manure, as a rotation crop and as ground cover (Small, Citation2011). Its characteristics make it a valuable crop for supporting biodiversity and agroecosystem services (Putnam, Citation2001). In Australia, alfalfa has been used to reduce soil salinization (Robertson, Citation2006), and in the United States, cultivars have been developed to support bioremediation of high-nitrogen soils (Russelle, Citation2007). Alfalfa is also consumed directly by humans as alfalfa sprouts, juice and powder. Potential new uses include biofuel and the production of industrial enzymes, such as peroxidase, alpha-amylase, cellulase, and phytase (Small, Citation2011).
Conventional alfalfa breeding programs generally identify useful germplasm in nurseries or pest and disease screening trials. Selections are then incorporated into elite populations using phenotypic or genotypic recurrent selection. Synthetic cultivars are then developed by intercrossing individual plants and harvesting equal quantities of seed from each parent, which is bulked to form the Synthetic 1 generation. However, MAS and genetic engineering are being adopted, especially in private industry. Reich (Citation2012) describes efforts to develop cultivars resistant to saline soils using conventional breeding techniques along with marker-assisted selection and genetic engineering to reach their goals. Genetic engineering efforts are also focused on developing alfalfa that is more nutritious by decreasing lignin production and increasing tannin production (McCaslin and Reisen, Citation2012).
B. Genus Trifolium L.
Trifolium is one of the largest genera in the tribe, with about 255 species (Zohary and Heller, Citation1984; Gillett and Taylor, Citation2001). The genus is cosmopolitan, with species that occur mostly in the northern hemisphere. Primary centers of diversity are Eurasia (150-160 species), North America (60-65 species) and Africa (25-30 species). Over half of its species originated in the Mediterranean region. While most species occupy temperate and subtropical regions, some occur in the Tropics of West Africa and South America, where they are generally restricted to montane and alpine zones (Raven and Polhill, Citation1981; Zohary and Heller, Citation1984; Small, Citation1989). The genus includes annuals and perennials (Watson et al., Citation2000). Trifolium species occur in a wide range of habitats, including meadows and prairies, open woodlands, semi-deserts, mountains, and alpine peaks. A common feature of these diverse habitats is high solar radiation; few clover species tolerate shade.
In some studies (Roskov, Citation1989), the genus is divided into three separate genera: Chrysaspis Desv. (pavis free, wings and keel knitted only in the basal part), Amoria C. Presl. (pavis free, wings and keel knitted into a tube) and Trifolium s. str. (all petals knitted into a tube). The genus Chrysaspis is very close to the genus Melilotus (Roskov, Citation1989). The genus name refers to the distinctive leaves, which are usually composed of three leaflets (trifoliolate). Trifolium is a member of a large, monophyletic clade of 45 genera – mostly the temperate and herbaceous (Polhill, Citation1981; Doyle, Citation1995). Trifolium belongs to the vicioid subclade and is closely related to Medicago, Melilotus, and Trigonella. Altogether they comprise Trifolieae s. str. or subtribe Trigonellinae of Trifolieae s. l. (Zohary, Citation1972). Trifolium is the only one of these four genera with some species restricted to the New World. Trifolium differs from these allied genera by ovule morphology and by the position of seeds in the pod (Heyn, Citation1981). Wings are clawed, and keels are adnate to the staminate fascicle (Hossain, Citation1961).
There are some species that appear to be intermediate between all four closely allied genera (Heyn, Citation1981). Nuclear and chloroplast sequences from a variety of genes and genic regions support the monophyly of Trifolium (Steele et al., 1998, 1999). Within Trifolieae tribe s. str., Trifolium is more derived because of a trend towards reduction of pod size, reduction of the number of seeds per pod, and loss of the pod septum, resulting in indehiscence and unique fruit dispersal accessories (Zohary and Heller, Citation1984; Small, Citation1989). Most contemporary classifications recognize Trifolium as one large genus of eight sections: Lotoidea, Paramesus, Mistyllus, Vesicaria, Chronosemium, Trifolium, Trichocephalum and Involucrarium (Hossain, Citation1961; Zohary and Heller, Citation1984; Ellison et al., 1996). Bobrov (Citation1967) recognized nine to eleven smaller, segregated genera. Biogeography, morphological evolution, and the existing classification for Trifolium were examined by Watson et al. (Citation2000). Trifolium was found to be monophyletic. The two largest sections of the genus, sections Lotoidea and Trifolium, are not monophyletic; only one small section (Chronosemium) is. Sect. Lotoidea is the largest section and is considered ancestral to all other sections on the basis of a worldwide distribution, large size (over 90 species), and morphological heterogeneity among species (Zohary, Citation1972; Zohary and Heller, Citation1984). Six sections occur in the Old World, primarily in Eurasia, with some species extending into Africa. One section (Involucrarium) is restricted to the New World and occurs in both North and South America. The monophyly of a clade with New World species of sections Involucrarium and Lotoidea is confirmed by molecular data, even if no section is considered to be monophyletic (Steiner et al., Citation1997).
Some contrasting views have been proposed on the origin and radiation of Trifolium. Most frequently mentioned is the Mediterranean origin of the genus, probably in the Early Miocene. A single origin of all North and South American species is hypothesized, while the species of sub-Saharan Africa may originate from three separate dispersal events (Ellison et al., Citation2006). Gillett (Citation1952) and Taylor (Citation1985) suggested a Mediterranean origin on the basis of species diversity, morphological heterogeneity, and chromosome number. Raven and Polhill (198l) agreed that Trifolium is of Eurasian origin and suggested that repeated migrations to North America occurred. These migrations were followed by multiple radiations and dispersal events. Old World origin for Trifolium is in contrast to the hypothesis of Zohary (Citation1972) that the genus originated in North America and that migration and colonization led the species via the Bering Strait to Asia, followed by a series of secondary radiations in the Mediterranean region and dispersal into Africa. The molecular data support a Mediterranean origin of Trifolium, with the New World clade deeply nested among the Old World species. Zohary and Heller (Citation1984) indicated two regions of interest for genus evolution: the Mediterranean region, with 110 species belonging to seven sections, and the Californian region, which includes a smaller number of species.
A hypothetical ancestral form for Trifolium was described as a perennial with large flowers on bracteate pedicels, with a simple calyx, choripetalous corolla, and numerous ovules (Celakovsky, Citation1874; Bobrov, Citation1967). Some species of section Lotoidea are similar to this proposed archetype (Zohary and Heller, Citation1984). The annual habit evolved from a perennial repeatedly. Most species of Trifolium are diploid, with approximately 20% polyploidy occurrence. Most of the polyploid species are Old World perennials, with 65% in section Lotoidea (Zohary and Heller, Citation1984). Hybridization among Trifolium species is rare or non-existant (Wexelsen, Citation1928; Zohary, Citation1972). Chromosome numbers are known for at least 184 species of Trifolium (Zohary and Heller, Citation1984). Over 80% of the examined species are 2n = 16, and x = 8 is the basal number of the genus (Goldblatt, Citation1981). Aneuploidy (2n = 10, 12, or 14) has been identified in 31 species, eleven of which have both aneuploid and diploid (2n = 16) or polyploid counts. Polyploidy has been identified in 24 species, of which six are exclusively tetraploid, two are hexaploid, and one is dodecaploid. Eleven species have both diploid and polyploid counts, while three have multiple polyploid counts at the tetraploid level and above. Nitrogen-fixing root nodules have been reported in over 125 species of clover (Sprent, Citation2001).
1. Crop clover (Trifolium)
The genus Trifolium includes about ten species of agricultural significance. The most important are white clover (Trifolium repens L.) and red clover (T. pratense L). Clovers are of global agricultural significance as forage species, particularly important in temperate areas, both for direct grazing and for conserved forage (Zohary and Heller, Citation1984). At least 16 species of Trifolium are actively cultivated (Gillett and Taylor, Citation2001, ), a fairly large number for a single genus. Many native species are also used for animal grazing (Crampton, Citation1985). Overall, approximately 74,000 accessions of Trifolium are held in global ex situ collections, 53% of which are wild, 14% of which are cultivated and 33% are of unknown improvement status (FAO, Citation2010, ). The largest collections are held by Australia, New Zealand, and ICARDA (FAO, Citation2010). The Australian Temperate Forage Legume Center (Trifolium), Perth, Western Australia, has 119 species and 11,000 accessions. The Genebank of the VIR, Saint Petersburg, Russian Federation, has 4,605 accessions of the clover genus. ICARDA conserves 4,536 accessions. USDA conserves 6,229 accessions of the genus: 815 accessions of T. repens, 1,367 accessions of T. pratense, and 4,047 accessions of other Trifolium species. The Leibniz Institute of Plant Genetics and Crop Plant Research has collectively 1,657 accessions, of which 136 acc. are of T. repens, 572 acc. are of T. pratense and 949 acc. are of other Trifolium species. Thanks to the efforts of Dr. Norman Taylor, the U.S. collection houses samples of almost all species in the genus and has accessions from 74 countries (GRIN, 2013). The International Livestock Research Institute genebank, in Addis Ababa, Ethiopia, holds an important collection of wild clovers native to Ethiopia and other African countries. Significant collections of breeding lines and cultivars are also held by AgResearch, New Zealand (Margot Forde Forage Germplasm Center). Like most germplasm collections of crop species, agriculturally important clover species are well represented in genebanks, while wild species frequently are represented by only a few accessions. In reviewing the U.S. clover collection, Morris and Greene (2001) concluded that it contained gaps for (i) cultivars and landraces of red and white clover originating from China, Japan, South America, and South Africa; (ii) obsolete cultivars developed in the USA; (iii) minor-use species; (iv) related wild species; and (v) germplasm distinguished by traits that may be of value to the nutritional supplement or bioremediation industries and that may convey adaptation to abiotic stress or be supportive of sustainable agriculture. Other important questions that need to be examined in order to determine the global ex situ coverage of Trifolium include the extent to which current collections represent an appropriate level of geographical coverage around the globe and to what degree the major collections are sampling independent geographical regions (Abberton and Thomas, Citation2011). The study of variation in clover germplasm collections based on morphological, phenological, and agronomical characters and molecular markers has resulted in the development of core collections (Kouame and Quesenberry, Citation1993; Vymyslický et al., 2010, 2012). No concordance between morphologic and RAPD marker classification of wild red clover populations was also reported by Greene et al. (Citation2004). White clover wild relatives often display contrasting phenotypes for agriculturally important traits, such as drought tolerance (T. occidentale D. E. Coombe), cold tolerance (T. pallescens Schreb.), high inflorescence and seed set (T. nigrescens Viv.), presence/absence of stolons (T. occidentale vs. T. pallescens and T. nigrescens), and annual/perennial growth habit (T. nigrescens vs. T. occidentale). Generation of segregating progenies from these crosses would enable mapping of these traits and the discovery of the hidden genes. This could have dramatic consequences on clover breeding programs (Ravagnani et al., Citation2012).
White clover (T. repens L.) is a perennial legume and the primary legume found in grazed pastures in most parts of the world. It could be also used for silage. Most often, it is used in mixed swards with grasses – mostly perennial ryegrass (Lolium perenne L.). A very important attribute of white clover is its nitrogen fixation, contributing about 250 kg N/ha per year in mixed grassland. White clover develops a dense network of stolons, which enhances grazing tolerance, winter hardiness and persistence. Resistance to pests and diseases, efficient use of water and nutrients, and compatibility with grass are important targets of white clover breeding (Abberton and Thomas, Citation2011). Breeding programs for white clover are carried out throughout the world: New Zealand (Williams et al., Citation2007), Australia (Lane et al., Citation1997; Jahufer et al., Citation2002), the United States (Taylor, Citation2008) and the UK (Abberton and Marshall, Citation2010). They are focused on particular environments and management systems but share the objectives of more fully realizing the potential of white clover to contribute to livestock nutrition and soil fertility (Abberton and Thomas, Citation2011).
Red clover (T. pratense) is an important perennial legume in many parts of the world with oceanic climate: Western and Northern Europe, parts of Russia, Japan and the USA (Taylor and Smith, Citation1995). It has erect stems emerging from a meristematic ‘crown.’ Red clover is a high-yielding forage crop under optimal climates, is typically used for silage and is not tolerant to intensive grazing. Red clover cultivars are classified by ploidy level and by maturity. Tetraploid cultivars are artificially produced by chromosome doubling of diploid lines. Early flowering or ‘double cut’ cultivars are widely grown and give two more or less equal conservation cuts and subsequent lower yielding cuts. Late flowering or ‘single cut’ types give a greater proportion of their yield at the first cut. Red clover is traditionally important in organic farming systems, where it is used as a source of nitrogen in crop rotation and for its high protein content in modern silage technologies. Red clover is an important feed resource for pollinating insects (Abberton and Thomas, Citation2011).
Important breeding objectives include yield, persistence and pest and disease resistance (Boller et al., Citation2010). Programs of interspecific hybridization between red clover and related species were reviewed by Abberton (Citation2007). The main emphasis has been increasing longevity through crosses with more persistent species, particularly T. medium L. (Jakesova et al., Citation2011). Many other clover species are used in agriculture as minor crops with different purposes. Clovers are very popular for pollinators, especially T. hybridum L., T. resupinatum L., T. pannonicum Jacq., T. alexandrinum L., and T. incarnatum L. (Ishii, Citation2013). A number of minor clover species may also be important with respect to future breeding efforts. Among these are the species most closely related to the putative diploid ancestors of white clover, namely T. pallescens and T. occidentale. Other species, such as T. elegans Savi, are of local agricultural importance, and consideration with respect to conservation and use needs to be given to all of them. T. diffusum Ehrh. has been used in programs of interspecific hybridization with red clover (Strzyzewska, Citation1995).
V. TRIBE CICEREAE
A. Genus Cicer L.
The genus Cicer was transferred from the tribe Vicieae Alefeld to its own tribe, Cicereae Alef., due to some morphological differences (Kupicha, Citation1977). Currently, Cicer includes 44 taxa, 9 annuals and 35 perennials (van der Maesen et al., Citation2007; Davies et al., Citation2007). The following taxa, C. uludereensis Dönmez (Dönmez, Citation2011), C. floribundum Fenzl. var. amanicola M. Öztürk & A. Duran (Öztürk et al., 2011), C. heterophyllum Contandr., Pamukc. & Quezel var. kassianum M.Öztürk & A.Duran (Öztürk, 2011) and C. incisum (Willd.) K. Maly subsp. serpentinica M. Öztürk & A. Duran (Öztürk et al., 2013) have recently been discovered in Turkey. The most widely known species in the genus Cicer is the cultivated chickpea (Cicer arietinum L.), with 2n = 2x = 16 chromosomes and a genome size of ∼738 Mb (Varshney et al., Citation2013a). The ‘macrosperma’ or ‘kabuli’ and ‘microsperma’ or ‘desi’ chickpeas are distinguished on the basis of size and coloration of seeds and flowers and pigmentation on plants (Muehlbauer and Singh, Citation1987). Both of the cultivated chickpeas are thought to be derived from C. reticulatum (Ladizinsky and Adler, 1976; Toker, Citation2009), which is native to southeastern Turkey and northern Syria (Ladizinsky and Adler, 1976; Zohary and Hopf, Citation2000, Toker, Citation2009). According to the classical definition of Harlan and deWet (1971), there is a primary (C. arietinum and C. reticulatum), secondary (C. echinospermum P. H. Davis), and tertiary gene pool (C. pinnatifidum Jaub. & Spach., C. bijugum Rech. f., C. judaicum Boiss, C. yamashitae Kitam., C. chorassanicum Popow, and C. cuneatum Hochst. ex A. Rich. and perennial wild Cicer species) (Ahmad et al., 1988; ).
1. Crop chickpea (Cicer arietinum L.)
The ex-situ collections of chickpea landraces and wild relatives are stored in 44 genebanks worldwide. These collections hold a combined 98,313 accessions, with largest collections at the International Crop Research Institute for the Semi-Arid Tropics (ICRISAT) in India (20,140 accessions) and ICARDA in Syria (13,818 accessions) (Genesys Citation2013, ). Other genebanks with over 1,000 accessions include: the USDA in Pullman, Washington, USA (6,789 acc.); Aegean Agricultural Research Institute in Izmir Turkey (2,075 acc.); the Australian Temperate Field Crops Collection (ATFCC) in Horsham, Australia (8,655 acc.); the National Plant Gene Bank of Iran (5,700 acc.); the Vavilov Institute in Russia (2,091 acc.); and the Institute for Agrobotany Tápiószele, Hungary (1,170 acc.). Significant source countries for chickpea landrace accessions are India (10,526), Iran (8,912), Turkey (4,927), Syria (2,517), Afghanistan (1,949), Spain (1,494), Pakistan (1,272), and Ethiopia (1,175), plus over 60 additional countries that are also sources of additional cultivars and landraces (Redden et al., Citation2007; Genesys, Citation2013). Over 75% of the ICRISAT collection consists of desi-type accessions, 21% are kabuli type and 4% are intermediate, while the ICARDA collection consists mainly of kabuli types (Redden et al., Citation2007). There may be over 15% overlap between the ICRISAT and ICARDA collections; much higher levels of duplication occur between these two collections and national collections around the world, as well as among various national collections. The ATFCC has 4,001 landraces, cultivars and wild relatives, the latter numbering 241 accessions, all of which are duplicated in the ICRISAT, ICARDA and USDA collections. The total ex-situ holdings of the 27 species of wild Cicer are 1,105 accessions, which included 147 accessions of uncertain identification. There are 166 accessions of the progenitor C. reticulatum and 64 of C. echinospernum (Genesys, Citation2013, plus the ATFCC collection), although the number of unique accessions is reduced due to repeated subsampling of the original accessions (Berger et al., Citation2003). For example, 43 subsequent accessions have been subsampled from C. reticulatum ILWC 21, originally collected from a single site along the Savur-Midyat road in southeastern Anatolia (Berger et al., Citation2003). Fortunately, the numbers of unique C. reticulatum and C. echinospernum accessions will more than double as a result of a 2013 joint Turkish-USA-Australian collecting expedition in Turkey that specifically targets Cicer wild relatives (Berger, personal communication).
Chickpea ranks second among food grain legumes in the world after common bean with production of 11,308 Kt and mean yield of 931 kg/ha (FAOSTAT, Citation2012). Chickpea is grown in over 50 countries, ranging from subtropical and temperate regions of the world, for its protein-rich seeds. Chickpea seeds are a rich source of minerals, fiber, unsaturated fatty acids, β-carotene and do not contain any antinutritional factors (Jukanti et al., Citation2012). Due to its high nutritional value, chickpea is considered one of the most nutritious food grain legumes and serves as an important protein source for humans who consume vegetarian diets. Because chickpea plants are efficient symbiotic nitrogen fixers, chickpea fits well in crop rotation, improving soil fertility and playing an important role in the sustainability of farming systems.
While loss of genetic diversity is a universal phenomenon among crops (Tanksley and McCouch, Citation1997), in chickpea there has been a particularly drastic narrowing of genetic diversity due to a series of bottlenecks unique to this crop (Abbo et al., Citation2003a). Consequently, chickpea displays a lack of adaptive diversity for a range of biotic and abiotic stresses. Unlike cultivated chickpea, wild Cicer species possess useful variation for morphological traits (Robertson et al., Citation1995), protein content (Ocampo et al., Citation1998), and genetic sources for resistance to both biotic (Di Vito et al., Citation1996; Collard et al., Citation2001; Ansari et al., Citation2004; Rubiales et al., Citation2004) and abiotic stresses (Singh et al., Citation1990, 1998; Croser et al., Citation2003; Toker, Citation2005; Toker et al., Citation2007aCitationb; Canci et al., Citation2009). Wild Cicer species were identified as sources of resistance to a number of diseases and pests such as ascochyta blight [Ascochyta rabiei (Pass.) Labr.], fusarium wilt [Fusarium oxysporum Schlechtend. Fr. f. sp. ciceris (Padwick) Matua & K. Sato], botrytis gray mold (Botrytis cinera Pers. ex. Fr.), rust (Uromyces ciceris-arietini (Grognot) Jacz. & Boyd), pod borer (Helicoverpa armigera Hubner), leaf miner (Liriomyza cicerina Rond.), seed-beetles (Callosobruchus Pic. sp.) and nematodes (Di Vito et al., Citation1996; Collard et al., Citation2001; Croser et al., Citation2003; Ansari et al., Citation2004; Sharma et al., Citation2005; Sillero et al., Citation2012). By far the most pressing issue relating to chickpea genetic resources is the urgent need to collect and characterize annual wild relatives, particularly those that are readily crossable with domestic chickpea (Ben-David et al., Citation2010). Numbers of independent accessions in the primary genepool are <20 per species, as indicated more than a decade ago (Berger et al., Citation2003). Even with such an extremely limited collection, the utility of annual wild Cicer genetic resources for providing useful adaptive variation and genetic diversity is unparalleled, as outlined in previous sections.
VI. TRIBE PHASEOLEAE (BRONN) DC.
A. Genus Phaseolus L.
According to the most recent monograph of the genus (Freytag and Debouck, Citation2002), there are 76 species of Phaseolus, all distributed in the New World with a center of diversity in Mexico. Recent phylogenetic analyses of chloroplast and nuclear ribosomal DNA regions (Delgado-Salinas et al., Citation1993, 1996, Citation2006) revealed that the genus in its current circumscription is monophyletic and that all species can be grouped in two clades. Clade A comprises the three well-supported groups Pauciflorus, Pedicellatus, and Tuerckheimii, plus a few species of unclear affinity (P. glabellus Piper, P. macrolepis Piper, and P. oaxacanus Rose), and is mainly Mexican. Clade B comprises five groups (Filiformis, Vulgaris, Lunatus, Leptostachyus, and Polystachios) and has a much broader distribution range, including the Andes and several islands, such as the Galapagos-endemic P. mollis Hook. f. (Delgado-Salinas et al., Citation2006; Porch et al., Citation2013). Freytag and Debouck (Citation2002) prefer to group not by molecular phylogeny but by taxonomic characteristics and describe 16 sections of the genus. The five most important crop species of the genus, P. acutifolius (tepary bean), P. coccineus (scarlet runner bean), P. lunatus L. (lima bean), P. polyanthus Greenm. (year bean), and P. vulgaris L. (common bean) belong in two clades only, the Vulgaris and the Lunatus groups (Delgado-Salinas et al., Citation1999), which also agree in the sections defined by Freytag and Debouck (Citation2002). Molecular clock analyses revealed a stem age estimate for the Phaseolus clade of c. 8 million years and a crown age of max. 6 million years (Delgado-Salinas et al., Citation2006).
The basic chromosomal number for Phaseolus species is n = 11. Despite all the cultivated species in the genus Phaseolus having the same number of chromosomes, many of the species are difficult to cross and are organized into primary, secondary, tertiary and quaternary genepools relative to the P. vulgaris (). In this regard, P. coccineus (scarlet runner bean) and P. dumosus (year-long bean) are in the secondary genepool, with simple crosses possible but with some difficulties for F1 recovery as well as for introgression of genomic segments when backcrossing or deriving inter-specific lines. Cytoplasmic effects are important, and introgressions tend to be small, partial segments of the genome. P. acutifolius (tepary bean) and its close wild relatives from P. parvifolius Freytag (Blair et al., Citation2012a) are in the tertiary genepool of common bean, principally crossable only with embryo rescue and with congruent backcrossing to avoid cytoplasmic effects and to increase the rate of introgression (Muñoz et al., Citation2006). Meanwhile, P. lunatus (Lima bean) and the related species of P. augusti Harms and P. bolivianus Piperare in the quaternary genepool of common bean, and no confirmed crosses among these species have been possible (Porch et al., Citation2013). The primary centers of origin for the Phaseolus cultigens are in the New World; however, for each crop there has been a different spread outside the original range, with greater spread for common bean than for any of the other species. Common bean has secondary centers of diversity in Africa, Brazil, the Caribbean, China, Europe and India (Zhang et al., Citation2008; Asfaw et al., Citation2009; Blair et al., Citation2010; Sharma et al., Citation2013), while tepary bean has spread very little outside its original range in northern Mexico, spreading only to parts of Central America (Blair et al., Citation2012a). Similarly, scarlet runner beans have spread ancestrally from Central America only to northern South America, while year-long beans for the most part remain in the Guatemalan highlands. Overall, this reflects the original diversity of the wild species in the genus, which are more diverse in the Mesoamerican center than in the South American centers. The genome sizes of the Phaseolus cultigens are small, ranging between 450 and 650 Mb (Pedrosa Harand et al., 2006, 2013).
B. Crop Common bean (Phaseolus vulgaris L.)
Several large germplasm collections for Phaseolus exist around the world, holding 261,968 accessions (FAO, Citation2010, ). The largest, with 36,000 accessions, is at the International Center for Tropical Agriculture (CIAT) based in Cali, Colombia. This collection includes a duplicate of the USDA collection, which consists of almost 18,000 accessions when considering all the species of Phaseolus, including wild and cultivated. Common beans make up the majority of the collection, and the representation of other species is limited in all of the collections. Other important in situ collections are found in Bolivia, Brazil and Mexico. Core collections have been made in both the CIAT and the USDA collections, while working collections are found in many other countries. A reference collection with representative sampling of the core collection was described by Blair et al. (Citation2013). Apart from this, collections of common bean and, many times, scarlet runner beans are found in many European countries. A recent EU-funded project, PHASEOLEU, compiled data on the common beans in Europe. The state of collections in African countries are often precarious, although collections exist in Burundi, Democratic Republic of Congo, Ethiopia, Kenya, Republic of South Africa, Rwanda, Tanzania, Uganda and Zimbabwe. These countries should place a greater emphasis on protecting the genetic wealth of beans in Africa, since many of the African countries are large per capita consumers of beans. Additional collections of common bean are found in Asia at the Indian Council for Agricultural Research (ICAR) and at the Chinese Academy of Agricultural Sciences (CAAS). Collections of beans in Bhutan, Laos, Myanmar, Nepal, Pakistan, Thailand and Vietnam would be of interest, as these are not represented in the international collections at USDA based in Fort Collins, Colorado or at CIAT. The recent improvements to genebanks in many provinces of China as well as in Beijing have created good conditions for the storage of landraces and cultivars, especially of snap bean types. Worldwide, there may be over 100,000 landraces of Phaseolus preserved in genebanks, but the documentation of these needs to be improved and shared among countries that are part of the FAO treaty on genetic diversity, under which beans is a species with prioritized access as a food crop. Increased collections of wild relatives are needed, as shown by the low representation of wild P. vulgaris in the CIAT collection (1,300 accessions with gaps in the regions sampled). The loss of scarlet runner bean and tepary bean genotypes due to changes in local agriculture in producing countries also argues for greater preservations efforts. An interesting model used for common bean and some lima beans has been the ex situ collection system found in such organizations as the Seed Savers Exchange in the United States or the seed fairs for farmer-to-farmer exchange of seed found in Colombia and other countries with a wealth of landraces.
A core collection of 1,400 genotypes has been created from the FAO-protected germplasm and has been characterized at the phenotypic level for basic traits (flower color, growth habit, maturity date, etc.). About half of this collection (the reference collection referred to above) has been genotyped with multi-copy AFLP and RAPD markers and more recently with single-copy SSR and SNP markers (Blair et al., Citation2009, 2013). Apart from this, there are core collections for the CAAS, EMBRAPA and USDA genebanks, with the latter one holding 700 genotypes out of a total of 16,000 accessions of common bean. The FAO collection was prepared with the intent to cover all geographical regions where common beans are grown and does represent the current races of common bean well (Blair et al., Citation2009). It is also important for national collections to be represented by a core set of germplasm that has been well characterized and genotyped, such as in the CFP collection center in Bolivia (Avila et al., Citation2012). Core collections for the other species of cultivated Phaseolus have not been made, perhaps because there are so few actually collected that it is impossible to sample randomly from within the genebanks. Genotyping has proved useful for identifying duplicates in the tepary bean collection, where monomorphism is high in the cultivars but low in the wild accessions (Blair et al., Citation2012a). Genetic mapping in common bean is quite advanced (Galeano et al., Citation2011), however, the other species of Phaseolus still have no genetic maps. In some cases, such as for tepary beans, inter-specific crosses may be necessary for polymorphism mapping, but in common bean, crosses between races or between Andean and Mesoamerican genepools often suffice. Therefore, in P. vulgaris, many traits have been tagged, and several reviews describe the genes or QTL involved in controlling certain insect-, disease- and stress-resistance mechanisms.
The most important of all the Phaseolus cultigens, the common bean, spread from its origins in the New World throughout Africa, Asia and Europe to become a leading food crop as a dry grain of a multitude of shapes, sizes and color and as a vegetable favored for its lightly flavored pods. Although far less important than cereals, common bean is a cheap source of vegetable proteins, calories and micronutrients. Like other legumes, the major limitations are the low content of sulphur-amino acids and the presence of antinutritional compounds. Common beans are primarily grown for home or local consumption in the developing world, while in the developed world they are primarily grown for processing uses and for exports. Brazil and Mexico are the largest producers of dry grain, while China is the largest producer of common beans as a vegetable. Argentina and Canada are considered among the largest exporters, along with China and the United States (FAOSTAT, Citation2012). Vegetable production of common beans is common in Kenya as well as around the Mediterranean for the winter diet of Europeans. Even parts of West Africa produce snap or dry beans. Dry bean production then continues on into many countries in Southern Africa. In Asia, common beans are found in India, along the western Bhat range and in the Himalayas. Neighboring Nepal also produces many different landraces of common bean. In China, red dry beans are used for pastries, but northern production of the crop for the export market has become significant and competes with soybean in rotation with maize.
Lima bean improvement programs are independent of common bean improvement. Scarlet runner bean and tepary bean accessions have been used to obtain resistance genes and other traits for common bean. Wild relatives of common bean from the same species are an important source of diversity for the crop as well (Blair et al., Citation2012b; Blair and Izquierdo, Citation2012). Tepary beans are by far the least consumed of the five cultivars, although tepary beans are still grown in northern Mexico and among native peoples in the Southwestern US. Scarlet runner bean and year-long bean are grown locally in parts of Central and South America, Asia and Africa, and some fetch a high price as an export crop for European markets. Lima beans are mainly a food security crop in dryland areas of the Caribbean, Mexico and Peru but are also an important fresh vegetable.
Early maturity, adaptation to higher altitude, upright plant type, high pod quality and seed yield, and some resistances to diseases such as viruses and rust, insect pests, and drought and abiotic constraints such as deficiency of nitrogen, phosphorus and zinc or tolerance to aluminium and manganese toxicity have been bred into common bean cultivars (reviewed in Singh and Schwartz, Citation2010, Citation2011).
Common bean is by far the most widely grown of the Phaseolus, as it is the most important legume for direct human consumption, grown on over 20 M ha worldwide and having both a dry grain (seed) and vegetable (snap pod) market. However average yield is low, 804 kg/ha, compare to other legumes (FAOSTAT, Citation2012). It is cultivated extensively in the five continents and spans from 52°N to 32°S latitude, and from near sea level in the continental USA and Europe to elevations of more than 3000 m above sea level in Andean South America. In summary, common beans are one of the most widespread crops of the world and deserve their position as the most commonly grown legume for direct human consumption as food.
C. Genus Vigna Savi
The genus Vigna contains c. 150 species distributed throughout both the Old and New Worlds, species that can be grouped into the six subgenera Vigna, Ceratotropis, Plectotropis, Sigmoidotropis, Lasiosporon, and Haydonia (Vaillancourt et al., Citation1993; Vijaykumar et al., Citation2010). The genus is polyphyletic, with one clade comprised of New World species and the genera Ramirezella and Oxyrhynchus (Delgado-Salinas et al., Citation1993). The Old World species, however, seem to form a monophyletic group, with the possible exception of V. frutescens A. Rich. (Vaillancourt et al., Citation1993). The Vigna species grow in warm temperate and tropical regions globally. It is most closely related to Phaseolus, and Asian Vigna (subgenus Ceratotropis) was categorized as Phaseolus until 1970 (Verdcourt, Citation1970). Vigna differs from Phaseolus in biochemistry and pollen structure and in the details of its style and stipules (Verdcourt, Citation1970). The subgenus Vigna or African Vigna comprises c. 40 species, among them the agriculturally important species V. unguiculata (cowpea or black-eyed pea), V. mungo (black gram) V. radiata (mungbean), and V. subterranea (Bambara groundnut), along with related wild species and accessions (Tomooka et al., Citation2002, 2005, 2006; Vijaykumar et al., Citation2010). The subgenus Ceratotropis currently consists of 16 (Verdcourt, Citation1970) to 17 (Maréchal et al., Citation1978) recognized species, which are naturally distributed across Asia and thus are often called Asiatic or Asian Vigna (Singh et al., Citation2006). Tomooka et al. (Citation2002) describes 21 species of Asian Vigna, 8 of which are used for human food or animal feed. This is in contrast to the African Vigna (the subgenus Vigna), out of whose 36 species only two have been domesticated (Maréchal et al., Citation1978, ). V. lancoelata is endemic to Australia, and four others are also distributed in Africa or Asia (Lawn, 2014). Chromosome complements in Vigna species are 2n = 2x = 22, with the exception of V. glabrescens (2n = 4x = 44). Chromosome rearrangements play a significant role in the genetic differentiation of Asian Vigna species. Even the two close relatives V. radiata and V. mungo have some structural differentiation of their genomes (Bisht and Singh, Citation2013). The progenitor of cowpea is V. unguiculata var. spontanea (formerly var. dekindtiana), whose habitat has been found in all lowland areas of Subsaharan Africa, outside the high rain forests and deserts. However, southern Africa has been suggested as the center of origin for wild cowpea (Padulosi and Ng, Citation1997). The restricted distribution of these primitive forms of wild cross-compatible cowpea relatives in this part of southern Africa provides strong evidence that the region is probably the centre of origin of wild cowpea. The existence of substantial variation among traditional cowpea cultivars grown by farmers in western and central Africa confirms that the region is the possible center of diversity for cowpea. The revision of subgenus Ceratotropis by Tateishi (Citation1985) is the most comprehensive one to date. The eight cultivated species of the subgenus Ceratotropis as described by Tomooka et al. (Citation2002) are Vigna radiata (green gram or mungbean), V. mungo (black gram or urd bean), V. angularis (small red bean or azuki/adzuki bean), V. umbellata (rice bean or red bean), V. aconitifolia (moth bean), V. reflexopiloxa var. glabra (Creole bean), V. trilobata (wild bean) and V. trinervia (Tooapée).
1. Crop cowpea (Vigna unguiculata (L.) Walp.)
Cultivated cowpea is divided into four cultivar groups: Biflora, Sesquipedalis, Textilis and Unguiculata. Cowpea belongs to culti-group unguiculata, while the yard-long bean or asparagus bean belongs to sesquipedalis. While cowpea is grown mainly for its dry grains in sub-Saharan Africa, South and Central America, the southern United States and Europe, the yard-long bean is commonly grown in Southeast Asia for the long, green, fleshy pods consumed as a vegetable. Because of its drought tolerance, cowpea is well adapted to the dry savanna; consequently, it is probably the most commonly grown and consumed legume in the dry savanna regions of sub-Saharan Africa. Most wild species of Vigna have adapted to various environments through the evolutionary process of diversification or specialization. They can provide an important gene pool for cultivated crops of Vigna. For example, some wild Vigna species can grow in extreme or marginal environments and are therefore believed to harbor interesting genetic information. However, wild Vigna species are rarely collected, with the exception of some efforts undertaken in the past decade by the the National Institute of Agrobiological Sciences genebank in Japan and Kasetsart University, Thailand to collect Asian species. The most comprehensive collection of V. vexillata (L.) A. Rich. is in the seed bank of the Royal Botanic Gardens of Belgium. According to the study of Maxted et al. (Citation2004), more than 20 species of African Vigna species are not conserved in any ex-situ collection. However, there are many collections of Vigna subgenus Cerototropis germplasm. Most of these collections consist primarily of accessions of the cultigens in this subgenus, and most of the accessions conserved were evaluated on basic agronomic traits. The main collections are at the International Institute for Tropical Agriculture (IITA) in Nigeria mainly for Vigna unguicalata, and for predominantly Vigna spp. mungo & radiata Asian Vegetables Research and Development Center (AVRDC), Taiwan and the National Board for Plant Genetic Resources, India for Vigna spp. angularis & umbellata at the Institute of Crops Sciences (ICS), Beijing, China; and for a range of Vigna spp. in the Plant Genetic Resources Conservation Unit, Georgia, USA. Wild Vigna species of subgenus Cerototropis are poorly represented in world genebanks. Some countries have comprehensive collections of their own indigenous Vigna genetic resources, such as V. radiata var. sublobata (Roxb.) Verdc. in the CSIRO collections (Lawn and Cottrell, Citation1988; Tomooka et al., Citation2002).
Several studies have focused on the genetic diversity of Vigna (Kaga et al., 1993; Wang et al., 2008; Undal et al., Citation2011; Kumar et al., Citation2012; Kaewwongwal et al., Citation2013) and QTL analysis (Young et al, 1993; Tomooka et al., Citation2002, 2005, 2006; Sholihin and Hautea, 2002; Humphry et al., 2005; Kasettranan et al., 2010; Kongjaimun et al., Citation2012; Chankaew et al., 2014; Kajonphol et al., 2012). To better characterize the cowpea germplasm, a core collection of 2,062 accessions was defined based on geographical, agronomical and botanical descriptors (Mahalakshmi et al., Citation2007). A mini-core set of 374 accessions was further defined and are being used intensively in several cowpea breeding programs. The main objectives are to evaluate the entire cowpea germplasm for priority traits and to complete the agro-morphological description of wild Vigna accessions. Primary production constraints, include drought and heat stresses, insects (flower thrips, pod-sucking bugs, cowpea aphid), diseases (viral, fungal, bacterial and nematode) and Alectra and Striga parasitic weeds. Research has been intensified in recent times to develop cowpea cultivars with enhanced levels of drought tolerance (Adegbite and Amusa, Citation2008). A few accessions of the wild Vigna species have also been screened for resistance to insect pests of cowpea. Many accessions of V. vexillata were found to show high levels of resistance to pod-sucking bugs and storage weevils and moderate resistance to maruca pod borers (Singh et al., Citation1992). However, the basic need for exploiting the wild relatives is its cross compatibility with cultivated cowpea. It is possible that some of the available wild cowpea lines belong to the same or different gene pools. The subspecies or cultivars that constitute the primary and secondary gene pools for cowpea are not yet well defined. Cross compatibility studies have shown that lines that can hybridize successfully with cultivated species are found only among members of the subspecies unguiculata, i.e. those belonging to section Catiang in the genus Vigna (Tomooka et al., Citation2002).
Cowpea is among the top five food legumes or pulses grown worldwide and has a presence on every continent except Oceania and Australia. The West African subregion contributes to about 95% of global cowpea production, with Nigeria being the largest producer of cowpeas in the world (FAOSTAT, Citation2012). The crop's reputation as very adapted to drought conditions makes it ideal for rotations and inter-cropping with sorghum and millets in these regions, but it is also grown in wetter areas, along with maize. Cowpeas are intermediate in nitrogen fixation, fixing more than common bean and less than soybean. It is the most important legume for cereal legume rotations in the world. Of growing importance to food security in many parts of Eastern Africa, South Asia and especially Southern Africa, the cowpea deserves more investment in agronomic and breeding activities. Bambara groundnuts, which set seed under ground, are of limited importance but are interesting for their high level of disease resistance compared to Arachis groundnut. They are found mostly in Southern, Eastern and Western Africa in a range extending from Malawi to Senegal, but they are not consumed outside of the Sub-Saharan region.
2. Crop mungbean (Vigna radiata (L.) R. Wilczek)
Mungbean is a photo- and thermosensitive crop. The best temperature for its cultivation is 30–35°C with good atmospheric humidity. It is cultivated throughout South and Southeast Asia, including India, Pakistan, Bangladesh, Sri Lanka, Myanmar, Thailand, Philippines, Laos, Cambodia, Vietnam, Indonesia, Malaysia, South China and Taiwan. It is also grown to a lesser extent in many parts of Africa, the United States (especially in Oklahoma), and has been recently introduced in parts of Australia. Black gram (Vigna mungo) is also an important pulse crop of India. Black gram is widely adapted both to semi-arid and subtropical areas. Black gram is a protein- rich food (about 26% protein) that consequently legume by India's vegetarian population. In addition to being an important source of human food and animal feed, black gram also plays an important role in sustaining soil fertility by improving soil physical properties and fixing atmospheric nitrogen. Also, as a drought-resistant crop, it is suitable for dryland farming and is predominantly used as an intercrop with other crops.
Meanwhile, mungbeans and black gram are found mainly in Asia and retain importance at their centers of origin (China and India). Southeast Asia, from Myanmar to the Philippines, also produces a large number of mungbeans where they can be double-cropped after rice. They are fast-growing, early maturing and drought-tolerant and can therefore be grown on residual moisture after a crop of vegetables and cereals or at the end of the rainy season. Other Asian species of legumes little known in the West are urd beans and moth beans, but these are adaptable outside their current range and fit into additional agro-ecological niches.
The nitrogen fixation potential of the Asian Vigna species has been poorly studied. The mining of elite genes in wild Vigna species will be a great genetic resource for Vigna crops. At present, the production of mungbean, adzuki bean and cowpea is being seriously damaged by different diseases or pests, especially bruchid, a pest that occurs frequently among stored Vigna seeds. However, the wild types of Vigna have resistant genes, which have proved to be transferable into cultivated crops by direct crosses or by using bridge plants (Tomooka et al., Citation2008; Pandiyan et al., Citation2008). Disease and insect resistance breeding has been a priority at IITA (Smithson et al., 1980). Curiously, disease resistance breeding has not been a priority in cowpea, perhaps because of its origin in the drier parts of West Africa, while insects, nematodes and viruses are important constraints which so far have had few resistance sources.
A total of 10,551 accessions of various Vigna species comprised of mungbean (3,704), urd bean (3,131), moth bean (1,486), rice bean (2,045) and azuki bean (185) have been stored at −18°C in the long-term repository of the national gene bank at NBPGR, New Delhi. Green gram germplasm accessions are maintained by more than 35 institutions globally, which hold a total of more than 25,000 accessions. IITA maintains over 15,000 accessions of cowpea, the Asian Vegetable Crops Research Centre, AVRDC at Taiwan, maintains 5,616 accessions of mungbean, and over 12,000 various Vigna are held in the Conservation Unit in Georgia. Limited germplasm accessions of moth bean are also available in several countries, including Bangladesh, Belgium and Kenya.
D. Genus Glycine Willd.
The genus Glycine contains c. 22 species, which can be grouped in two subgenera. The first, subgenus Soja, includes two Asian annuals: G. max (the cultivated soybean) and the similar wild species G. soja Siebold & Zucc. (Doyle et al., Citation1990). Glycine max is thought to be derived from a common ancestor with the wild G. soja lineage (Kim et al., Citation2010; Li et al., Citation2013b). Domestication of soybean was initiated c. 6,000-9,000 years ago in Asia and has resulted in considerable genetic differences between G. max and G. soja, as revealed by a comparison of whole-genome sequences (Kim et al., Citation2010). The second clade, subgenus Glycine, comprises c. 20 wild species. They are all perennials and restricted in their distribution to the Australian continent (Doyle et al., Citation1990). G. soja and G. max both have 20 chromosomes (2n = 40) and can easily interbreed. The split between G. max and G. soja was estimated to have occurred c. 3,000-5,000 years ago (Carter Jr. et al., 2004; Lee et al., Citation2010) based on molecular phylogenetic analyses and historical documents and 270,000 years ago based on comparative analysis of re-sequenced wild soybean genome (Kim et al., Citation2010) using cultivated soybean (Glyma1.0, var. Williams 82) as the reference genome. This suggests that the domestication time of soybean remains to be ascertained.
1. Crop soybean (Glycine max L.)
As the center of cultivated soybean (G. max), China has the most abundant genetic resources for soybean, with >23,000 cultivated soybean accessions and >7,000 wild soybean (Glycine soja) accessions conserved at the Chinese National Soybean GeneBank (CNSGB) (Qiu et al., Citation2013) and replicated at the National Germplasm Storage Facility in Qinhai (Qiu et al., Citation2011).
In order to efficiently analyze and utilize this large ex-situ collection, a series of the core collections, including primary core, core, mini-core and integrated applied core collections―has been constructed based on the study of phenotypic and genotypic (SSR markers) datasets (Qiu et al., Citation2013). In the past decades, these soybean core collections were widely used in the genomic study, molecular evolution clarification, elite genetic resources discovery, gene identification, elite lines development, and so on. For example, the analysis of the mini-core collection using SSR markers and allelic variation of the soybean determinate growth habit regulated gene GmTfl1 revealed that human selection for determinacy took place at early stages of landrace radiation (Tian et al., 2010). Twenty-one SSR markers were identified in the soybean applied core collection as associated with important agronomy traits, including high oil content, high protein content, drought tolerance, soybean cyst nematode resistance. Guo et al. (2013) identified three new low-frequency alleles of GmF3′H and GmF3′5′H in the mini-core collection. This indicated that this series of core collections with concentrated genetic diversity will play an important role in soybean molecular breeding (Qiu et al., Citation2013). The second largest collection of soybean accessions is conserved by the USDA: 19,557 cultivated soybean accessions, derived from 87 countries, 1,181 wild soybean accessions and 1,038 representatives of the 20 perennial species. In recent years, Brazil has conserved 2,000 special accessions, with the exception of the USDA soybean accessions introduced. Most of the cultivated soybean accessions are from China, Japan or Korea; therefore, redundancy in the global collections may be as high as 70% (Nelson, Citation2009). In Japan, approximately 11,300 soybean accessions are conserved at the National Institute of Agrobiological Sciences (NIAS) Genebank. These accessions include local landraces collected in Japan and overseas, as well as cultivars and breeding lines developed by regional Japanese agricultural research institutes (Kaga et al., Citation2012).
A large-scale evaluation of 17 traits was conducted for >20,000 soybean accessions conserved in CNSGB, but none of the 17 traits were completely identified in all the accessions. The accession evaluation rate was different for various traits, but the average was 35% (Qiu et al., Citation2011). In order to characterize phenotype and genotype of soybean accessions efficiently, additional core collections have been developed from the whole collection of soybean accessions (Brown et al., Citation1987; Yaklich et al., Citation1999; Zhao et al., Citation2005; Wang et al., Citation2006; Cho et al., Citation2008; Oliveira et al., Citation2010; Kaga et al., Citation2012). Due to their reduced size, these core collections could be studied extensively, and the information derived can be used to guide more efficient utilization of the much larger reserved collection (Qiu et al., Citation2013).
The soybean is classified as an oilseed rather than as a pulse by the FAO, due to the 20-25% oil content of the seeds. Soybean seed is also rich in protein content (40%), higher than that of all the pulses. Its cultivation and production is 2.5x that of all other grain legumes taken together, with a world production of 253,137 Kt with an average yield of 2,374 Kg/ha. Due to its amino acid composition, soybean is considered a source of complete protein. The remaining proportions are soluble or starch-converted sugars (35%) and minerals. The seeds also contain important isoflavones, such as genistein, daidzein and glycitein, which act as phytoestrogens. Traditional nonfermented food uses of soybean include soy milk, tofu and tofu skin. Fermented foods include soy sauce, fermented bean paste, natto, and tempeh, among others. The oil is used in many industrial applications. Only a very small proportion of the crop is consumed directly by humans; however, it is used in a large variety of processed foods. The grain is also used as animal feed. Interestingly, soybean did not become an important crop outside of Asia until about 1910. The main world producers of soy are the United States (35%), Brazil (27%), Argentina (19%), China (6%) and India (4%), with world total production at 249 million metric tons (FAOSTAT, Citation2012). The breeding effort is largely at private companies that focus on yield stability, resistances to pests and diseases (Wilcox, Citation1983) and altered oil composition. Breeding of low-phytate soybeans are desirable from both a nutritional and environmental standpoint and also provide an economic advantage to producers (Maroof et al., Citation2009). It is notable that a large proportion (up to 81% in 2011) of globally cultivated soybeans is transgenic, mainly for herbicide (Roundup Ready®, Liberty Link®) and pest (Bt) resistances, but also for altered oil composition (gene silencing to suppress the GmFAD3 gene family in Plenish® soybean) (www.gmo-compass.org).
E. Genus Cajanus L.
The pigeonpea (Cajanus cajan L.) belongs to the legume subtribe Cajaninae. This subtribe contains a total of 13 genera. Until recently, the genera Atylosia Wight & Arn. and Cajanus were considered closely related genera, but van der Maesen (Citation1990) merged the genus Atylosia with Cajanus. In total, the combined genus Cajanus now has 32 species. These species are endemic to Asia (18), Australia (13) and western Africa (1) (van der Maesen, Citation1990). The primary gene pool consists of the cultivated species and its landraces, whereas the secondary gene pool consists of ten wild relative species. There are 20 wild species in the tertiary gene pool (). Wild species placed in the quaternary gene pool of Cajanus belong to different genera, such as Flemingia Roxb. ex W. T. Aiton, Rhynchosia Lour., Dunbaria Wight & Arn., and Eriosema (DC.) Desv. The genus Cajanus has the same chromosome number (2n = 22) in all its species (Deodikar and Thakar, 1956; Dundas, Citation1990). The genome size of cultivated pigeonpea has been estimated as 833 Mb (Varshney et al., Citation2012). India is the primary center of pigeonpea diversity, while East Africa is considered the secondary center of diversity (Songok et al., 2010).
1. Crop pigeonpea (Cajanus cajan L.)
A total of 40,820 Cajanus accessions, comprising landraces, modern cultivars and CWR, have been conserved in ex-situ genebanks. The ICRISAT genebank holds 13,771 accessions, including 8,315 landraces, 4,830 breeding lines, 71 improved cultivars, and 555 accessions of wild relatives from 74 countries (). Other genebanks conserving pigeonpea germplasm are the National Bureau of Plant Genetic Resources (11,427 accessions), New Delhi, India; All India Coordinated Research Project on Pigeonpea (5,195 accessions); Indian Agricultural Research Institute (IARI), New Delhi (1,500 accessions); and the Crop Plant Genetic Resources Center, Muguga (1,380 accessions), Kenya. Based on available passport and morpho-agronomic data of the entire pigeonpea collection at ICRISAT, a “core collection” of 1,290 accessions was developed. This core collection was designed to represent the genetic variability of the entire collection and was further evaluated for various morphological, agronomic, and quality traits (Reddy et al., Citation2005). In addition to field evaluation, the diversity in the core collection was estimated using SSR markers. Furthermore, a subset of about 10% of the accessions from the core collection was selected. This subset contained 146 accessions and represented more than 80% of the diversity of the entire pigeonpea collection (Upadhyaya et al., 2010). A number of marker systems have been used in Cajanus to detect polymorphism. Initially, biochemical markers were used to analyze the relationships of wild relatives with the cultivated pigeonpea and identified C. cajanifolius (Haines) Maesen as the closest relative to pigeonpea (Krishna and Reddy, 1982). RFLP markers were also used to determine phylogenetic relationships among 12 Cajanus species (Nadimpalli et al., 1994). Two species, C. cajanifolius and C. scarabaeoides (L.) Thouars, showed a close relationship with each other; however, C. cajanifolius was closest to C. cajan. Other marker systems used to estimate the polymorphism in Cajanus were RAPD (Ratnaparkhe et al., 1995), AFLP (Punguluri et al., 2006), DArT (Yang et al., 2006), SSR (Saxena et al., 2010a, b; Bohra et al., Citation2011a), and, recently, single-nucleotide polymorphism (Saxena et al., 2012; Roorkiwal et al., 2013). All marker-based studies have revealed that a very low level of diversity is present in cultivated pigeonpea, whereas the wild relatives of pigeonpea showed enormous diversity (Ratnaparkhe et al., 1995; Punguluri et al., 2006; Saxena et al., 2010a, b; Bohra et al., Citation2011a). These studies also revealed that two of the wild relatives, C. cajanifolius and C. scarabaeoides, are closely related to pigeonpea. Cytological studies have also proved that C. cajanifolius is the progenitor species of C. cajan, as both species have similar karyotypes, and the hybrids produced from crossing the two species have normal meiosis with high pollen fertility (Pundir and Singh, 1985; Mallikarjuna et al., Citation2006).
Pigeonpea (Cajanus cajan) is a short-lived perennial shrub that is cultivated as an annual grain legume crop in tropical and subtropical regions. Its cultivation area is 4.64 M ha, with an annual production of 3.43 million tonnes and a mean productivity of 780 kg/ha, making it the sixth most important legume food crop in the world (FAOSTAT, Citation2012). It is primarily grown for dry, dehulled, split seeds, green seeds and pods as vegetables. It can also be used as forage, fodder, fuel and medicine (Saxena et al., Citation2010). The deep roots of pigeonpea help recycle minerals from deep soil and make them available to other intercropping plants. Pigeonpea has several unique characteristics that render it an ideal crop for sustainable agricultural systems. Its partial out-crossing nature affects its breeding and selection efficiency and makes research activities more difficult in comparison to other food legumes. However, the presence of both additive and non-additive genetic variations allows for the development of both high-yielding, pure-line cultivars and hybrids (Saxena, Citation2008). Pigeonpea improvement programs have evolved around long-duration, photo-sensitive types and earliness, whereas dwarfness, disease resistance (mainly fusarium wilt, sterility mosaic disease and phytophthora blight), insect resistance (pod borers, Helicoverpa armigera and Maruca vitrata, and pod fly, Melanagromyza obtusa), abiotic stress tolerance (soil salinity and water logging), fodder, yield, and yield-related traits are the prime targets for pigeonpea improvement (Saxena, Citation2008).
VII. TRIBE AESCHYNOMENEAE
A. Genus Arachis L.
The genus Arachis is divided into nine sections (Arachis, Trierectoides, Erectoides, Extranervosae, Triseminatae, Heteranthae, Caulorrhizae, Procumbentes, and Rhizomatosae) based on morphological, cross-compatibility and geographic origin/distribution and has a total of 80 species. All of the species are diploid in nature, except two tetraploids, A. hypogaea L. and A. monticola Krapov. & Rigoni. Of the nine sections, Arachis is the largest section, comprised of 32 species, including the cultivated groundnut (A. hypogaea) (Krapovickas and Gregory, 1994; Valls and Simpson, 2005). All species within the Arachis sections are found mostly in Brazil, followed by Paraguay, Argentina and Uruguay (Upadhyaya et al., Citation2011). Arachis species can be grouped into nine sections comprised of 80 species with both annual and perennial life cycles (Krapovickas and Gregory, 1994; Valls and Simpson, 2005). Of these nine sections, the most important section is Arachis, which includes the cultivated and domesticated groundnut (Arachis hypogaea). The cultivated groundnut is amphidiploid (tetraploid) and originated through a single hybridization event between two diploid wild species, A. duranensis Krapov. & W. C. Greg. (A-genome) and A. ipaënsis Krapov. & W. C. Greg. (B-genome), followed by a spontaneous chromosome duplication (Halward et al., 1991). Because of its uncommon origin, the resulting cultivated tetraploid (A. hypogaea, AABB genome) was reproductively isolated from its wild relatives.
Along with cultivated groundnut, A. monticola is another tetraploid species and seems to have been the intermediate species in the domestication of cultivated groundnut from diploid species. Cultivated groundnut species (A. hypogaea) were classified into two subspecies (A. hypogaea subsp. hypogaea and A. hypogaea subsp. fastigiata Waldron) based on differences in growth habit, reproductive modes, flowering on mainstem, seed size, and maturity duration, with a total of six botanical cultivars (Krapovickas and Gregory, 1994). The subspecies hypogaea shows a spreading growth habit, alternating vegetative and reproductive nodes, absence of flowers on the mainstem, medium-to-large seeds and medium-to-late maturity. The botanical variety A. hypogaea var. hypogaea (Virginia and Runner market types) is the most cultivated group. Subspecies fastigiata shows an erect growth habit, sequential reproductive nodes, the presence of flowers on the mainstem, small seeds, and early maturity. It can be divided into the botanical cultivars fastigiata (Valencia), vulgaris Krapov. & W. C. Greg. (Spanish), peruviana Krapov. & W. C. Greg., and aequatoriana Krapov. & W. C. Greg. (Krapovickas and Gregory, 1994; Burrow et al., 2013).
Based on compatibility features and genetic variability, Singh and Simpson (1994) have classified the genus Arachis into four gene pools (). The first gene pool includes two tetraploid species (A. hypogaea and A. monticola) from section Arachis. The secondary gene pool includes the remaining diploid species of section Arachis that show strong cross-compatibility with A. hypogaea. The tertiary gene pool includes species from section Procumbentes, which show weak cross-compatibility with A. hypogaea. The quaternary gene pool prescribes the most distantly related wild relatives to A. hypogaea and includes all species from the remaining seven sections of the genus Arachis. Despite the availability of broad genetic variations among species of the tertiary and quaternary (fourth) gene pools, the breeding community has been unable to exploit them because of incompatibility problems; thus, efforts need to be undertaken in finding efficient allele sharing methodologies for further improvement of cultivated groundnut. The hybrid origin of cultivated groundnut, followed by reproductive isolation and further sections during domestication, left groundnut's primary gene pool with very limited genetic diversity. Earlier, genetic diversity studies using a range of molecular markers reported a very low level of diversity in the primary gene pool (Kochert et al., 1996; Subramaninan et al., 2000; Herselman, 2003). Nevertheless, in the few other studies in which large germplasm sets were used reported low levels of diversity in primary gene pools, while better genetic diversity still exists within the wild relatives (Varshney et al., 2009a; Koppolu et al., 2010; Khera et al., 2013). Similarly, diversity array technology (DArT) and kompetitive allele specific PCR (KASP) markers showed very low polymorphism in cultivated genotypes and moderate polymorphism in diploid wild relatives (see Varshney et al., Citation2013b).
1. Crop groundnut (Arachis hypogaea L.)
Ex situ germplasm collections for groundnut are maintained in India, China, United States, Argentina and Brazil, holding all together 128,435 accessions (FAO, Citation2010, ). The largest collection for groundnut is held at ICRISAT in India, where a total of 15,445 accessions representing 93 countries have been conserved. The other main institutes that conserve groundnut germplasm include the National Bureau of Plant Genetic Resources (14,585 accessions) and the Directorate of Groundnut Research of the Indian Council of Agricultural Research (9,024 accessions), both in India; the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (8,083 accessions) and the Crops Research Institute of the Guangdong Academy of Agricultural Sciences (4,210 accessions) in China; the United States Department of Agriculture (9,917 accessions), Empresa Brasiliera de Pesquisa Agropecuaris (EMBRAPA)-CENARGEN (2,420 accessions) and the Instituto Agronomico de Campinas (2,140 accessions) in Brazil; and Instituto Nacional de Technologia Agropecuaria (3,640 accessions) and Instituto de Botánica del Nordeste (472 accessions) in Argentina. As far as wild relatives are concerned, Texas A & M University (1,200 accessions) holds the largest collection, followed by USDA (607 accessions), North Carolina State University (406 accessions) in the United States; EMBRAPA-CENARGEN in Brazil (1,220 accessions), ICRISAT in India (477 accessions) and the Instituto de Botánica del Nordeste (472 accessions) in Argentina. To facilitate maintenance and especially phenotyping, core collections (i.e. 10% of the entire germplasm collection) were developed for USDA germplasm (831 accessions) (Holbrook et al., 1993) and ICRISAT (1,704 accessions) (Upadhyaya et al., 2003). In addition, a composite collection of 1,000 accessions was developed by ICRISAT based on phenotypic data, geographic origin and taxonomic data. To assist breeders in handling small sets of their working collection, a further, smaller germplasm set (reference set) comprising 300 genotypes was developed after screening 20 SSR markers in the composite set (Upadhyaya et al., 2002, 2003). An even smaller germplasm set called the ‘mini-core collection’ (i.e. 10% of the core collections and 1% of the entire germplasm collection) was constituted by ICRISAT (184 accessions) (Upadhyaya et al., 2002), USDA/ARS (112 accessions) (Holbrook and Dong, 2005) and China (298 accessions) (Jiang et al., 2008).All three mini-core collections have been well phenotyped over the years for several agronomic traits, and next-generation genotyping has been planned for the three above-mentioned germplasm sets for conducting genome-wide association analysis (GWAS).
Groundnut or peanut is a crop of global importance that supports the livelihood of millions of resource-poor farmers in the semi-arid tropics (SAT). Besides being an important crop for cooking oil, food, and feed, it also enriches soil by fixing nitrogen. Currently, this crop is grown in more than 100 countries in an area of 24 million ha with a total production of 38 million tonnes and mean yield of 1,675 kg/ha (FAOSTAT, Citation2012). The range of usage is increasing by the processing industry is continuously increasing the range of usage; thus the projected demand for groundnut is very high. This crop is highly regarded among all economic classes, especially among the poor, it serves as a good source of nutrition for people and their livestock. Groundnut kernels contain 48–50% oil and 25–28% protein, providing 564 kcal of energy for every 100 g (Jambunathan, Citation1991). In addition, groundnut contains several micronutrients and health-enhancing components, including minerals, antioxidants and vitamins, along with some biologically active polyphenols, flavonoids and isoflavones (see Janila et al., Citation2013). The breeding objectives for groundnut are based on consumer and local industry preferences. Nevertheless, the majority of the improvement programs across the world have similar objectives and are continuously working on yield enhancement, early maturity, biotic resistance, abiotic stress tolerance, pre-harvest dormancy, high oil or protein contents and high oleate trait. Among these, significant achievements have been made in improving biotic resistance and high oleate trait by developing and releasing improved cultivars. Breeders are still working to develop improved cultivars with increased pod yield and abiotic stress tolerance. The main biotic stresses include foliar fungal diseases (late leaf spot, early leaf spot and rust), soil-borne fungi, bacterial wilt, groundnut rosette virus (GRV), peanut bud necrosis (PBND), peanut stunt virus (PSV), peanut strip virus (PStV), tomato spotted wilt virus (TSWV), and nematodes. Terminal drought is the major abiotic stress, followed by acidic soil, low soil fertility, and low temperature. The breeding approaches used in genetic enhancement of groundnut are the same as those used with all other self-pollinated crops, including selection, pedigree, inter-mating, mutation and backcross breeding (Holbrook and Stalker, Citation2003).
VIII. TRIBE GENISTEAE
A. Genus Lupinus L.
The taxonomy of the Genisteae tribe and other Genistoid legumes has been an area of considerable confusion for many years (Percy and Cronk, Citation2002). Recent focused efforts on defining phylogenetic relationships within the Genisteae have substantially clarified the situation (Cardoso et al., Citation2012a, b, 2013; The Legume Phylogeny Working Group, Citation2013). The Genisteae tribe is currently considered to include 618 species in 25 genera (Cardoso et al., Citation2013), and its diversity is centered in the Mediterranean region and in the Canary Islands (Cristofolini, Citation1997; Cristofolini and Chiapella, Citation1984). Lupinus is the largest Genisteae genus, comprising 267 species, and appears to be monophyletic in origin (Ainouche and Bayer, Citation1999; Drummond et al., Citation2012). Chromosome numbers range between 2n = 24 to 2n = 52, and there are multiple lines of evidence showing that at least one polyploidy event has taken place since the divergence of Genisteae from other Papilionoid legumes (Wolko and Weeden, Citation1989; Gupta et al., Citation1996; Naganowska et al., Citation2003; Nelson et al., Citation2006; Parra-Gonzalez et al., 2012; Yang et al., Citation2013b; Kroc et al., 2014). The structural distinctiveness of Lupinus genomes from other Papilionoid genomes has been investigated by comparing genetic maps of L. angustifolius to the reference genome sequences of Medicago truncatula and Lotus japonicus (Nelson et al., Citation2006; Nelson et al., Citation2010) and by comparing the genetic map of L. albus to the genome of M. truncatula (Phan et al., Citation2007a). These studies revealed that Lupinus genomes are highly rearranged relative to other Papilionoid genomes, with regions of gene collinearity extending over relatively short distances. Comparison between Lupinus genomes has so far been limited by low numbers of shared genetic markers, but initial results suggest that genome rearrangements have occurred even between the relatively closely related L. angustifolius and L. albus (Wolko et al., Citation2011). A complete genome sequence for L. angustifolius is expected in the near future (Gao et al., Citation2011) and will serve as a valuable reference for genomic studies within Lupinus and between Lupinus and other sequenced legume genomes, such as Medicago truncatula, Lotus japonicus, soybean, chickpea and pigeonpea (Sato et al., Citation2008; Schmutz et al., Citation2010; Varshney et al., Citation2012, 2013a; Young et al., Citation2011).
Lupinus has centers of diversity in the Old World and New World (Gladstone, Citation1970). Old World lupins comprise 13 annual species and include rough-seeded and smooth-seeded types distributed around the Mediterranean region and North Africa (Mahé et al., Citation2011). Chromosome numbers range from 2n = 32 to 2n = 52, and nuclear DNA contents range from 2C = 0.97 pg to 2C = 2.44 pg (Naganowska et al., Citation2003), although rare autopolyploids have been observed with 2n = 100 and 2n = 104 chromosomes (Ainouche and Bayer, Citation1999). The most strongly supported clade within the Old World lupins is the rough-seeded type, with four less well-defined smooth-seeded sections recognized (Ainouche and Bayer, Citation1999). Three smooth-seeded Old World species have been used regularly in agriculture: L. albus, as long ago as 2,000 BC; and L. angustifolius and L. luteus by the 19th century (Gladstone, Citation1970). Several other Old World lupin species have been used sporadically in agriculture, including the rough-seeded L. pilosus L., L. atlanticus Gladstone and L. cosentinii Guss. (Gladstone, Citation1970; Wolko et al., Citation2011). Experimental hybridization studies have found that many rough-seeded types can intercross, albeit at a relatively low frequency, while crossing between smooth-seeded types (including the main cultivated species) is rarely successful (reviewed by Wolko et al., Citation2011). Given the great difficulty in interspecific crossing between the cultivated species of lupin, it is perhaps unsurprising that there has so far been no example of the successful transfer of useful genes into lupin crop species. Therefore, for practical breeding purposes, the gene pool of the cultivated Old World lupin species is restricted to the species themselves.
New World lupins comprise over 250 annual and perennial species with centers in western North America (c. 100 species) and the Andes of South America (c. 85 species) (Hughes and Eastwood, Citation2006). Despite the much larger number of Lupinus species in the New World, the diversity of chromosome numbers is remarkably less than in Old World species. Chromosome numbers are typically 2n = 48 for Andean and North American species and 2n = 36 for southeastern South American species, with a few exceptions, including putative autotetraploids (Conterato and Schifino-Wittmann, Citation2006). DNA contents range from 2C = 1.08 pg to 2C = 2.68 pg, with North American species showing the widest range of DNA contents (Naganowska et al., Citation2006). Lupinus in the Andes has among the highest known rates of species diversification for any angiosperm genus (Hughes and Eastwood, Citation2006). The vast range of morphological variation (from tiny herbs to large trees) and ecological adaptation (from coastal sand dunes to montane forests) pose challenges for resolving phylogenetic relationships using taxonomic methods (Drummond et al., Citation2012). Comparisons of nuclear and chloroplast DNA sequences (Ainouche and Bayer, Citation1999; Wink and Mohamed, Citation2003; Ainouche et al., Citation2004; Ree et al., 2004; Drummond and Hamilton, Citation2007; Drummond, Citation2008; Eastwood et al., Citation2008b; Drummond et al., Citation2012) have made some progress toward resolving Lupinus phylogeny, but ambiguities still remain. Current efforts are underway to sample large gene sets obtained by whole transcriptome sequencing across New World Lupinus (C. Hughes, G. Atchison, D. Filatov, personal communication), which should definitively answer these remaining questions. Transcriptome-based studies will also provide insights into the nature and age of polyploidy events that have shaped Genisteae genomes through the comparison of rates of synonymous and non-synonymous substitution in the coding regions of duplicated genes (Cannon et al., Citation2010). The only New World species to be adopted in arable agriculture was L. mutabilis (Andean lupin). It was domesticated around 2,000-1,000 BC in the central Andes, where it was used by the Chavinoid culture and later by Tihuanacoid and Inca civilizations in their crop rotations (Wolko et al., Citation2011).
1. Crop lupin (L. angustifolius, L. albus, L. luteus and L. mutabilis)
The world collection of Lupinus is estimated to comprise approximately 38,000 accessions, duplicates notwithstanding (). Ex-situ collections of Lupinus germplasm are extensive though focused primarily on the four cultivated species (Wolko et al., Citation2011; Berger et al., Citation2013). Lupin germplasm stands out in terms of the large proportion of wild (18%) compared to cultivated material. Due to its recent domestication the current focus is on collecting wild diversity. The largest repository exists in the Australian Lupin Collection (ALC), which holds 66 wild and 912 cultivated acc. of Lupinus albus, 1,327 wild and 729 cultivated acc. of L. angustifolius, 198 wild and 299 domesticated acc. of Lupinus luteus, 31 wild and 208 domesticated acc. of Lupinus mutabilis and 821 accessions of other Lupinus species. Significant collections are also found in the Russian Federation (2,450 acc.), USDA (1,183 acc.), Germany (1,969 acc.), and other European countries. New World species such as L. mutabilis are held in South American institutions (in Peru), but also in the USDA (79); the Leibniz Institute of Plant Genetics and Crop Plant Research, Germany (987); the Institute for Agrobotany, Hungary; and the Vavilov Institute, Russia (129). Molecular marker-based analyses of genetic diversity in geographically defined and/or small germplasm sets of Old World cultivated Lupinus species have been reported (Talhinhas et al., Citation2003, 2006; González-Andrés et al., Citation2007; Raman et al., Citation2008; Sbabou et al., Citation2010; Nelson et al., Citation2011; Berger et al., 2012a). Systematic investigations that integrate both molecular and ecophysiological information are required for all four cultivated Lupinus species.
The global production of lupin in 2012 was 1.29 M tonnes, of which Australia was the largest producer (0.98 M tonnes) with mean yield of 1,445 kg/ha (FAOSTAT, Citation2012). The main use of L. angustifolius grain in Australia is as a high-protein sheep feed. However, there is increasing interest in using L. angustifolius as a human health food. When the seed coat is removed, the kernel contains 40–45% protein and 25–30% dietary fiber and has a low fat and carbohydrate content (Lee et al., Citation2006). Including lupin kernel flour as part of a regular human diet could help address growing obesity and diabetes problems, since it has been shown to increase satiety, thereby reducing further caloric intake (Lee et al., Citation2006).
Research is underway to understand how seed storage proteins are produced in L. angustifolius (Foley et al., Citation2011), which may provide effective selection and/or transgenic tools for breeders to increase the quality and quantity of seed protein. A small proportion of the population is allergic to lupin kernel flour; consequently, research is underway to understand the components of lupin seed proteins associated with allergenicity (Goggin et al., Citation2008). Breeders successfully addressed problems of late maturity caused by a strong vernalization requirement, excessive indeterminate branching and excessive height (Wolko et al., Citation2011). Remaining constraints on the wider adoption of L. albus as a crop are susceptibility to anthracnose, BYMV, Pleiochaeta root rot, brown leaf spot, Fusarium wilt and grey mold. Most L. angustifolius cultivars are susceptible to a range of fungal diseases (anthracnose, phomopsis and pleiochaeta root rot) and viral pathogens (CMV, BYMC) but are resistant to aphids. As current cultivars are slow in establishment after sowing, herbicide tolerance is essential to reduce weed competition. L. angustifolius is tolerant of simazine and diflufenican, and some cultivars are partially tolerant to metribuzin (Si et al., Citation2006; Wolko et al., Citation2011). Breeding has achieved an estimated genetic gain in yield of 81% between the first early flowering variety, “Unicrop” (released in 1973), and “Mandelup” (released in 2004), a rate of gain of 2.6% per year (Stefanova and Buirchell, Citation2010). These yield gains were associated with increased main stem productivity and higher seed numbers (Berger et al., Citation2012b). However, intensive breeding and domestication bottlenecks have reduced genetic diversity and restricted phenological adaptation of L. angustifolius in Australia, thus limiting the potential of future genetic gains (Berger et al., Citation2012a). In Germany in the 1930s, von Sengbusch identified natural sweet-seeded mutants, which heralded the beginning of modern L. albus breeding (Gladstone, Citation1970). Modern L. luteus cultivars are resistant to pod shattering, though improvements are still necessary in Mediterranean-type environments, which experience hot, dry conditions at harvest time (Wolko et al., Citation2011). Soft-seededness, removal of the vernalization requirement for flowering and restricted branching traits were also introduced. There has been excellent progress in increasing grain yield in Polish cultivars over the last decade (Wolko et al., Citation2011).
Lupinus luteus has the highest protein content of the Old World cultivated lupin species (average 38.3%) with high S amino acids, which has driven its increased use in Chile's aquaculture industry (Wolko et al., Citation2011). Constraints on its wider use include the narrow edaphic adaptation, aphid susceptibility in very low-alkaloid cultivars, and susceptibility to CMV, BYMV and anthracnose.
Lupinus mutabilis prefers mildly acidic to neutral loamy sands and loams, is tolerant of water-logging and has very high P-use efficiency (Wolko et al., Citation2011). It has the highest grain quality of all the cultivated lupins, rivalling soybean with an average of 42% protein, 18% oil and a thin seed coat (Wolko et al., Citation2011). It was an important part of the diet and farming practices of pre-Columbian civilizations but was marginalized after the European invasion of the Inca (Gross et al., Citation1988). In recent years, efforts have been made to re-establish L. mutabilis as a crop and to adopt it outside South America (Caligari et al., Citation2000). Low-alkaloid forms were initially developed by von Sengbusch in 1942 as a new variety, with <0.05% alkaloids reported (Gross et al., Citation1988). However, the current adoption of L. mutabilis is limited by late maturity, low and unstable yields and frost susceptibility (Eastwood and Hughes, Citation2008a).
IX. ECO-GEOGRAPHICAL AND ECO-PHYSIOLOGICAL APPROACHES TO CONSERVING AND IDENTIFYING USEFUL GERMPLASM
Geographical and ecological information has been key to many successful germplasm-collecting forays, as well as to the preservation of extant diversity in ex-situ collections. Following collecting with the compiling of descriptor, disease and agronomic data adds value to germplasm collection and enables breeders to make more informed selections when requesting germplasm. Targeting germplasm for tolerance to abiotic stresses can be done through eco-geographical identification of the collection sites of landraces. By overlaying these with world climatic data to a resolution of 1 square km for the months corresponding to local vegetative and reproductive growth phases, landraces associated with sites highly stressed for frost, high temperature and drought can be identified. These landraces, being adapted to such sites, may have undergone natural selection for such stress tolerances. This knowledge enables specific targeting of germplasm for screening of tolerance to abiotic stresses, as conducted for the pea landraces from China (Li et al., Citation2013).
A. Chickpea
Global chickpea distributions and habitat characteristics have been analyzed in detail using passport data from the ICRISAT, ICARDA, ATFCC and USDA collections, augmented by feedback from regional breeders (Berger and Turner, Citation2007; Berger, Citation2007; Berger et al., 2012). Chickpea seasonal climates fall into two broad categories (Berger and Turner, Citation2007):
Mediterranean-type: cool, wet winters, where the crop is reliant on in-season rainfall, and the growing season is terminated by drought (Mediterranean Basin, western parts of Central Asia, southern Australia, western Americas (California, Mexico, Chile). The most arid Mediterranean production areas are found in central Iran, central Pakistan, parts of Afghanistan, the inner Eastern Mediterranean, parts of North Africa, California, northern Chile, and Western Australia (Berger and Turner, Citation2007).
Summer dominant rainfall: winter chickpea relies on stored soil moisture from the preceding monsoon, seasonal temperatures are relatively high, and growth is also terminated by drought (South Asia, East Africa). In South Asia, there is a very strong latitudinal winter temperature gradient, the south being considerably warmer than the north (22.1 and 16.8°C, respectively), leading to much shorter growing seasons (Berger and Turner, Citation2007).
In both climatic regions, chickpea reproduction is timed to avoid chilling stress (see later discussion). However, notable exceptions include southern Australia, the western Mediterranean and Americas, and northern India (Berger, Citation2007). The combination of regionally appropriate farming practices and phenology facilitates avoidance of the principal abiotic and biotic stresses in chickpea. In Mediterranean regions, Ascochyta blight and vegetative frost are both long-standing stresses for the crop, graphically illustrated by ICARDA screening trials in the 1980s (Singh, Citation1990). Of 15,000 lines screened against Ascochyta blight and 4,500 against winter cold, only 18 and 15 accessions, respectively, were resistant, and there was no evidence of combined resistance (Singh, Citation1990). The traditional Mediterranean spring-sowing regime avoids both these stresses but comes with a considerable yield potential opportunity cost (Singh et al., Citation1997). Terminal drought is an almost ubiquitous stress in both Mediterranean and South Asian-type climates. Although chickpea has deep roots (Saxena et al., Citation1994), is able to extract water at depth (Zhang et al., Citation2000), and is capable of osmotic adjustment (Morgan et al., Citation1991; Basu et al., Citation2007; Turner et al., Citation2007), its principal adaptive strategy appears to be drought escape through early phenology (Silim and Saxena, 1993; Siddique et al., Citation2001; Berger et al., Citation2004, 2006). However, this must be balanced against its considerable chilling sensitivity, which causes chickpea to delay pod set until temperatures are warm enough. Delays in pod set are strongest between 11°C and 16°C, and although tailing off after 17.5°C, remain statistically significant until 20.6°C (Berger et al., 2012). In much of its global distribution, chickpea is exposed to terminal drought stress because low chilling tolerance delays the onset of pod formation (Berger, Citation2007; Berger et al., 2012). Chickpea evolution has selected for regionally appropriate phenology to negotiate these various stresses (Berger, Citation2013). Thus, as seasonal temperatures increase from the Mediterranean through South Asia, chickpea cultivars become increasingly temperature responsive, flowering ever earlier to escape terminal drought, minimizing the risk of encountering sub-optimal chilling temperatures (Berger et al., Citation2011). Modern autumn-sown Mediterranean germplasm is relatively unresponsive to temperature and compensates by a strong photoperiod response, flowering comparatively late to minimize both early low temperature and late terminal drought stress. An inverse relationship between photoperiod and temperature response in Mediterranean material (Berger et al., Citation2011) made it possible for chickpea to colonize warmer areas to the south and southeast early in its domestication history, as photoperiod-initiated flowering is wholly maladaptive in low latitude, terminal drought-prone South Asian environments, where flowering occurs under reducing rather than increasing daylength.
B. Lupin
Collection site habitats of the Old World species housed in the Australian Lupin Collection (ALC) have been characterized by calculating site-specific bioclimatic variables, such as monthly mean rainfall; mean, minimum and maximum temperatures; relative humidity; rainy days per month; coefficients of variation for monthly precipitation; frost days per month; and sunshine percentage (Berger et al., Citation2008a, Citationb). Mapping the results of multivariate analysis demonstrates that in all species, accessions tend to align along latitudinal drought stress gradients. Thus, in the Mediterranean basin, cooler, higher rainfall/elevation sites are typically found in northerly locations, such as the Iberian Peninsula, while warmer, drier sites in are common in North Africa and the southern Levant. Material from these contrasting environments forms the basis of current efforts to understand specific adaptation in the genus, as outlined below. L. albus collection site climates are more complex than those of L. angustifolius and L. luteus, including low rainfall sites in central Anatolia, very warm irrigated locations along the Nile River Valley, and warm and wet locations in the Ethiopian highlands (Berger et al., Citation2008b). Recent genotype by environment interaction studies highlighted the limited adaptive and genetic diversity of modern elite cultivars, demonstrating that matching cultivar phenology to target environment (late for long season, early for short season) was not possible throughout much of the current production range because of a confounding between vernalization response and later flowering (Berger et al., Citation2012a, b). The idea that lupin yield could be increased by selecting appropriate adaptive traits for specific target environments has sparked interest in the adaptive strategies of wild germplasm, which, unlike domesticated material, has undergone natural selection in the contrasting Mediterranean environments described above. Long-season, high-rainfall habitats select strongly for delayed phenology, high above- and below-ground biomass production, high leaf area, seed yield and number, a combination of traits that leads to high water use, and the early onset of stress. L. luteus appears to ameliorate this aggressive competitive strategy with some degree of drought tolerance in high rainfall ecotypes, which can maintain higher leaf relative water content under lower critical leaf water potentials under water deficit. By contrast, lupins from terminal drought-prone environments are characterized by ruderal traits that facilitate drought escape/avoidance (early phenology, low biomass and water use, late stress onset) rather than tolerance which limit reproductive potential. Given that modern cultivars tend to express the ruderal traits of low-rainfall ecotypes, there appears to be considerable potential for lifting long-season productivity by introducing some of the competitive traits of high rainfall ecotypes.
C. Common Bean
The FAO-treaty germplasm-based core collection for cultivated common bean, housed at CIAT, was selected based on eco-geographical considerations, although not so formally as in other crops. The main purpose of the core collection was to sample the diversity across the two primary centers of origin for this crop, but only in the cultivated germplasm. Screening of this core collection for low phosphorus stress tolerance was successful, as was evaluation for some disease resistances, although many individual strain resistances or trait mechanisms are related to the overall differentiation of Andean and Mesoamerican phenotypes. This is especially clear when comparing the determinate plant type that occurs in the first genepool to indeterminate plants found in the both genepools, especially the bush beans of the second genepool. Wild germplasm of common bean has been used less often for screening but was the source of resistance to anthracnose and angular leaf spot in some cases. The multitude of pathogenic races for the fungi that cause these diseases makes it difficult to identify genes of any greater importance than those occurring in cultivated germplasm. The long history of introgression between wild and cultivated common beans and vice versa may explain why few novel disease-resistance genes have been found in the primary genepool. The use of secondary and tertiary genepool species, on the other hand, has proved promising for identifying resistances to both biotic and abiotic stresses. The sources of these traits are correlated with eco-geographical features even at the species level, since scarlet runner beans are from rain-leached, acid-soils of humid climates in Central America, and as such are resistant to fungal diseases and aluminum toxicity soil stress. In addition, tepary beans from the arid climates and sandy soils of northern Mexico and southwestern USA provide resistances to salinity and drought stress. Perhaps because of their evolution outside the wetter regions where bacterial and fungal pathogens occur, tepary beans also provide high levels of resistance to specific Xanthomonas and rust infections, respectively. Recently, Cortés et al. (2012a, 2012b and 2013) analyzed the drought tolerance of wild common beans based on climatic data for each accession's collection site and found a correlation with allelic diversity in candidate genes for drought tolerance, such as the ASR and DREB transcription factors.
X. WILD RELATIVES AS A SOURCE OF NOVEL VARIATION
Most researchers agree that wild relatives of crop plants are a useful source of novel variation for potential breeding (McCouch et al., Citation2013). The challenges now are to reintroduce traits that have been lost or not used during the domestication process and subsequent breeding, including disease and resistance genes to make use of the wild allelic diversity that exists in germplasm collections. Highly variable germplasm is found in the secondary and tertiary pools of crop plants. This exotic material has largely remained uncharacterized and underutilized. Fortunately, there is a rising concern surrounding CWR use, and it is now a priority for GCDT. Genetic improvement of many crop plants has already benefited from the incorporation of traits from related wild species and other exotic germplasm sources. The development of pre-bred lines has long been advocated as a means of facilitating the transfer of genes from wild species. However, the majority of published results have been achieved with dedicated crosses and specific selection; thus they need to be made in trait-by-trait manner, which is a time-consuming and expensive process. The synthesis of exotic libraries, such as introgression lines (IL) or chromosome segment substitution lines (CSSL) and near isogenic lines (NIL), containing chromosome segments defined by molecular markers from wild species in a constant genetic background of the related cultivated species, has made the use of alien genomes more precise and efficient (Zamir, Citation2001; McCouch, 2004; Gur and Zamir, Citation2004). Establishment of such a permanent introgression library with characterized genomic fragments of wild crop relatives in a defined genetic background will allow phenotypic characterization of an unlimited number of target traits, which, coupled with molecular tools, will provide a means of final gene identification and their subsequent incorporation, pyramiding in desired genotypes, ultimately leading to better performing commercial cultivars. So far, not many such series of lines have been developed in grain legumes, but there are several ongoing efforts to establish them in pea (Smýkal and Kosterin, Citation2010; Smýkal et al., unpublished), beans (Muñoz et al., Citation2004; Blair et al., Citation2006; Blair and Izquierdo, Citation2012), groundnut (Foncéka et al., Citation2009) and other legumes (reviewed in Upadhyaya et al., Citation2011). Intergeneric legume hybrids have been critically reviewed in McComb (1975), which found insufficient evidence for all reported crosses due to misleading paper titles, confusion of vegetative with generic hybrids, the occurrence of patrocliny, and the frequent occurrence of misplaced generic boundaries. Sobolev et al. (1970, 1971) even reported hybrids between Vicia faba and pea with chromosome numbers of 2n = 12 and 16, respectively. This result is doubtful today in light of unsuccessful hybridization attempts between Vicia faba and several of its relatives, such as V. narbonesis and V. johannis. By contrast, Golubev (Citation1990) reported a well-documented example of a successful intergeneric cross between Vavilovia formosa and Pisum sativum (reviewed in Mikič et al., Citation2013), which may not surprising given that they are sister lineages that diverged only c. 8 Mya ago (Schaefer et al., Citation2012). Ben Ze'ev and Zohary (Citation1973) were the first to perform systematic crosses within and between pea species and between subspecies and noted cytological behavior at meiosis. Hybrids between P. sativum subsp. sativum (P. humile) and P. sativum subsp. elatius had reduced fertility as a consequence of meiotic irregularities, and this was more pronounced in hybrids with P. fulvum. They noted in reciprocal crosses that it was only possible to use P. fulvum as the male parent. Due to translocation, the hybrids between cultivated P. sativum and P. sativum subsp. elatius, as well as southern “P. humile,” had also reduced fertility, while with northern “P. humile” were normal as a result of a standard karyotype. Two reciprocal translocations (T1-7) and (T3-5) account for reduced fertility and distorted segregation of hybrids between cultivated P. sativum and P. fulvum (Errico et al., 1991, Citation1996; Campbell Citation1997), together with different numbers of nucleolus-organizing chromosomes (De Martino et al., 2000). Durieu and Ochatt (2000) have tested protoplast fusion and regeneration of calli between Pisum sativum and Lathyrus sativus, and although heterokaryons were detected and up to 6 cell divisions were observed, no further growth or plant regeneration could be achieved. Pisum fulvum was used to introduce resistance to powdery mildew (Fondevilla et al., Citation2007), bruchid pests (Clement et al., Citation2002, 2009; Byrne et al., Citation2008) and Orobanche crenata (Rubiales et al., Citation2009), while primitive landraces were used in order to incorporate virus and Fusarium resistances (Providenti, 1990; McPhee et al., 1999). Wild accessions of P. sativum subsp. sativum or subsp. elatius (variously named P. humile or P. syriacum in papers) have often found to be resistant to various biotic stresses (reviewed in Smýkal et al., Citation2013). Attempts to cross Pisum with Lathyrus sativus did not result in fertile, viable plants (Ochatt et al., Citation2004), although the phylogenetically closest L. ochrus, L. clymenum, and L. neurolobus (Schaefer et al., Citation2012) have not been tested. The development of backcross recombinant inbred lines containing chromosome segments of the wild pea P. fulvum or P. sativum subsp. elatius in a cultivated pea (P. sativum subsp. sativum) genetic background defined by molecular markers is currently being performed by Smýkal and Kosterin (2010 and unpublished).
The wild Lens taxa are known to possess resistance to biotic and abiotic stresses (Bayaa et al., Citation1994, 1995; Hamdi and Erskine, Citation1996; Hamdi et al., Citation1996; Gupta and Sharma, Citation2006). Incorporation of diverse genetic material from wild relatives using intensive hybridization would make it possible to recreate some of the lost variability while still respecting productivity and other desirable traits in lentil. Intraspecific hybridization between cultivated lentil and L. culinaris subsp. orientalis, L. odemensis, L ervoides and L nigricans has been attempted in the past (Ahmad et al., Citation1996; Gupta and Sharma, Citation2007). Lens culinaris subsp. orientalis is readily crossable with the domesticated lentil, although the fertility of the hybrids depends on the chromosome arrangement of the wild parent. Pod abortion took place when the cultivated lentil was crossed with either L. ervoides or L. nigricans (Abbo and Ladizinsky, Citation1991). In vitro methods of embryo-ovule rescue are used overcome the post-fertilization interspecific barrier (Fratini and Ruiz, Citation2006; Citation2011).
In case of Lathyrus, among 1,555 accessions of 45 wild species conserved at ICARDA, a toxin-free gene has been identified in L. tingitanus, which can be used to develop toxin-free grass pea cultivars, providing its hybridization with L. sativus is possible. Lathyrus species, such as L. ochrus and L. clymenum and L. cicera (Fernández-Aparicio et al., Citation2009; 2010), have been identified as possessing resistance to Orobanche, a resistance that is not available within cultivated germplasm. Lathyrus cicera is also a good source for earliness and cold tolerance. However, alien gene transfer has hardly been attempted in grass pea in spite of the success of interspecific hybridization between L. sativus and two wild Lathyrus species (L. cicera and L. amphicarpos L.) with viable seeds (Yunus, Citation1991; Addis and Narayan, Citation2000). Other tested species formed pods but did not produce fully developed, viable seeds. It may be concluded that breeding strategies involving alien genetic transfer for the improvement of grasspea are possible through the readily crossable species L. cicera and L. amphicarpos, but biotechnology tools will be needed to assist in gene transfers among other species (Ochatt et al., 2004).
Numerous studies have attempted to facilitate the useful gene transfer from wild Cicer species to the cultivated chickpea and vice versa. Successful hybridizations between the cultivated chickpea and C. reticulatum or C. echinospermum and their reciprocals have been reported (Ladizinsky and Adler, Citation1976aCitationb; Jaiswal and Singh, Citation1986; Singh and Ocampo, Citation1993; Croser et al., Citation2003; Ahmad and Slinkard, Citation2004; Singh et al., Citation2005; Clarke et al., Citation2006; Knights et al., Citation2008; Malikarjuna et al., 2011; Thompson et al., Citation2012). Although some of the accessions of C. bijugum, C. judaicum and C. pinnatifidum used as pollen donors were crossed with the cultivated chickpea, hybrids were available via embryo rescue techniques (Ahmad and Slinkard, Citation2004). So far, there have been no successful gene transfers between the cultivated chickpea and perennial wild Cicer species due to post-zygotic hybridization barriers. Hybrids between C. arietinum and C. pinnatifidum (Badami et al., Citation1997; Mallikarjuna, Citation1999), C. arietinum and C. judaicum (Verma et al., Citation1995), and C. arietinum and C. bijugum (Mallikarjuna et al., Citation2007) were obtained via embryo rescue and tissue culture techniques. Some hybrids between C. judaicum and C. bijugum, as well as between C. cuneatum Hochst. ex A. Rich. and C. canariense A. Santos & G.P. Lewis, were produced by Abbo et al. (Citation2011). Other hybrids between C. arietinum and C. judaicum, C. arietinum and C. pinnatifidum, and reciprocal crosses were obtained by Clarke et al. (Citation2011).
As pigeonpea suffers limited genetic diversity within the cultivated gene pool, it is imperative to increase genetic diversity by using wild relatives from different gene pools. A number of species from the secondary gene pool (C. sericeus, C. albicans (Wight & Arn.) Maesen, C. lineatus (Wight & Arn.) Maesen, C. trinervius (DC.) Maesen, C. cajanifolius and C. scarabaeoides) have shown crossability with the cultivated type. Several inter-specific crosses have produced hybrids that showed shrivelled and non-viable seeds, proving that crossability barriers exist within the genus (Yadav and Padmaja, Citation2002). Some of these wild species have been found to be sources of resistance/tolerance to various biotic and abiotic stresses and of agronomically important traits, such as sterility mosaic disease resistance, high protein content, high fruit set, pod borer resistance, salinity tolerance, etc. (see Bohra et al., Citation2011b). Inter-specific hybridization has played an important role in the development of the cytoplasmic male sterility (CMS) system in pigeonpea (see Saxena et al., Citation2010). Therefore, some of the secondary gene pool species have been used successfully. However, few of the wild relative species from the tertiary gene pool have shown promising crossability and are difficult to use for pigeonpea improvement.
Even though wild soybean is considered the closest relative of the cultivated soybean (Hymowitz, Citation1970), it has significant phenotypic differences. The large phenotypic diversity in soybean is genetically controlled in both qualitative and quantitative aspects. For example, wild soybean has mainly tiny, black seeds in contrast to the large, yellow seeds of cultivated soybean. There are also significant differences in the seed oil and protein concentration between wild and cultivated soybeans (Xu and Gai, Citation2003; Chen and Nelson, 2004). Several studies suggest that wild soybean has important phenotypic characteristics and specific alleles that are not present in cultivated soybean (Carter et al., Citation2004). Major traits of agricultural importance, including yield and stress tolerance, are polygenic, and the presence of these favorable alleles in G. soja will help breeding programs introduce beneficial traits into soybean (Tanksley and McCouch Citation1997; Li et al., Citation2008). Therefore, wild soybeans are important sources of novel alleles that can be used to broaden the genetic base of cultivated soybean, which is necessary due to the fact that diversity in soybean has been greatly reduced by the genetic bottleneck of domestication (Guo et al., Citation2010; Lam et al., Citation2010; Li et al., Citation2010).
Cowpea has an intrinsically narrow genetic base that limits breeders’ progress today. However, there are few reports in published literature on the use of wild cowpea relatives for the genetic improvement of cultivated cultivars. The relatively low level of utilization of wild cowpea relatives in the development of improved cowpea cultivars may be due to factors like linkage drag. The basic need for exploiting the wild relatives is its cross compatibility with cultivated cowpea. It is possible that some of the available wild cowpea lines belong to the same or different gene pools. The subspecies or cultivars that constitute the primary and secondary gene pools for cowpea are not yet well defined ().
Among all the genetic barriers, the difference in ploidy level is cultivated groundnut's main obstacle in sharing alleles with its wild relatives. Even though there has been a continuous effort to tackle this genetic barrier via three main pathways (the hexaploid, the autotetraploid and the allotetraploid routes), very limited success has been achieved. The hexaploid route involves crossing between diploid and tetraploid genotypes, followed by chromosome doubling through colchicine treatment achieving hexaploid (60 chromosomes). This hexaploid was used for repeated backcrossing with cultivated species (A. hypogaea), and resultant progenies were used for several genetic and breeding applications, such as introgression lines/populations, genetic maps and even germplasm releases or cultivars with disease resistance (see Burrow et al., 2013). The second route involves the creation of synthetic autotetraploides (AAAA or BBBB) through colchicine treatment of diploid species (AA or BB genome) and their use in crossing with cultivated genotype (Singh, 1985; Mallikarjuna et al., Citation2011). The third route involves the creation of synthetic allotetraploides (AABB) through crossing two diploid species (AA and BB genomes), followed by colchicine treatment. Several allotetraploid synthetics were successfully developed using this method and were used for introgressing wild alleles into cultivated germplasm (Simpson, 1991; Simpson et al., 1993; Fávero et al., 2006; Mallikarjuna et al., Citation2011). The development and use of the allotetraploid “TxAG-6 ({A. batizocoi Krapov. & W.C. Greg. × [A. cardenasii Krapov. & W.C. Greg. × A. diogoi Hoehne]}4x)” presents one notable example for wide applications, such as introgression of resistance for root-knot nematode and genetic maps using mapping populations (cultivated Florunner × amphidiploid TxAG-6). Resistance to root-knot nematode and foliar diseases (rust and late leaf spot) was introduced into the cultivated genepool from A. cardenasii via the hexaploid route (Garcia et al., 1996; Gowda et al., 2002), while root-knot nematode resistance was introduced via the tetraploid route (Simpson, 1991). Development and release of root-knot nematode resistant cultivar “COAN” (Simpson and Starr, Citation2001) and foliar disease (rust and late leaf spot) resistant cultivar “GPBD 4” (Gowda et al., 2002) are other notable examples. TxAG-6 amphidiploid was used for developing root-knot nematode resistant cultivar “COAN” by crossing with cultivated Florunner (Simpson and Starr, 2001). Similarly, an interspecific line (CS 16 or ICGV 86855) derived from the cross between A. hypogaea and A. cardenasii was used as a parent in the development of GPBD-4 (KRG 1 × ICGV 86855) (Gowda et al., 2002). Thus far, 12 wild relatives have been deployed for the development of synthetics for enriching the primary gene pool in groundnut. These include A. cardenasii, A. diogoi, A. batizocoi, A. ipaënsis, A. duranensis, A. gregoryi C.E. Simpson, Krapov. & Valls, A. linearifolia Valls, Krapov. & C.E. Simpson, A. magna Krapov., W.C. Greg. & C.E. Simpson, A.valida Krapov. & W.C. Greg., A. kempff-mercadoi Krapov. & W.C. Greg., A. stenosperma Krapov. & W.C. Greg., and A. hoehnei Krapov. & W.C. Greg. In addition to above-mentioned cultivars, several other elite lines have been bred using wild relatives from across the world, and these elite lines possess resistances to different diseases and pests in groundnut (see Sharma et al., Citation2013).
Due to unique adaptations, alfalfa CWR have made substantial contributions to alfalfa breeding. Cold-hardy and drought-tolerant M. sativa subsp. falcata has been used to expand the adaptive range of alfalfa into colder and drier locations (Small, Citation2011; Barnes et al., Citation1977). There have also been breeding efforts to capitalize on heterosis between subsps. M. sativa subsp. sativa and falcata (Riday et el., 2002a,b; Riday and Brummer, Citation2005). The glandular hair trait found in M. sativa subsp. falcata var. viscosa, M. sativa subsp. glomerata and M. sativa subsp. sativa × M. sativa subsp. glomerata is considered an adaptation that conveys insect resistance (Small, Citation1986; Small and Brooks, Citation1986). In the United States, CWR introductions with glandular hairs have given rise to proprietary alfalfa cultivars that are resistant to potato leafhopper (Empoasca fabae Harris), a serious pest in the Eastern United States (Shockley, Citation2002). Fertile interspecific hybrids are difficult to obtain in Trifolium (Taylor et al., Citation1980), and generally success occurs between closely related taxa only (Taylor and Quesenberry, Citation1996). There are some exceptions: allotetraploid white clover originated as a hybrid between T. pallescens Schreb. and T. occidentale D. E. Coombe (Williams et al., Citation2012).
An introgressive crossing strategy was proposed by Cowling et al. (Citation2009) to increase genetic diversity in the Australian Lupinus angustifolius breeding program. The strategy involves crossing wild donor accessions with a domesticated variety three times, followed by single-seed descent. Only targeted selection of domestication traits is applied before the BC2S3 generation (two backcrosses of the F1, followed by three generations of single-seed descent) to maximize the probability that most of the wild-donor genome is represented. Early yield trial data indicated that this strategy is effective, with some introgression lines yielding almost 30% higher than the recurrent domesticated variety (Berger et al., Citation2013). A simplified version of this approach, whereby European cultivars acted as donors, appeared to be even more effective, with up to 44% higher yields than the recurrent Australian variety (Berger et al., Citation2013). However, given the close genetic relationship between Australian and European breeding material, it is likely that such gains will be less sustainable compared to introgressive crossing with the much more diverse wild germplasm (Berger et al., Citation2012a). In addition to their role in increasing yield, these genetically diverse populations will be grown in multi-environment trials to study specific adaptations in a domestic framework, linking adaptive traits to QTLs where possible. Interspecific crossing of L. mutabilis has been successfully achieved with other New World lupins with 2n = 48 chromosomes, most notably L. tomentosus DC., L. mexicanus Cerv. ex Lag, and L. hartwegii Lindl. (Clements et al., Citation2008). Indeed, the ability to intercross New World lupin species was the basis of the development of ornamental Russell lupins with a wide variety of flower colors (Wolko et al., Citation2011). Therefore, unlike the Old World lupin crop species, where interspecific crossing is extremely challenging, L. mutabilis breeders have access to a broad secondary genepool of related species for introgressing traits not available within the primary genepool.
Two novel techniques for introgression of wild germplasm diversity into breeding programs have been pioneered in common bean. P. vulgaris was the first legume in which the advanced backcross (AB)-QTL method was applied to incorporate agrononomically valuable alleles from the wild into the cultivated form (Blair et al., Citation2006). In another study, Blair and Izquierdo (Citation2012) found that genes from the small-seeded, wild common beans can increase the seed concentration of mineral elements of nutritional importance, such as iron and zinc, in an AB-QTL breeding program of large-seeded Andean beans. However, further research summarized in Blair (Citation2013) found that wild beans concentrate many minerals in their seed coats, and that this is the mechanism of higher mineral concentration in the chromosome segment substitution lines developed by the AB breeding method. These results build on the transfer of other seed characteristics, such as the arcelin and APA cotyledonary proteins that confer insect resistance from wild to cultivated beans by marker-assisted selection, even in regions of low linkage disequilbrium (Blair et al., Citation2010). In terms of inter-specific crosses, certain common bean advanced lines have been improved from embryo-rescued hybrids between P. vulgaris and P. acutifolius, with introgression confirmed by AFLP analysis of congruity backcross derived lines compared with standard backcross lines (Muñoz et al., Citation2004, 2006). A similar program, but with limited backcrossing, has been initiated for common bean using P. coccineus and P. dumosus accessions and has resulted in limited introgression in need of confirmation through marker analysis. More focused and concentrated introgression of P. acutifolius or P. parvifolius genes may be useful for incorporating drought and heat tolerance into common beans. Meanwhile, P. lunatus and its relatives have never been used for common bean improvement, although they may be valuable for climate change adaptation. Another alternative is to use lima beans and tepary beans in place of common beans. This will require breeding of these wilder species into more widely adapted modern crops for a range of climates and markets. One major goal of breeding in tepary beans is to increase seed size and to produce more variable seed colors, while the bush bean habit still needs to be improved in lima beans.
The wild, related species and other cultigens of Vigna do not form a particularly extensive or accessible gene pool (Smartt, Citation1990). Even the two closest relatives, V. radiata and V. mungo, have some structural differentiation among their genomes. Despite the phylogenetic proximity of V. vexillata and cowpea, there exists a strong barrier to cross compatibility between them (Fatokun, Citation2002). Lawn (Citation1995) proposed that the Asian Vigna consists of three more or less isolated genepools, based on cross-compatibility studies, which correspond with groups based on seedling characteristics proposed by Tateishi (Citation1996).
XI. IMPACT OF GENOMICS FOR CROP LEGUME GERMPLASM UTILIZATION
Until recently, a very limited number of genomic resources―—a few hundred molecular markers, some fragmentary genetic maps―were available in most of the legumes. Over the last decade, various types of genomic resources, such as microsatellites or simple sequence repeats (SSR), expressed sequence tags (ESTs), single nucleotide polymorphism (SNP), cleaved amplified polymorphic sequences (CAPS), conserved intron spanning primers and diversity array technology (DArT) markers have been developed. Molecular marker technologies, however, are currently undergoing a transition from largely serial technologies based on separating DNA fragments according to their size (SSR, AFLP) to highly parallel, hybridization-based technologies that can simultaneously assay hundreds to tens of thousands of variations, especially in genes. With completed and annotated genomes of model legumes, such as the 373 Mb genome of Medicago truncatula (Young et al., Citation2011), the 472 Mb genome of Lotus japonicus (Sato et al., Citation2008), and of three legume seed crops: the 1,112 Mb genome of Glycine max (Schmutz et al., Citation2010), the 833 Mb genome of pigeonpea (Cajanus cajan) (Varshney et al., Citation2012), the 738 Mb genome of Cicer aerietinum (Jain et al., Citation2013; Varhsney et al., 2013), and the ongoing genome sequencing efforts in Phaseolus vulgaris (550 Mb), Pisum sativum (4,600 Mb), Lupinus angustifolius (924 Mb), Trifolium praetense (440 Mb) and Arachis hypogaea (2,800 Mb). There is strong potential for comparative genomics and its applications, including specific gene/allele mining and deeper diversity studies of legume germplasm collections. One example is that the sequencing of 90 chickpea accessions, composed of landraces and five wild species, has provided information related to domestication and diversification (Varhsney et al., 2013). This information and many other re-sequencing efforts in chickpea and pigeonpea will be used to gather insight into genome evolution and phylogeny and gene-to-trait identification. Moreover, comparative sequencing of wild crop progenitors, such as studies involving Glycine and Cicer, should provide clues to the domestication process and enable practical exploration of CWR.
A chromosome-scale draft sequence of cultivated soybean (var. Williams 82) with 46,430 deduced protein-coding genes has been available since 2010 (Schmutz et al., Citation2010). Using the genome of Williams 82 as a reference, a wide range of nucleotide and structural variations between wild and domesticated soybean have been catalogued (Kim et al., Citation2010; Lam et al., Citation2010; Li et al., Citation2013b). However, some genomic regions present in wild soybean but absent in the cultivated reference need to be uncovered by de novo sequencing of wild soybean (Stupar, 2010). Therefore, it is necessary to sequence a set of diverged wild soybean accessions and build up a pan-genome of wild soybean for uncovering their specific genes for soybean improvement (Qiu et al., Citation2013). TILLING populations have also been made for this crop in order to discover mutant alleles in the small-seeded common bean advanced line BAT93 (Blair et al., Citation2008; Porch et al., Citation2009).
The reference genome of an Andean genepool common bean (Phaseolus vulgaris) G19833 line has been obtained by re-sequencing various other genotypes, which will lead to faster gene discovery or characterization and development of markers for the selection of specific genes with known functions (Blair et al., Citation2013). The missing ingredients for rapid advances in breeding of common bean are i) the lack of genomic or transcriptomic sequences for the other cultigens within the genus, such as lima bean, scarlet runner bean and tepary bean; ii) the small number of accessions of wild germplasm collected for each of the cultivated species and their fast disappearance in regions of heavy urbanization across the mid-elevation valleys of Latin America; and iii) the small number of inter-specific and even inter-varietal crosses that have been made for each of the cultivated groups (Blair et al., Citation2012a, 2012b). Phenotyping, while challenging to carry out on a large scale, is quite advanced in common bean; therefore, common bean is not subject to the tremendous limitation predicted for other legume species. However, some of the cultivated species with long-season production cycles are indeed difficult to phenotype. These include climbing (or pole) common and lima beans, as well as scarlet runner beans, which are, for the most part, very late maturing and limited in adaptability.
Comparably, genomic study of Vigna crops have has lagged. The utilization of cowpea germplasm has gradually been strengthened through the application of molecular breeding technology (Undal et al., Citation2011; Kumar et al., Citation2012; Kaewwongwal et al., Citation2013) and QTL analysis (Sholihin and Hautea, 2002; Humphry et al., 2005; Kasettranan et al., 2010; Kongjaimun et al., Citation2012; Chankaew et al., 2014; Kajonphol et al., 2012). The similarity of the cowpea and common bean genomes is well documented, and this should help to transfer genetic knowledge between genera and from one crop to the other. Sequencing of the cowpea genome is underway, and large transcriptome, SNP marker and physical mapping resources are available for the crop. Through the Tropical Legumes I project in the Generation Challenge Program at the University of California, Riverside, cowpea genomics activities are being conducted, and the tools developed there will be used in cowpea breeding programs. A high-throughput SNP genotyping platform based on Illumina 1536 GoldenGate Assay was developed. The result was a cowpea consensus map containing 928 SNP markers on 619 unique map positions distributed over 11 LGs, covering a total genetic distance of 680 cM (Muchero et al., Citation2009). This offers the framework for QTL identification, map-based cloning, and assessment of genetic diversity, association mapping and applied breeding.
Until recently, lentil molecular breeding relied on other legume species’ genomic resources for the development of new markers (Pandian et al., Citation2000; Reddy et al., Citation2010; Alo et al., Citation2011; Datta et al., Citation2011). This was a successful strategy because of the synteny between lentil and the model legume Medicago truncatula (Phan et al., Citation2007b). With the publication of three transcriptomes of lentil (Sharpe et al., Citation2013; Verma et al., Citation2013; Kaur et al. Citation2011), this is rapidly changing. One transcriptome targeted SNP discovery, resulting in the publication of a SNP-dense genetic linkage map (Sharpe et al., Citation2013). New EST-SSRs (2,393 and 8,722) were discovered using unigene sets of 20,419 and 20,009 (Kaur et al., Citation2011; Verma et al., Citation2013). Many of the main lentil breeding objectives are quantitative (yield, quality, disease and stress tolerances), and the development of useful maps can assist in effective QTL identification for marker-assisted selection. Moreover, mapping the new SNPs and EST-SSRs moves lentil a step closer to genome wide association studies (GWAS) (Sharpe et al., Citation2013).
Yang et al. (2012) reported a total of 162,448,842 base pairs of genomic sequences from SSR enriched libraries constructed with genomic DNA from 247 faba bean accessions. Next-generation sequence technology has been applied to generate faba bean genomics resources for large-scale SSR identification (Yang et al., 2012). A high throughput SNP genotyping array targeting 887 loci has been developed for genomic-assisted breeding in faba bean (Cottage et al., Citation2012). Limited gene sequence homologies and synteny to Medicago truncatulla have been applied to anchor gene-based markers in the faba bean and lentil linkage groups (Ellwood et al., Citation2008). However, genome-wide comparison between these two species has not yet been carried out due to the scarcity of faba bean genome sequence data.
In groundnut, identified linked markers to root-knot nematode were used to transfer resistance from amphidiploid to cultivated groundnut, which resulted in the development of the first molecular breeding product in groundnut, NemaTAM (reviewed in Varshney et al., Citation2013b). The use of markers proved helpful in selecting plant progenies under varied soil and fluctuating environmental conditions. Parallel efforts also led to the development of cleaved amplified polymorphic sequence (CAPS) markers for the high-oleate trait, which provided an opportunity to improve both traits (nematode and high oleate), together leading to the development of another molecular breeding product, “Tifguard High O/L” (reviewed in Varshney et al., Citation2013b). Superior lines with desirable yield and higher rust resistance were identified and subjected to yield evaluation in replication for further multiplication and multilocation trails (Varshney et al., Citation2014). However, in the case of drought tolerance and yield components, several QTLs contributing only small phenotypic variation were identified using family-based mapping populations. Recent advances in genomic technologies have opened avenues of research and marker development in ‘orphan’ legume species that were previously the preserve of well-resourced model species (Varshney et al., Citation2009b).
In case of alfalfa, genomics can use the most directly knowledge of Medicago truncatula model. Exploiting genetic diversity in alfalfa genetic resources is being advanced by the development of molecular markers for important abiotic and agronomic traits, such as aluminium tolerance (Ku et al., 2013), biomass (Robins et al., Citation2007a), persistence (Robins et al., Citation2008), yield, plant height and regrowth (Robins et al., Citation2007b). More recently, a large number of genome-wide EST and SNP markers have been developed using transcriptome sequencing (Li et al., Citation2012). These markers, available through the Legume Information System (http://medsa.comparative-legumes.org/), will support marker-assisted breeding efforts and may be helpful in guiding introgression efforts.
Large insert clone libraries of the Lupinus angustifolius genome have been developed from European and Australian cultivars (Kasprzak et al., Citation2006; Gao et al., Citation2011). Transcriptome sequencing has been reported for L. albus and L. luteus (Parra-Gonzalez et al., 2012; O’Rourke et al., 2013) and is underway for L. angustifolius and a range of New World lupin species. A reference genome sequence assembly for L. angustifolius is currently under construction (Gao et al., Citation2011), and a genome survey has been reported (Yang et al., Citation2013b). Genotyping-by-sequencing has been used to develop improved markers for anthracnose and phomopsis resistance in L. angustifolius (Yang et al., Citation2012; Yang et al., Citation2013a). These resources are being used to address basic questions relating to genome evolution, Lupinus phylogeny, seed storage protein synthesis, specific adaptation and identification of domestication genes, as well as developing markers for applied breeding purposes. Knowing the genes underlying important agronomic and quality traits paves the way for refining traits by reverse genetic and transgenic approaches (Berger et al., Citation2013).
With advances in model legume sequencing and genomic knowledge, there has been a switch to gene-based markers in pea (Aubert et al., Citation2006; Jing et al., Citation2007; Deulvot et al., Citation2010; Bordat et al., Citation2011). Recently, a comprehensive transcriptome of pea was published (Franssen et al., Citation2011), and another RNA-seq atlas is being established at INRA, France (http://bios.dijon.inra.fr). This trend can be expected to further proliferate in conjunction with rapid advances in high-throughput single-nucleotide polymorphism (SNP) generation and detection assays (Deulvot et al., Citation2010; Bordat et al., Citation2011). In pea, a transcription atlas (RNAseq) and gene-based maps (Bordat et al., Citation2011) will aid translation of genomic knowledge to practical breeding. Moreover, the pea variety Cameor was used to develop the TILLING mutant population (Dalmais et al., Citation2008) and to construct a BAC library, both essential tools for positional cloning and pea genome sequencing (reviewed in Smýkal et al., Citation2012; Smýkal and Konečná, Citation2014). Increased knowledge of the pea genome has not only a scientific but also a great educational and social impact, owing to seminal work of J. G. Mendel (1865–66) (Schwarzbach et al., 2014).
Because of the limited amount of genomic resources and low polymorphism in cultivated germplasm, initial genetic mapping studies were restricted to inter-specific mapping populations. For trait mapping, however, it is important to develop genetic maps based on intra-specific mapping populations. However, the translation of genomics (QTLs/ markers identified) to legume breeding is still in its infancy, and, despite the efforts and progress made in developing molecular resources, their use in breeding has been limited (see Varshney et al., Citation2013b, this issue). Several factors limit the direct application of QTLs and their associated markers, including: i) high genotype x environment interactions on expression; ii) the necessity of testing the polymorphism of the molecular markers in different genetic backgrounds; iii) large (5–10 cM on average) genetic distances between markers and the QTLs; iv) imprecise phenotypic description that has resulted in inaccurate marker-trait associations; v) the use of small mapping populations (50–200 individuals) that has resulted in limited genetic resolution; vi) the lack of common reference markers across QTL studies; vii) the limited range of variation in the cultivated genepool; viii) trait and marker validation in different genetic backgrounds; and ix) inadequate investment in many of legume crops, which has created a lag in the development of molecular tools for breeding (Smýkal et al., Citation2012). The availability of a high-throughput genotyping platform on the appropriate germplasm collections mentioned will facilitate the use of the association genetics approach for identification of genes/markers associated with traits of interest to breeders. However, the association of genomic sequences/haplotypes with traits of interest to breeders would require multi-location and precise phenotyping data, as well as appropriate analytical tools on a high-computing bioinformatics platform.
Genetic characterization raises new issues for the management of genetic diversity within accessions, since preserving the original genetic composition of sample accessions is usually required. In many cases, particularly for wild relatives and traditional cultivars, this involves conserving genetically heterogeneous populations in a form that is difficult to use for gene discovery. It has been recommended that wherever possible, single plants should be used as the source of DNA for sequencing, and seed derived from these single plants should be set aside as “reference seed stocks.” These stocks will also serve as the source of material for phenotyping and ensure that phenotypic information can be associated with the sequence information in a meaningful way. However, creating a new accession for each genotyped accession has significant consequences over the cost and size of germplasm maintenance. A coordinated effort to characterize germplasm collections is needed, along with advanced analytical methods that allow for three-way testing of diversity in genotypes, locations and quantitative traits to provide dynamic characterization of genotypic and phenotypic diversity. Such dynamic characterization could be used to study adaptation across a range of different ecological locations. Such a core set would provide a useful and powerful resource for next-generation markers, such as SNPs or whole-genome sequencing (WGS), and, more importantly, for phenotypic analysis of agronomic traits. Recent advances in genomic technology, the impetus to exploit natural diversity, and the development of robust statistical analysis methods make association mapping affordable for most legumes (reviewed in Smýkal et al., Citation2012). Genomics-assisted breeding has already proved to be a very useful approach, which provides much-needed precision in the selection of target traits and significantly reduces the duration of cultivar development. Compared to conventional linkage-mapping based on time-consuming biparental mapping population development, linkage disequilibrium (LD)-mapping using the non-random associations of loci in haplotypes is a powerful, high-resolution tool for complex quantitative traits.
XII. FUTURE OUTLOOK
Next-generation sequencing and high-throughput genotyping platforms promise to further revolutionize our understanding of genetic diversity and to assist in designing strategies to utilize the genomic information for legume crop improvement. It is possible to sequence large-scale germplasm collections of legumes held in genebanks of CGIAR centers and national genebanks of different countries. Genome-wide sequence data for these germplasm collections will provide an opportunity to develop a “hapmap” for the given species. These “hapmaps,” on one hand, can be used for genome wide association analysis if, provided precise, large-scale phenotyping data for traits of interest to breeders is available. Therefore, there is an urgent need to establish a global phenotyping network for comprehensive and efficient characterization of legume crop germplasm for an array of target traits, particularly for biotic and abiotic stress tolerance and nutritional quality. Genome-wide sequence data can be stored in user-friendly databases that can serve as “digital genebanks.” Such “digital genebanks” can be helpful in designing the primer pairs and amplifying gene(s) of interest. Low utilization of germplasm use, despite its accessibility, in crop breeding has always been debated, and it is anticipated that genome-wide sequence information and, most importantly, the association of alleles with targeted phenotyping traits potentially will provide sufficient knowledge to the crop community to decide which accession(s) and which genomic segment(s) they need to target for improving a given trait in a particular legume crop species. It is also important to mention that information generated regarding germplasm of, whether genotyping, genome sequencing or phenotyping, should be made available as open access data.” This would link seeds and genetic stocks directly to passport, genomic and phenotypic information, thereby engaging the creativity of geneticists and breeders. This will help crop communities across the world make the best use of the information generated/ available for crop improvement.
Additional information
Funding
REFERENCES
- Abberton, M.T. 2007. Interspecific hybridisation in the genus Trifolium. Plant Breeding 127: 597–601.
- Abberton, M.T. and Marshall, A.H. 2010. White clover. In: Fodder Crops and Amenity Grasses. Handbook of Plant Breeding. vol. 34., pp. 457–476. Boller, B., Posselt, U., and Veronesi, F., Eds., Springer, New York.
- Abberton, M.T. and Thomas, I. 2011. Genetic resources in Trifolium and their utilization in plant breeding. Plant Genet. Res. Charact. Util. 9: 38–44.
- Abbo, S. and Ladizinsky, G. 1991. Anatomical aspects of hybrid embryo abortion in the genus Lens L. Bot. Gaz. 152: 316–320.
- Abbo, S., Berger, J. and Turner, N.C. 2003a. Evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation. Funct. Plant Biol. 30: 1081–1087.
- Abbo, S., Lev-Yadun, S., and Gopher A. 2012. Plant Domestication and Crop Evolution in the Near East: On Events and Processes. Crit. Rev. Plant Sci. 31: 241–257.
- Abbo, S., Lev-Yadun, S., and Gopher, A. 2010. Agricultural origins: centers and noncenters; a Near Eastern reappraisal. Crit. Rev. Plant Sci. 29: 317–328.
- Abbo, S., Lev-Yadun, S., Heun, M., and Gopher, A. 2013. On the ‘lost’crops of the neolithic Near East. J. Exp. Botany 64: 815–822.
- Abbo, S., Mesghenna, Y.T., and Van Oss, H. 2011. Interspecific hybridization in wild Cicer sp. Plant Breeding 130: 150–155.
- Abbo, S., Shtienberg, D., Lichtenzveig, J., Lev-Yadun, S., and Gopher, A. 2003b. The Chickpea, Summer Cropping, and a New Model for Pulse Domestication in the Ancient Near East. The Quart. Review Biol. 78: 435–438.
- Abbo, S., Zezak, I., Schwartz, E., Lev-Yadun, S., and Gopher, A. 2008. Experimental harvesting of wild peas in Israel: implications for the origins of Near East farming. J. Arch. Sci. 35: 922–929.
- Abo-Elwafa, A., Murai, K., and Shimada, T. 1995. Intra-and inter-specific variations in Lens revealed by RAPD markers. Theor. Appl. Genet. 90: 335–340.
- Addis, G. and Narayan, R.K. J. 2000. Inter-specific hybridization of Lathyrus sativus (Guaya) with wild Lathyrus species and embryo rescue. African Crop Sci. J. 8: 129–136.
- Adegbite, A.A. and Amusa, N.A. 2008. The major economic field diseases of cowpea in thehumid agro-ecologies of South-western Nigeria. African J. Biotech. 7: 4706–4712.
- Ahmad, F. and Slinkard, A.E. 2004. The extent of embryo and endosperm growth following interspecific hybridization between Cicer arietinum L. and related annual wild species. Genet. Res. Crop Evol. 51: 765–772.
- Ahmad, F., Slinkard, A.E., and Scoles, G.J. 1988. Investigations into the barrier(s) to interspecific hybridization between Cicer arietinum L. and eight other annual Cicer species. Plant Breeding 100: 193–198.
- Ahmad, M., McNeil, D.L., Fautrier, A.G., Armstrang, K.F., and Paterson, A.M. 1996. Genetic relationships in Lens species and parentage determination of their interspecific hybrids using RAPD markers. Theor. Appl. Genet. 92: 1091–1098.
- Ainouche, A.K. and Bayer, R.J. 1999. Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on internal transcribed spacer sequences (ITS) of nuclear ribosomal DNA. Amer. J. Bot. 86: 590–607.
- Ainouche, A.K., Bayer, R.J., and Misset, M.T. 2004. Molecular phylogeny, diversification and character evolution in Lupinus (Fabaceae) with special attention to Mediterranean and African lupines. Plant Syst. Evol. 246: 211–222.
- Al-Atawneh, N., Shehadeh, A., Amri, A., and Maxted, N. 2009. Conservation Field Guide to Medics of the Mediterranean Basin. pp. 1–214. ICARDA, Syria.
- Ali, Z., Qureshi, A.S., Ali, W., Gulzar, H., Nisar, M., and Ghafoor, A. 2007. Evaluation of genetic diversity present in pea (Pisum sativum L.) germplasm based on morphological traits, resistance to powdery mildew and molecular characteristics. Pakistan J. Botany 39: 2739–2747.
- Alo, F., Furman, B.J., Akhunov, E., Dvorak, J., and Gepts, P. 2011. Leveraging genomic resources of model species for the assessment of diversity and phylogeny in wild and domesticated lentil. J. Heredity 102: 315–329.
- Ambrose, M.J. 1995. From Near East centre of origin the prized pea migrates throughout world. Diversity 11: 118–119.
- Ambrose, M.J. and Ellis, T.H. N. 2008. Ballistic seed dispersal and associated seed shadow in wild Pisum germplasm. Pisum Genet. 40: 5–9.
- Amri, A., Ajlouni, M., Assi, R., Sbeih, Y., Saad, A., Khoury W., and Khnifes A. 2008a. Major Achievements of the West Asia Dryland Agrobiodiversity Conservation Project. In: Proceedings of the International Conference on promoting community-driven in situ conservation of dryland agrobiodiversity. ICARDA, Aleppo, Syria.
- Amri, A., Monzer, M., Al-Oqla, A., Atawneh, N., Shehadeh, A. and Konopka, J. 2008b. Status and Treats to Natural Habitats and Crop Wild Relatives in Selected Areas in West Asia Region. In: Proceedings of the International Conference on promoting community-driven in situ conservation of dryland agrobiodiversity. ICARDA, Aleppo, Syria.
- Ansari, M.A., Patel, B.A., Mhase, N.L., Patel, D.J., Douaik, A., and Sharma, S.B. 2004. Tolerance of chickpea (Cicer arietinum L.) lines to root-knot nematode, Meloidogyne javanica (Treub) Chitwood. Genet. Resour. Crop Evol. 51: 449–453.
- Apaydin, H., Ertan, S., and Ozekmekçi, S. 2000. Broad bean (Vicia faba)—a natural source of L-DOPA—prolongs “on” periods in patients with Parkinson's disease who have “on-off “ fluctuations. Move Disord. 15: 164–166.
- Arbaoui, M., Balko, C., and Link, W. 2008a. Study of faba bean (Vicia faba L.) winter hardiness and development of screening methods. Field Crops Res. 106: 60–67.
- Arbaoui, M., Link, W., Satovic, Z., and Torres, A.M. 2008b. Quantitative trait loci of frost tolerance and physiologically related trait in faba bean (Vicia faba L.). Euphytica 164: 93–104.
- Arese, P. and De Flora, A. 1990. Pathophysiology of hemolysis in glucose-6-phosphate dehydrogenase deficiency. Sem. Hematol. 27: 1–40.
- Arumuganathan, K. and Earle, E.D. 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218.
- Asfaw, A., Blair, M.W., and Almekinders, C. 2009. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African Highlands. Theor. Appl. Genet. 120: 1–12.
- Aubert, G., Morin, J., Jacquin, F., Loridon, K., Quillet, M.C., Petit, A., Rameau, C., Lejeune-Hénaut, I., Huguet, T., and Burstin, J. 2006. Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor. Appl. Genet. 112: 1024–1041.
- Avagyan, A. 2008. Crop wild relatives in Armenia: diversity, legislation and conservation issues. In: Crop Wild Relative Conservation and Use. pp. 58–68. Maxted, N., Ford-Lloyd, B.V., Kell, S.P., Iriondo, J., Dulloo, E., and Turok, J., Eds., CABI Publishing, Wallingford.
- Avila, T., Blair, M.W., Reyes, X., and Bertin, P. 2012. Genetic diversity of bean (Phaseolus) landraces and wild relatives from the primary centre of origin of the Southern Andes. Plant Genet. Resour. Charact. Util. 10: 83–92.
- Azmat, M.A., Khan, A.A., Saeed, A., Ashraf, M., and Niaz, S. 2012. Screening pea germplasm against Erysiphe polygonifor disease severity and latent period. Inter. J. Veget. Sci. 18: 153–160.
- Babayeva, S., Akparov, Z., Abbasov, M., Mammadov, A., Zaifizadeh, M., and Street, K. 2009. Diversity analysis of Central Asia and Caucasian lentil (Lens culinaris Medik.) germplasm using SSR fingerprinting. Genet. Res. Crop Evol. 56: 293–298.
- Badami, P.S., Mallikarjuna, N., and Moss, J.P. 1997. Interspecific hybridization between Cicer arietinum and C. pinnatifidum. Plant Breeding 116: 393–395.
- Baginsky, C., Peña-Neira, A., Cáceres, A., Hernández, T., Estrella, I., Morales, H., and Pertuzé, R. 2013. Phenolic compound composition in immature seeds of fava bean (Vicia faba L.) cultivars cultivated in Chile. J. Food Compos. Analysis 31: 1–6.
- Balachandra, R., Redden, B., and Enneking, E. 2006. Development of an integrated database system to facilitate the storage and retrieval of germplasm data as well as sourcing the germplasm. In: 13th Australasian Plant breeding Conference Proceedings, Mercer C.F., Ed., NZ Grassland Association, Christchurch, New Zealand.
- Baldev, B. 1988. Origin, distribution, taxonomy, and morphology. In: Pulse crops. pp. 3–51. B. Baldev, S. Ramanujam, and Jain, H.K., Eds., New Delhi, India: Oxford and IBH Publishing Co.
- Bao, S., Yuhua, H., Zong, X., Wang, L., Lichi, L., Enneking, D., Rose, I.A., Leonforte, T., Redden, R.J., and Paull, J. 2008. Collection of pea (Pisum sativum) and faba bean (Vicia faba) germplasm in Yunnan. Plant Genet. Res News. 156: 11–22.
- Baranger, A.G., Aubert, G., Arnau, G., Lainé, A.L., Deniot, G., Potier, J., Weinachter, C., Lejeune-Hénaut, J., Lallemand, J., and Burstin, J. 2004. Genetic diversity within Pisum sativum using protein- and PCR-based markers. Theor. Appl. Genet. 108: 1309–1321.
- Bari, A., Street, K., Mackay, M., Endresen, D., DePauw, E., and Amri, A. 2011. Focused identification of germplasm strategy (FIGS) detects wheat stem rust resistance linked to environmental variables. Genet. Res. Crop Evol. 59: 1–17.
- Barilli E., Sillero, J.C., Fernández-Aparicio, M., and Rubiales, D. 2009. Identification of resistance to Uromyces pisi (Pers.) Wint. in Pisum spp. germplasm. Field Crops Res. 114: 198–203.
- Barnes, D.K., Bingham, E.T., Murphy, R.P., Hunt, O.J., Beard, D.F., Skrdla, W.H., and Teuber, L.R. 1977. Alfalfa germplasm in the United States: genetic vulnerability, use, improvement and maintenance. Agricultural Research Service Tech. Bull. No. 1571. USDA, Hyattsville, MD. pp. 21.
- Barrow, M.V., Simpson, C.F., and Miller, E.J. 1974. Lathyrism: A Review. Quart. Review Biol. 49: 101–128.
- Barulina, H. 1930. Lentils of the USSR and other countries. Bull. Appl. Genet. Plant Breeding 40: 1–319. [In Russian]
- Basu, P.S., Berger, J.D., Turner, N.C., Chaturvedi, S.K., Ali, M., and Siddique, K.H. M. 2007. Osmotic adjustment of chickpea (Cicer arietinum) is not associated with changes in carbohydrate composition or leaf gas exchange under drought. Ann. Appl. Biol. 150: 217–225.
- Bauchan G. and Greene, S.L. 2002. Status of the USDA Medicago germplasm collection. Plant Genet. Res. News. 129: 1–8.
- Baudoin, J.P., Rocha, O.J., Degreef, J., Zoro, BiI., Ouédraogo, M., Guarino, L., and Toussaint, A. 2008. In situ conservation strategy for wild Lima bean (Phaseolus lunatus L.) populations in the Central Valley of Costa Rica: a case study of short-lived perennial plants with a mixed mating system. In: Crop Wild Relative Conservation and Use. pp. 364–379. Maxted, N., Ford-Lloyd, B.V., Kell, S.P., Iriondo, J., Dulloo, E. and Turok, J., Eds., CABI Publishing, Wallingford.
- Bayaa, B., Erskine, W., and Hamdi, A. 1994. Response of wild lentil to Ascochyta fabae f. sp. lentis from Syria. Genet. Res. Crop Evol. 41: 61–65.
- Bayaa, B., Erskine, W., and Hamdi, A. 1995. Evaluation of a wild lentil collection for resistance to vascular wilt. Genet. Res. Crop Evol. 42: 231–235.
- Bena, G., Lejeune, B., Prosperi, J.M., and Olivieri, I. 1998. Molecular phylogenetic approach for studying life-history evolution: The ambiguous example of the genus Medicago L. Proc. R. Soc. Lond. Biol. 265: 1141–1151.
- Ben-David, R., Abbo, S., and Berger, J.D. 2010. Stress gradients select for ecotype formation in Cicer judaicum Boiss., a wild relative of domesticated chickpea. Genet. Res. Crop Evol. 57: 193–202.
- Bennett, S.J., Broughton, D.A., and Maxted, N. 2006. Ecogeographical analysis of the perennial Medicago. CRC Salinity Bull. 1: 1–62.
- Ben-Ze’ev, N. and Zohary, D. 1973. Species relationships in the genus Pisum L. Israeli J. Botany 22: 73–91.
- Berger, J.D. 2007. Ecogeographic and evolutionary approaches to improving adaptation of autumn-sown chickpea (Cicer arietinum L.) to terminal drought: The search for reproductive chilling tolerance. Field Crops Res. 104: 112–122.
- Berger, J.D. 2013. An evolutionary perspective on the role of phenology in the specific adaptation of chickpea. Legume Perspectives 4.
- Berger, J.D. and Turner, N.C. 2007. The ecology of chickpea: evolution, distribution, stresses and adaptation from an agro-climatic perspective. In: Chickpea Breeding and Management. S.S. Yadav, R. Redden, W. Chen, and B. Sharma, Eds., CABI, Wallingford, UK. pp. 47–71.
- Berger, J.D., Abbo, S., and Turner, N.C. 2003. Ecogeography of annual wild Cicer species: The poor state of the world collection. Crop Sci. 43: 1076–1090.
- Berger, J.D., Adhikari, K.N., Wilkinson, D., Buirchell, B.J., and Sweetingham, M.W. 2008a. Ecogeography of the Old World lupins. 1. Ecotypic variation in yellow lupin (Lupinus luteus L.). Aus. J. Agric. Res. 59: 691–701.
- Berger, J.D., Ali, M., Basu, P.S., Chaudhary, B.D., Chaturvedi, S.K., Deshmukh, P.S., Dharmaraj, P.S., Dwivedi, S.K., Gangadhar, G.C., Gaur, P.M., Kumar, J., Pannu, R.K., Siddique, K.H. M., Singh, D.N., Singh, D.P., Singh, S.J., Turner, N.C., Yadava, H.S., and Yadav, S.S. 2006. Genotype by environment studies demonstrate the critical role of phenology in adaptation of chickpea (Cicer arietinum L.) to high and low yielding environments of India. Field Crops Res. 98: 230–244.
- Berger, J.D., Buirchell, B., Luckett, D., and Nelson, M. 2012a. Domestication bottlenecks limit genetic diversity and constrain adaptation in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 124: 637–652.
- Berger, J.D., Buirchell, B.J., Luckett, D.J., Palta, J.A., Ludwig, C., and Liu, D.L. 2012b. How has narrow-leafed lupin changed in its 1st 40 years as an industrial, broad-acre crop? A G x E-based characterization of yield-related traits in Australian cultivars. Field Crops Res. 126: 152–164.
- Berger, J.D., Clements, J.C., Nelson, M.N., Kamphuis, L.G., Singh, K.B., and Buirchell, B. 2013. The essential role of genetic resources in narrow-leafed lupin improvement. Crop Pasture Sci. 64: 361–373.
- Berger, J.D., Kumar, S., Nayyar, H., Street, K., Sandhu, J.S., Henzell, J.M., Kaur, J., and Clarke, H.C. 2012c. Temperature-stratified screening of chickpea (Cicer arietinum L.) genetic resource collections reveals very limited reproductive chilling tolerance compared to its annual wild relatives. Field Crops Res. 126: 119–129.
- Berger, J.D., Ludwig, C., and Buirchell, B.J. 2008b. Ecogeography of the old world lupins: characterising the habitat range. In: Lupins for health and wealth. Proceedings of the 12th International Lupin Conference, pp. 355–361, Fremantle, Western Australia, 14-18 September 2008. Palta, J.A. and Berger, J.D., Eds., International Lupin Association.
- Berger, J.D., Milroy, S.P., Turner, N.C., Siddique, K.H. M., Imtiaz, M., and Malhotra, R. 2011. Chickpea evolution has selected for contrasting phenological mechanisms among different habitats. Euphytica 180: 1–15.
- Berger, J.D., Turner, N.C., Siddique, K.H. M., Knights, E.J., Brinsmead, R.B., Mock, I., Edmondson, C., and Khan, T.N. 2004. Genotype by environment studies across Australia reveal the importance of phenology for chickpea (Cicer arietinum L.) improvement. Aus. J. Agric. Res. 55: 1–14.
- Bhatty, R.S. 1988. Composition and Quality of Lentil (Lens culinaris Medik): A Review. Can. Inst. Food Sci. Technol. J. 21: 144–160.
- Bisby, F.A., Buckingham, J., and Harborne, J.B. 1994. Phytochemical dictionary of the Leguminosae. Vol. 1: Plants and their constituents. Chapman and Hall, London.
- Bisht I.S. and Singh, M. 2013. Asian Vigna. In: Genetic and Genomic Resources for Grain Legume Improvement. pp. 237–267. Singh, M. and Bisht I.S., Eds., Elsevier Insights, London, UK.
- Blair, M.W. 2013. Mineral Biofortification Strategies for Staples: The Example of Common Bean. J. Agric. Food Chem. 61: 8287–8294.
- Blair, M.W., Cortés, A., Penmetsa, R.V., Carrasquilla-Garcia, N., Farmer, A., and Cook, D. 2013. Development of high throughput SNP markers for parental polymorphism screening and diversity analysis in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 126: 535–548
- Blair, M.W., Díaz, L.M., Buendía, H.F., and Duque, M.C. 2009. Genetic diversity, seed size associations and population structure of a core collection of common beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 119: 955–73.
- Blair, M.W., Gonzales, L.F., Kimani, P., and Butare, L. 2010. Inter-genepool introgression, genetic diversity and nutritional quality of common bean (Phaseolus vulgaris L.) landraces from Central Africa. Theor. Appl. Genet. 121: 237–248.
- Blair, M.W., Iriarte, G., and Beebe, S. 2006. QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean x wild common bean (Phaseolus vulgaris L.) cross. Theor. Appl. Genet. 112: 1149–1163.
- Blair, M.W., Pantoja, W., and Muñoz, L.C. 2012a. First use of microsatellite markers in a large collection of cultivated and wild accessions of tepary bean (Phaseolus acutifolius A. Gray). Theor. Appl. Genet. 125: 1137–1147.
- Blair, M.W., Porch, T., Cichy, K., Galeano, C.H., Lariguet, P., Pankurst, C., and Broughton, W. 2008. Induced mutants in common bean (Phaseolus vulgaris), and their potential use in nutrition quality breeding and gene discovery. Israel J. Plant Sci. 55: 191–200.
- Blair, M.W., Soler, A., and Cortés, A.J. 2012b. Diversification and population structure in common beans (Phaseolus vulgaris L.). PLOS One 7: e49488.
- Blair, M.W. and Izquierdo, P. 2012. Use of the advanced backcross-QTL method to transfer seed mineral accumulation nutrition traits from wild to Andean cultivated common beans. Theor. Appl. Genet. 25: 1015–1031.
- Bobrov, E.G. 1967. On the span of the genus Trifolium s.l. Bot. Zurn., S.S.R. 52: 1593–1599. (Russian with English summary).
- Bogdanova, V.S. 2007. Inheritance of organelle DNA markers in a pea cross associated with nuclear-cytoplasmic incompatibility. Theor. Appl. Genet. 114: 333–339.
- Bogdanova, V.S., Galieva, E.R., and Kosterin, O.E. 2009. Genetic analysis of nuclear-cytoplasmic incompatibility in pea associated with cytoplasm of an accession of wild subspecies Pisum sativum subsp. elatius (Bieb.) Schmahl. Theor. Appl. Genet. 118: 801–809.
- Bohra, A., Dubey, A., Saxena, R.K., Penmetsa, R.V., Poornima, K.N., Kumar, N., Farmer, A.D., Srivani, G., Upadhyaya, H.D., Gothalwal, R., Ramesh, R., Singh, D., Saxena, K.B., Kavikishor, P.B., Town, C.D., May, G.D., Cook, D.R., and Varshney, R.K. 2011a. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.). BMC Plant Biol. 11: 56.
- Bohra, A., Mallikarjuna, N., Saxena, K.B., Upadhyaya, H.D., Vales, I., and Varshney, R.K. 2011b. Harnessing the potential of crop wild relatives through genomics tools for pigeonpea improvement. J. Plant Biol. 37: 83–98.
- Boller, B., Schubiger, F.X., and Kolliker, R. 2010. Red clover. In: Fodder Crops and Amenity Grasses. Handbook of Plant Breeding. pp. 439–456. Boller, B., Posselt, U., and Veronesi, F., Eds., Springer, New York, USA.
- Bolton, J.L. 1962. Alfalfa- botany, cultivation and utilization. Interscience, NY.
- Bond, D.A., and Poulsen, M.H. 1983. In: The Faba Bean (iVicia faba L.). pp. 77–101. Hebblethwaite P.D., Ed., Butterworths, London.
- Bordat, A., Savois, V., Nicolas, M., Salse, J., Chauveau, A., Bourgeois, M., Potier, J., Houtin, H., Rond, C., Murat, F., Marget, P., Aubert, G., and Burstin, J. 2011. Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. G3. Genes-Genomes-Genetics 1: 93–103.
- Brown, A.H. D. and Spillane, C. 1999. Implementing core collections-principles, procedures, progress, problems and promise. In: Core collections for today and tomorrow. pp. 1–9. Johnson R.C. and Hodgkin T., Eds., International Plant Genetic Resources Institute, Rome, Italy.
- Brown, A.H. D., Grace, J.P., and Speer, S.S. 1987. Designation of a ‘‘core’’ collection of perennial Glycine. Soyb. Genet. Newsl. 14: 59–70.
- Brummer, E.C., Cazcarro, P.M., and Luth, D. 1999. Ploidy determination of alfalfa germplasm accessions using flow cytometry. Crop Sci. 39: 1202–1207.
- Buchwaldt, L., Anderson, K.L., Morrall, R.A. A., Gossen, B.D., and Bernier, C.C. 2004. Identification of lentil germ plasm resistant to Colletotrichum truncatum and characterization of two pathogen races. Phytopathology 94: 236–243.
- Bunsupa, S., Saito, K., and Yamazaki, M. 2013. Molecular biology and biotechnology of quinolizidine alkaloid biosynthesis in Leguminosae plants. In: Biotechnology for Medicinal Plants. pp. 263–273. Suman, C., Hemant, L., and Ajit, V., Eds., Springer, Heidelberg, Germany.
- Byrne, O.M., Hardie, D.C., Khan, T., and Yan, G. 2008. Genetic analysis of pod and seed resistance to pea weevil in a Pisum sativum × P. fulvum interspecific cross. Aust. J. Agric. Res. 59: 854–862.
- Caldas, G.V. and Blair, M.W. 2009. Inheritance of condensed tannin content and relationship with seed color and pattern genes in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 119: 131–142.
- Caligari, P.D. S., Römer, P., Rahim, M.A., Huyghe, C., Neves-Martins, J., and Sawicka-Sienkiewicz E.J. 2000. The potential of Lupinus mutabilis as a crop. In: Linking Research and Marketing Opportunities for Pulses in the 21st Century. Current Plant Science and Biotechnology in Agriculture. Volume 34, pp. 569–573. Knight, R., Ed. Kluwer Academic Publishers.
- Campbell, C.G. 1997. Grass pea. Lathyrus sativus L. Promoting the conservation and use of underutilized and neglected crops. International Plant Genetic Resources Institute, Rome, Italy.
- Canci, H. and Toker, C. 2009. Evaluation of annual wild Cicer species for drought and heat resistance under field conditions. Genet. Res. Crop Evol. 56: 1–6.
- Cannon, S.B., Ilut, D., Farmer, A.D., Maki, S.L., May, G.D., Singer, S.R., and Doyle, J.J. 2010. Polyploidy did not predate the evolution of nodulation in all legumes. PLoS ONE 5: e11630.
- Cardoso, D., de Lima, H.C., Rodrigues, R.S., de Queiroz, L.P., Pennington, R.T., and Lavin, M. 2012a. The Bowdichia clade of Genistoid legumes: Phylogenetic analysis of combined molecular and morphological data and a recircumscription of Diplotropis. Taxon 61: 1074–1087.
- Cardoso, D., de Queiroz, L.P., Pennington, R.T., de Lima, H.C., Fonty, É., Wojciechowski, M.F., and Lavin, M. 2012b. Revisiting the phylogeny of papilionoid legumes: New insights from comprehensively sampled early-branching lineages. Amer. J. Bot. 99: 1991–2013.
- Cardoso, D., Pennington, R.T., de Queiroz, L.P., Boatwright, J.S., Van Wyk, B.E., Wojciechowski, M.F., and Lavin, M. 2013. Reconstructing the deep-branching relationships of the papilionoid legumes. South African J. Botany. in press.
- Carter, Jr T.E., Nelson, R.L., Sneller, C.H., Cui, Z. 2004. Genetic diversity in soybean. Soybeans: Improvement, Production, and Uses. Madison, Wisconsin, USA: ASA, CSSA, and SSSA.
- CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 106: 12794–12797.
- Celakovsky, L. 1874. Uber den Aufbau der Gattung Trifolium. Osterr. Bot Z. 24: 37–45, 75–82.
- Chankaew, S., Isemura, T., Naito, K., Ogiso-Tanaka, E., Tomooka, N., Somta, P., Kaga, A., Vaughan, D.A., and Srinives, P. 2014. QTL mapping for salt tolerance and domestication-related traits in Vigna marina subsp. oblonga, a halophytic species. Theor Appl Genet. 127: 691–702.
- Chen, W., Basandrai, A.K., Basandrai, D., Banniza, S., Bayaa, B., Buchwaldt, L., Davidson, J., Larsen, R., Rubiales, D., and Taylor, P.W. J. 2009. Diseases and their management. In: The Lentil Botany, Production and Uses. pp. 262–281. Erskine, W., Muehlbauer, F.J., Sarker, A., and Sharma, B., Eds., CAB International, Wallingford, UK.
- Chen, W., Sharma, H.C., and Muehlbauer, F.J. 2011. Compendium of Chickpea and Lentil Diseases and Pests. The American Phytopathological Society, St. Paul, MN, USA.
- Chimwamurombe, P.M., and Khulbe, R.K. 2011. Domestication. In: Biology and Breeding of Food Legumes. pp. 19–34. Pratap, A. and Kumar J., Eds., CABI, Cambridge, MA, USA.
- Cho, G.T., Yoon, M.S., Lee, J., Baek, H.J., Kang, J.H., Kim, T.S., and Paek, N.C. 2008. Development of a core set of Korean soybean landraces [Glycine max (L.) Merr.]. J. Crop Sci. Biotech. 11: 157–162.
- Chowdhury, M.A., and Slinkard, A.E. 2000. Genetic diversity in grasspea (Lathyrus sativus L.). Genet. Res. Crop Evol. 47: 163–169.
- Clarke, H.J., Kumari, M., Khan, T.N. and Siddique, K.H. M. 2011. Poorly formed chloroplasts are barriers to successful interspecific hybridization in chickpea following in vitro embryo rescue. Plant. Cell. Tiss. Organ. Cult. 106: 465–473.
- Clarke, H.J., Wilson, J.G., Kou, I., Lulsdorf, M.M., Mallikarjuna, N., Kou, J. and Siddique, K.H. M. 2006. Embryo rescue and plant regeneration in vitro of selfed chickpea (Cicer arietinum L.) and its wild annual relatives. Plant. Cell. Tiss. Org. Cult. 85: 197–204.
- Clement, S.L., Hardie, D.C., and Elberson, L.R. 2002. Variation among accessions of Pisum fulvum for resistance to pea weevil. Crop Sci. 42: 2167–2173.
- Clement, S.L., McPhee, K.E., Elberson, L.R., and Evans, M.A. 2009. Pea weevil, Bruchus pisorum L. (Coleoptera: Bruchidae), resistance in Pisum sativum × Pisum fulvum interspecific crosses. Plant Breeding 12: 478–486.
- Clements, J., Prilyuk, L., Quealy, J., and Francis, G. 2008. Interspecific crossing among the New World lupin species for Lupinus mutabilis crop improvement. In: Lupins for Health and Wealth: Proceedings of the 12th International Lupin Conference. pp. 324–327. Palta, J.A. and Berger, J.B. D., Eds., International Lupin Association, Fremantle, Western Australia.
- Collard, B.C. Y., Ades, P.K., Pang, E.C. K., Brouwer, J.B., and Taylor, P.W. J. 2001. Prospecting for sources of resistance to ascochyta blight in wild Cicer species. Australas. Plant Pathol. 30: 271–276.
- Conterato, I.F. and Schifino-Wittmann, M.T. 2006. New chromosome numbers, meiotic behaviour and pollen fertility in American taxa of Lupinus (Leguminosae): contributions to taxonomic and evolutionary studies. Bot. J. Linn. Soc. 150: 229–240.
- Cook, D.R. 1999. Medicago truncatula a model in the making. Curr. Opinion Plant Biol. 2: 301–304.
- Cortés, A.J., Chavarro, M., Madriñan, S., This, D., and Blair, M.W. 2012. Molecular ecology and selection of drought related Asr gene polymorphisms in wild and cultivated common bean (Phaseolus vulgaris L.). BMC Genetics 13: 58.
- Cortés, A.J., Monserrate, F.A., Ramírez-Villegas, J., Madriñán, S., and Blair, M.W. 2013. Drought tolerance in wild plant populations: The Case of Common Beans (Phaseolus vulgaris L.). PLoS ONE 8: e62898.
- Cortés, A.J., This, D., Chavarro, M.C., Madriñan, S., and Blair, M.W. 2012. Nucleotide diversity patterns at the drought related DREB encoding genes in wild and cultivated common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 125: 1069–1085.
- Cottage, A., Webb, A., Hobbs, D., Khamassi, K., Maalouf, F., Ogbannaya, F., Stodard, F., Duc, G., Link, W., Thomas, J.E., and O’Sullivan, D.M. 2012. SNP discovery and validation for genomic-assisted breeding of faba bean (Vicia faba L.). In: Proc VI International Conference on Legume Genetics and Genomics. October 2-7, 2012, Hyderabad, India
- Cowling, W.A., Buirchell, B.J., and Falk, D.E. 2009. A model for introducing novel genetic diversity from wild relatives into elite crop populations. Crop Pasture Sci. 60: 1009–1015.
- Coyne, C.J. and McGee, R.J. 2013. Lentil. In: Genetic and Genomic Resources for Grain Legume Improvement. Chapter 7, pp. 157–180. Singh, M. and Bisht I.S., Eds., Elsevier Insights, London, UK.
- Coyne, C.J., McGee, R.J., Redden, R.J., Ambrose, M.J., Furman, B.J., and Miles, C.A. 2011. Genetic Adjustment to Changing Climates: Pea. In: Crop Adaptation to Climate Change. pp. 238–249. Yadav, S.S., Redden, B., Hatfield, J.L., and Lotze-Campen H., Eds., Wiley-Blackwell, Ames, IA.
- Crampton, B., 1985. Native range clovers. In: Clover Science and Technology. pp. 579–590. Taylor, N.L., Ed., American Society of Agronomy, Madison, WI, USA.
- Cristofolini, G. and Chiapella L.F. 1984. Origin and diversification of Genisteae (Fabaceae): a serosystematic purview. Webbia 38: 105–122.
- Cristofolini, G. 1997. The biodiversity of the “Leguminosae-Genisteae” and its genesis. Lagascalia 19: 121–128.
- Crop Wild Relatives and Climate Change. 2013a. Online resource. Accessed on 22-09-2013. www.cwrdiversity.org
- Croser, J.S., Ahmad, F., Clarke, H.J., and Siddique, K.H. M. 2003. Utilization of wild Cicer in chickpea improvement progress, constraints, and prospects. Austral. J. Agric. Res. 54: 429–444.
- Cubero, J.I. 1981. Origin, domestication and evolution. In: Lentils. pp. 15–38. Webb, C. and Harwtin, G.C., Eds., Commonwealth Agricultural Bureau, Slough.
- Cubero, J.I. 1973. Evolutionary trends in Vicia faba. Theor. Appl. Genet. 43: 59–65.
- Cubero, J.L., Perez de la Varga, M., and Fratini, R. 2009. Origin, phylogeny and spread. In: The Lentil, Botany, Production and Uses. pp. 13–33. Erskine, W., Muehlbauer, F.J., Sarker, A., and Sharma, B., Eds., CABI, Wallingford.
- Czefranova Z. 1971. Review of species in the genus Lens Mill. Novosti Systematischeski Vyssich Rastenii 8: 184–191 (In Russian).
- Dahl, W.J., Foster, L.M., and Tyler, R.T. 2012. Review of the health benefits of peas (Pisum sativum L.). Brit. J. Nutrit. 108(S1): S3–S10.
- Dalmais, M., Schmidt, J., Le Signor, C., Moussy, F., Burstin, J., Savois, V., Aubert, G., Brunaud, V., de Oliveira, Z., Guichard, C., Thompson, R., and Bendahmane, A. 2008. UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 9: R43.
- Datta, S., Gangwar, S., Kumar, S., Gupta, S., Rai, R., Kaashyap, M., Singh, P., Kumar Chaturvedi, S., Singh, B.B., and Nadarajan, N. 2012. Genetic Diversity in Selected Indian Mungbean [Vigna radiata (L.) Wilczek] Cultivars Using RAPD Markers. Am. J. Plant Sci. 3: 1085–1091.
- Datta, S., Tiwari, S., Kaashyap, M., Gupta, P.P., Choudhury, P.R., Kumari, J., and Kumar, S. 2011. Genetic similarity analysis in lentil using cross-genera legume sequence tagged microsatellite site markers. Crop Sci. 51: 2412–2422.
- Davies, A.M. R., Maxted, N., and van der Maesen, L.J. G. 2007. A natural infrageneric classification for Cicer (Leguminosae, Cicereae). Blumea 52: 379–400.
- Davis, P.H. 1970. Pisum L. In: Flora of Turkey and East Aegean Islands. Vol. 34, pp. 370–373. Davis, P.H., Ed., Edinburg University Press.
- De Candolle, A. 1884. Origin of cultivated plants. Kesinger Publishing LCC, Whitefish, MT, 2006
- De Martino, T., Errico, A., Lassandro, A., and Conicella, C. 2000. Distorted segregation resulting from pea chromosome reconstructions with alien segments from Pisum fulvum. J. Hered. 91: 322–325.
- Delgado-Salinas, A., Bibler, R., and Lavin, M. 2006. Phylogeny of the genus Phaseolus (Leguminosae): A recent diversification in an ancient landscape. Syst. Bot. 31: 779–791.
- Delgado-Salinas, A., Bruneau, A., and Doyle, J.J. 1993. Chloroplast DNA phylogenetic studies in New World Phaseolinae (Leguminosae: Papilionoideae: Phaseoleae). Syst. Bot. 18: 6–17.
- Delgado-Salinas, A., Thulin, M., Pasquet, R., Weeden, N., and Lavin, M. 2011. Vigna (Leguminosae) sensu lato: the names and identities of the American segregate genera. Am. J. Bot. 98: 1694–715.
- Delgado-Salinas, A., Turley, T., Richman, A., and Lavin, M. 1999. Phylogenetic analysis of the cultivated and wild species of Phaseolus (Fabaceae). Syst. Bot. 24: 438–460.
- Deodikar, G.B., and Thakar, C.V. 1956. Cytotaxonomic evidence for the affinity between Cajanus indicus Spreng. and certain erect species of Atylosia W. and A. Proc. Indian Acad. Sci. 43: 37–45.
- Deulvot, C., Charrel, H., Marty, A., Jacquin, F., Donnadieu, C., Lejeune-Henaut, I., Burstin, J., and Aubert, G. 2010. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics 11: 468.
- Di Vito, M., Singh, K.B., Greco, N., and Saxena, M.C. 1996. Sources of resistance to cyst nematode in cultivated and wild Cicer species. Genet. Resour. Crop Evol. 43: 103–10.
- Díaz, A.M., Caldas, G.V., and Blair M.W. 2010. Concentrations of condensed tannins and anthocyanins in common bean seed coats. Food Res. Internat. 43: 595–601.
- Dombrowski, J. 1970. Preliminary report on excavation in Lalibela and Natchabiet caves, Begemeder. Annales d’ Ethiopie 8: 21–29.
- Dönmez, A.A. 2011. Cicer uludereensis Donmez: a new species of Cicer (Chickpea) (Fabaceae) from around the Fertile Crescent, SE Turkey. Turk. J. Bot. 35: 71–76.
- Doyle, J.J. 1995. DNA data and legume phylogeny: a progress report. In: Advances in Legume Systematics. pp. 11–30. Part 7: Phylogeny. Crisp, M. and Doyle, J.J., Eds., Royal Botanic Gardens, Kew.
- Doyle, J.J. and Egan, A.N. 2009. Dating the origins of polyploidy events. New Phytol. 186: 73–85.
- Doyle, J.J., Doyle, J.L., and Brown, A.H. D. 1990. A chloroplast DNA phylogeny of the wild perennial relatives of soybean (Glycine subgenus Glycine): congruence with morphological and crossing groups. Evolution 44: 371–389.
- Drummond, C.S. 2008. Diversification of Lupinus (Leguminosae) in the western New World: Derived evolution of perennial life history and colonization of montane habitats. Mol. Phyl. Evol. 48: 408–421.
- Drummond, C.S. and Hamilton, M.B. 2007. Hierarchical components of genetic variation at a species boundary: population structure in two sympatric cultivars of Lupinus microcarpus (Leguminosae). Mol. Ecol. 16: 753–769.
- Drummond, C.S., Eastwood, R.J., Miotto, S.T. S., and Hughes, C.E. 2012. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): Testing for key innovation with incomplete taxon sampling. Syst. Biol. 61: 443–460.
- Duc, G., Bao, S., Baum, M., Redden, B., Sadiki, M., Suso, M.J., Vishniakova, M., and Zong, X. 2010. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop Res. 115: 270–278.
- Duc, G., Sixdenier, G., Lila, M., and Furstoss V. 1989. Search of genetic variability for vicine and convicine content in Vicia faba L. A first report of a gene which codes for nearly zero-vicine and zero-convicine contents. In: Recent Advances of Research in Antinutritional Factors in Legume Seeds. pp. 305–313. Huisman, J., van der Poel, A.F. B., Liener, I.E., Eds., Wageningen. NL, Pudoc.
- Dulloo, M.E., Labokas, J., Iriondo, J.M., Maxted, N., Lane, A., Laguna, E., Jarvis, A., and Kell, S.P. 2008. Genetic reserve location and design. In: Plant Genetic Population Management. pp. 23–64. Iriondo, J.M., Maxted, N. and Dulloo, E., Eds., CAB International, Wallingford.
- Dundas, I.S. 1990. Pigeonpea: cytology and cytogeneticsperspectives and prospects. In: The Pigeonpea. pp 117–136. Nene, Y.L., Hall, S.D., and Sheila, V.K., Eds., CABI Publishing, Wallington.
- Durán, Y., and Pérez de la Vega, M. 2004. Assessment of genetic variation and species relationships in a collection of Lens using RAPD and ISSR [Inter-simple sequence repeats]. Spanish J. Agric. Res. 2: 538–544.
- Durieu, P., and Ochatt, S.J. 2000. Efficient intergeneric fusion of pea (Pisum sativum L.) and grass pea (Lathyrus sativus L.) protoplasts. J. Exp. Botany 51: 1237–1242.
- Eastwood, R.J. and Hughes, C.E. 2008a. Origins of domestication of Lupinus mutabilis in the Andes. In: Lupins for Health and Wealth. pp. 373–379. Palta, J.A., and Berger, J.D., Eds., Proceedings of the 12th International Lupin Conference. Fremantle, Western Australia.
- Eastwood, R.J., Drummond, C.S., Schifino-Whittmann, M.T., and Hughes, C.E. 2008b. Diversity and evolutionary history of lupins - insights from new phylogenies. In: Lupins for Health and Wealth. pp. 346–354. Palta, J.A. and Berger, J.D., Eds., Proceedings of the 12th International Lupin Conference. Fremantle, Western Australia.
- Ellis, T.H. N. 2011. Pisum. In: Wild Crop Relatives: Genomic and Breeding Resources. pp. 237–248. Kole, C., Ed., Springer, Heidelberg, Germany.
- Ellis, T.H. N., Poyser, S.J., Knox, M.R., Vershinin, A.V., and Ambrose, M.J. 1998. Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 260: 9–19.
- Ellison, N.W., Liston, A., Steiner, J.J., Williams, W.M. and Taylor, N.L. 2006. Molecular phylogenetics of the clover genus (Trifolium - Leguminosae). Mol. Phyl. Evol. 39: 688–705.
- Ellwood, S.R., Phan, H.T. T., Jordan, M., Hane, J., Torres, A.M., Avila, C.M., Cruz-Izquierdo, S., and Oliver, R.P. 2008. Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genomics 9: 380
- Errico, A., Conicella, C., De Martino, T., Ercolano, R., and Monti, L.M. 1996. Chromosome reconstructions in P. sativum through interspecific hybridisation with P. fulvum. J. Genet. Breed. 50: 309–313.
- Erskine, W. 1983. Relationship between the yield of seed and straw in lentil. Field Crops Research 7: 115–121.
- Erskine, W. 1985. Selection for pod retention and pod indehiscence in lentils. Euphytica 34: 105–112
- Erskine, W. 2009. Global production, supply and demand. In: The Lentil: Botany, Production and Uses. pp. 4–12. Erskine, W., Muehlbauer, F., Sarker, F., and Sharma, F., Eds. CABI, Wallingford, UK.
- Erskine, W. and Muehlbauer, F.J. 1991. Allozyme and morphological variability, outcrossing rate and core collection formation in lentil germplasm. Theor. Appl. Genet. 83: 119–125.
- Erskine, W., Adham, Y., and Holly, L. 1989. Geographic distribution of variation in quantitative traits in a world lentil collection. Euphytica 43: 97–103.
- Erskine, W., Ellis, R.H., Summerfield, R.J., Roberts, E.H., and Hussain, A. 1990. Characterization of responses to temperature and photoperiod for time to flowering in a world lentil collection. Theor. Appl. Genet. 80: 193–199.
- Erskine, W., Hussain, A., Tahir, M., Bahksh, A., Ellis, R.H., Summerfield, R.J., and Roberts, E.H. 1994. Field evaluation of a model of photothermal flowering responses in a world lentil collection. Theor. Appl. Genet. 88: 423–428.
- Erskine, W., Saxena, N.P., and Saxena, M.C. 1993. Iron deficiency in lentil: Yield loss and geographic distribution in a germplasm collection. Plant Soil 151: 249–254.
- FAO. 2010. The second report on the state of the world's Plant Genetic Resources for food and agriculture, Rome.
- FAO. 2013. Towards the establishment of a global network for in situ conservation and on-farm management of PGRFA. Report of Technical Workshop held in Rome, Italy 13th November, 2012. Food and Agriculture Organisation of the UN, Rome, Italy.
- FAOSTAT. 2012. Final 2011 data and preliminary 2012 data for 5 major commodity aggregates.
- FAOSTAT. 2013. http://faostat.fao.org/site/567/default.aspx#ancor. Accessed October 2013.
- Fatokun, C.A. 2002. Breeding cowpea for insect pests: Attempted crosses between cowpea and Vigna vexillata. In: Challenges and opportunities for enhancing sustainable cowpea production. Proceedings of the world cowpea conference, pp. 52–61. Fatokun, C.A., Tarawali, S.A., Singh, B.B., Kormawa, P.M., and Tamo, M., Eds., Ibadan, Nigeria: International Institute of Tropical Agriculture IITA.
- Fedorov A.A. 1939. Wild high-mountain peas of Caucasus. Trans. Biol. Inst. Arm. SSR 1: 39–79.
- Ferguson, M.E. and Erskine, W. 2001. Lentils (Lens L.). In: Plant Genetic Resources of Legumes in the Mediterranean. pp. 132–157. Maxted, N., and Bennett, S.J., Eds., Kluwer Academic Publishers, Dordrecht.
- Ferguson, M.E., and Robertson, L.D. 1996. Genetic diversity and taxonomic relationships within the genus Lens as revealed by allozyme polymorphism. Euphytica 91: 163–172.
- Ferguson, M.E., Ford-Lloyd, B.V., Robertson, L.D., Maxted, N., and Newbury, H.J. 1998. Mapping the geographical distribution of genetic variation in the genus Lens for the enhanced conservation of plant genetic diversity. Mol. Ecol. 7: 1743–1755.
- Ferguson, M.E., Maxted, N., Slageren, M.V., and Robertson, L.D. 2000. A re‐assessment of the taxonomy of Lens Mill. (Leguminosae, Papilionoideae, Vicieae). Bot. J. Linn. Soc. 133: 41–59.
- Ferguson, M.E., Robertson, L.D., Ford-Lloyd, B.V., Newbury, H.J., and Maxted, N. 1998. Contrasting genetic variation amongst lentil landraces from different geographical origins. Euphytica 102: 265–273.
- Fernandez, M., Polanco, C., Ruiz, M.L., and Perez de la Vega, M. 2000. A comparative study of the structure of the rDNA intergenic spacer of Lens culinaris Medik., and other legume species. Genome 43: 597–603.
- Fernández, M., Ruiz, M.L., Linares, C., Fominaya, A., and Pérez de la Vega, M. 2005. 5S rDNA genome regions of Lens species. Genome 48: 937–942.
- Fernández-Aparicio, M., and Rubiales, D. 2010. Characterisation of resistance to crenate broomrape (Orobanche crenata Forsk.) in Lathyrus cicera L. Euphytica 173: 77–84.
- Fernández-Aparicio, M., Flores, F., Rubiales, D. 2009b. Field response of Lathyrus cicera germplasm to crenate broomrape (Orobanche crenata). Field Crops Res. 113: 321–327.
- Fernández‐Aparicio, M., Sillero, J.C., and Rubiales, D. 2009a. Resistance to broomrape in wild lentils (Lens spp.). Plant Breeding 128: 266–270
- Fernández-Aparicio, M., Sillero, J.C., Pérez-de-Luque, A., and Rubiales, D. 2008. Identification of sources of resistance to crenate broomrape (Orobanche crenata) in Spanish lentil (Lens culinaris) germplasm. Weed Res. 48: 85–94.
- Fernández-Aparicio M., Sillero, J.C., and Rubiales, D. 2009. Resistance to broomrape in wild lentils (Lens spp.). Plant Breeding 128: 266–270.
- Fikiru, E., Tesfaye, K., and Bekele, E. 2010. Genetic diversity and population structure of Ethiopian lentil (Lens culinaris Medikus) landraces as revealed by ISSR marker. African J. Biotech. 6: 1460–1468.
- Fikiru, E., Tesfaye, K., and Bekele, E. 2011. Morphological and molecular variation in Ethiopian lentil (Lens culinaris Medikus) cultivars. African J. Biotech. 3: 60–67
- Fiocchetti, F., Laddomada, B., Roselli, M., Crinò, P., and Lucretti, S. 2009. Fingerprinting of three typical macrosperma Italian lentil (Lens culinaris Medik.) landraces using fluorescence-based AFLP markers. Sci. Hortic. 121: 383–387.
- Flamand, M.C., Duc, G., Goblet, J.P., Hong, L., Louis, O., Briquet, M., and Boutry, M. 1993.Variant mitochondrial plasmids of broad bean arose by recombination and are controled by the nuclear genome. Nucleic Acid Res. 21: 5468–5473.
- Flight, C. 1976. The Kimtampo culture and its place in the economic prehistory of West Africa. In: Origin of African plant domestication. pp. 212–221. Harlan, J.R., de Wet, J.M. J., and Stemler, A.B. L., Eds., Mouton, Hague, the Netherlands.
- Flores, F., Hybl, M., Knudsen, J.C., Marget, P., Muel, F., Nadal, S., Narits, L., Raffiot, B., Sass, O., Solis, I., Winkler, J., Stoddard, F.L., and Rubiales, D. 2013. Adaptation of spring faba bean types across European climates. Field Crops Res. 145: 1–9.
- Flores, F., Nadal, S., Solis, I., Winkler, J., Sass, O., Stoddard, F.L., Link, W., Raffiot, B., Muel, F., and Rubiales, D., 2012. Faba bean adaptation to autumn sowing under European climates. Agron. Sustain. Dev. 32: 727–734
- Foley, R., Gao, L.L., Spriggs, A., Soo, L., Goggin, D., Smith, P., Atkins, C., and Singh, K. 2011. Identification and characterisation of seed storage protein transcripts from Lupinus angustifolius. BMC Plant Biol. 11: 59.
- Foncéka, D., Hodo-Abalo, T., Rivallan, R., Faye, I., Sall, M.N., Ndoye, O., Fávero, A.P., Bertioli, D.J., Glaszmann, J.C., Courtois, B., and Rami, J.F. 2009. Genetic mapping of wild introgressions into cultivated peanut: a way toward enlarging the genetic basis of a recent allotetraploid. BMC Plant Biol. 9: 103.
- Fondevilla, S., Almeida, N.F., Satovic, Z, Rubiales, D., Vaz Patto, M.C., Cubero, J.I., and Torres, A.M. 2011. Identification of common genomic regions controlling resistance to Mycosphaerella pinodes, earliness and architectural traits in different pea genetic backgrounds. Euphytica 18: 43–52.
- Fondevilla, S., Avila, C.M., Cubero, J.I., and Rubiales, D. 2005. Response to Mycosphaerella pinodes in a germplasm collection of Pisum spp. Plant Breeding 124: 313–315.
- Fondevilla, S., Carver, T.W. L., Moreno, M.T., and Rubiales, D. 2007. Identification and characterisation of sources of resistance to Erysiphe pisi Syd. in Pisum spp. Plant Breeding 126: 113–119.
- Fondevilla, S., Rubiales, D., Moreno, M.T., and Torres, A.M. 2008. Identification and validation of RAPD and SCAR markers linked to the gene Er3 conferring resistance to Erysiphe pisi DC in pea. Mol. Breeding 22: 193–200.
- Fondevilla, S., Torres, A.M., Moreno, M.T., and Rubiales, D. 2007. Identification of a new gene for resistance to powdery mildew in Pisum fulvum, a wild relative of pea. Breeding Sci. 57: 181–184.
- Frankel, O.H. and Brown, A.H. D. 1984. Plant genetic resources today: A critical appraisal. In: Crop Genetic Resources Conservation and Evaluation. pp. 249–257. Holden, J.H. W. and Williams, J.T., Eds., Winchester, Allen and Unwin, Massachusetts, USA.
- Franssen, S.U., Shrestha, R.P., Bräutigam, A., Bornberg-Bauer, E., and Weber, A.P. M. 2011. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12: 227.
- Fratini, R. and Ruiz, M.L. 2006. Interspecific hybridization in the genus Lens applying in vitro embryo rescue. Euphytica 150: 271–280.
- Fratini, R. and Ruiz, M.L. 2011. Wide crossing in lentil through embryo rescue. Methods Mol. Biol. 710: 131–139.
- Freytag, G.F. and Debouck, D.G. 2002. Review of Taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae Papilionoideae) in North America, Mexico, and Central America. Bot. Miscell. 23: 1–30.
- Furman, B.J., Coyne, C., Redden, B., Sharma, S.K., and Vishnyakova, M. 2009. Genetic Resources: Collection, Characterization, Conservation and Documentation. In: The Lentil, botany, production and uses. Ch 6, pp. 64–75. Erskins, W., Muelbauer, F.J., Sarker, A., and Sharma, B., Eds., CABI International, Wallingford, Oxfordshire U.K.
- Furman, B.J. 2006. Methodology to establish a composite collection: Case study in lentil. Plant Genet. Resour. Charact. Util. 4: 2–12.
- Galeano, C.H., Fernandez, A.C., Franco-Herrera, N., Cichy, K.A., McClean, P.E., Vanderleyden, J., and Blair, M.W. 2011. Saturation of a unified intra- and inter-genepool common bean consensus linkage map in common bean for fine-mapping and synteny analysis. PLoS ONE 6: e28135.
- Gao, L.L., Hane, J., Kamphuis, L., Foley, R., Shi, B.J., Atkins, C., and Singh, K. 2011. Development of genomic resources for the narrow-leafed lupin (Lupinus angustifolius): construction of a bacterial artificial chromosome (BAC) library and BAC-end sequencing. BMC Genomics 12: 521.
- Garcia, G.M., Stalker, H.T., Shroeder, E., and Kochert, G. 1996. Identification of RAPD, SCAR, and RFLP markers tightly linked to nematode resistance genes introgressed from Arachis cardenasii into Arachis hypogaea. Genome 39: 836–845.
- GENESYS. 2013. Gateway to genetic resources, www.genesys-pgr-org/
- Gepts, P., Aragao, F., Barros, E., Blair, M.W., Brondani, R., Broughton, W., Hernández, G., Kami, J., Lariguet, P., McClean, P., Melotto, M., Miklas, P., Pedrosa-Harand, A., Porch, T., and Sánchez, F. 2008. Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In: Genomics of Tropical Crops. Chp 5. pp. 113–143. Moore, P.H. and Ming, R., Eds., Springer, Heidelberg, Germany.
- Gillett, J.B. 1952. The genus Trifolium in southern Arabia and in Africa south of the Sahara. Kew Bull. 7: 367–404.
- Gillett, J.M. 1985. Taxonomy and morphology. Clover Science and Technology, Agronomy Monograph 34: 7–69.
- Gillett, J.M. and Taylor, N.L., 2001. The World of Clovers. Iowa State University Press, Ames, Iowa, USA.
- Gladstone, J.S. 1970. Lupins as crop plants. Field Crop Abst. 23: 123–148.
- Gnanasambandam, A., Paull, J., Torres, A., Kaur, S., Leonforte, T., Li, H., Zong, X., Yang, T., and Materne, M. 2012. Impact of molecular technologies on Faba Bean (Vicia faba L.) breeding strategies. Agronomy 2: 132–166.
- Goggin, D.E., Mir, G., Smith, W.B., Stuckey, M., and Smith, P.M. C. 2008. Proteomic analysis of lupin seed proteins to identify conglutin β as an allergen, Lup an 1. J. Agric. Food Chem. 56: 6370–6377.
- Goldblatt, P. 1981. Cytology and phylogeny of Leguminosae. In: Advances in Legume Systematics. pp. 427–463. Part 2. Polhill, R.M. and Raven, P.H., Eds., Royal Botanic Gardens, Kew, UK,
- Golubev, A.A. 1990. Habitats, collection, cultivation and hybridization vavilovia (Vavilovia formosa Fed.). Bull. Appl. Bot. Genet. Plant Breed. 135: 67–75. [in Russian]
- González-Andrés, F., Casquero, P., San-Pedro, C., and Hernández-Sánchez, E. 2007. Diversity in white lupin (Lupinus albus L.) landraces from northwest Iberian plateau. Genet. Res. Crop Evol. 54: 27–44.
- Govorov, L.I. 1937. Pisum. In: Flora of cultivated plants IV: Grain leguminosae. pp. 231–336. Vavilov, N.I. and Wulff, E.V., Eds., State Agricultural Publishing Company, Moscow.
- Gowda, M.V. C., Motagi, B.N., Naidu, G.K.B. Diddimani, S.B., and Sheshagiri, R. 2002. GPBD 4: a Spanish bunch groundnut genotype resistant to rust and late leaf spot. Int. Arachis. Newsl. 22: 29–32.
- Greene, S.L., Afonin, A., Dzyubenko, E., and Dzyubenko, N. 2012. Crop wild relatives of Medicago in Russia and neighboring countries: gap analysis for effective conservation. In: Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces. pp. 82–90. Maxted, N., Dullo, M.E., Ford-Lloyd, B.V. , Frese, L., Iriondo, J., and Pinheiro de Carvalho, M.A., Eds., Cab International, UK.
- Greene, S.L., Gritsenko, M., and Vandermark, G. 2004. Relating morphological and RAPD marker variation to collection site environment in wild populations of red clover (Trifolium pratense L.). Genet. Res. Crop Evol. 51: 643–653.
- Gross, R., von Baer, E., Koch, F., Marquard, R., Trugo, L., and Wink, M. 1988. Chemical composition of a new variety of the Andean lupin (Lupinus mutabilis cv. Inti) with low-alkaloid content. J. Food Compos. Analysis 1: 353–361.
- Guan, R.X., Liu, X.M., Chang, R.Z., Ning, H.X., Yuan, C.P., Liu, Z.X., and Qiu, L.J. 2006. Genetic diversity analysis of wild soybean (Glycine soja Sieb & Zucc.) from in-situ conserved population in Xinbin County of Liaoning Province. High Technology Letters 16: 67–72.
- Guarino, L. and Lobell, D.B. 2011. A walk on the wild side. Nature Clim. Change 1: 374–375.
- Gunn, C., and Kluve, R.J. 1976. Androecium and pistil characters for the tribe Vicieae (Fabaceae). Taxon 25: 563–575.
- Guo, J., Wang, Y., Song, C., Zhou, J., Qiu, L., Huang, H., and Wang, Y. 2010. A single origin and moderate bottleneck during domestication of soybean (Glycine max): implications from microsatellites and nucleotide sequences. Ann. Bot. 106: 505–514.
- Guo, Y. and Qiu, L.J. 2013. Allele-specific marker development and selection efficiencies for both flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes in soybean subgenus soja. Theor. Appl. Genet. 126: 1445–1455.
- Gupta, D. and Sharma, S.K. 2007. Widening the gene pool of cultivated lentil through introgression of alien chromatin from wild lentil subspecies. Plant Breeding 126: 58–61.
- Gupta, D. and Sharma, S.K. 2006. Evaluation of wild Lens taxa for agro-morphological traits, fungal diseases and moisture stress in North Western Indian Hills. Genet. Res. Crop Evol. 53: 1233–1241.
- Gupta, M., Verma, B., Kumar, N., Chahota, R.K., Rathour, R., Sharma, S.K., Bhatia, S., and Sharma, T.R. 2012. Construction of intersubspecific molecular genetic map of lentil based on ISSR, RAPD and SSR markers. J. Genet. 91: 279–287.
- Gupta, S., Buirchell, B.J., and Cowling, W.A. 1996. Interspecific reproductive barriers and genomic similarity among the rough-seeded Lupinus species. Plant Breeding 115: 123–127.
- Gur, A. and Zamir, D. 2004. Unused natural variation can lift yield barriers in plant breeding. PLoS Biology 2: e245.
- Gutierrez, N., Avila, C.M., Duc, G., Marget, P., Suso, M.J.T. Moreno, M.T., and Torres, A.M. 2006. CAPs markers to assist selection for low vicine and convicine content in faba bean (Vicia faba L.). Theor. Appl. Genet. 114: 59–66.
- Gutierrez, N., Avila, C.M., Moreno, M.T., and Torres, A.M. 2008. Development of SCAR markers linked to zt-2 one of the genes controlling absence of tannins in faba bean. Aust. J. Agric. Res. 59: 62–68.
- Gutierrez, N., Avila, C.M., Rodriguez-Suarez, C., Moreno, M.T., and Torres, A.M. 2007. Development of SCAR markers linked to a gene controlling absence of tannins in faba bean. Mol. Breeding 19: 305–314.
- Hamdi, A. and Erskine, W. 1996. Reaction of wild species of the genus Lens to drought. Euphytica 91: 173–179.
- Hamdi, A., Küsmenoĝlu, I., and Erskine, W. 1996. Sources of winter hardiness in wild lentil. Genet. Res. Crop Evol. 43: 63–67.
- Hammer, K. 1984. Das Domestikationssyndrom. Kulturpflanze 11: 11–34.
- Hamwieh, A., Udupa, S.M., Sarker, A., Jung, C., and Baum, M. 2009. Development of new microsatellite markers and their application in the analysis of genetic diversity in lentils. Breeding Sci. 59: 77–86.
- Hancock J.F. 2012. Plant Evolution and Origin of Species. 3rd edition. CABI, Wallingford, Oxfordshire, UK.
- Hanelt, P. and Mettin, D. 1989. Biosystematics of the genus Vicia L. (Leguminosae). Annu. Rev. Ecol. Syst. 20: 199–223.
- Harlan, J.R. and de Wett, J.M. J. 1971. Toward a rational classification of cultivated plants. Taxon 20: 509–517.
- Havey, M.J. and Muehlbauer, F.J. 1989. Variability for restriction fragment lengths and phylogenies in lentil. Theor. Appl. Genet. 77: 839–843.
- He, C., Liu, Y., Wu, K., Yuan, M., Feng, Q., Liu, Y., Yan, Q., Guan, J., Rose, I.A., Redden, R.J., and Enneking, D. 2008. Collecting and surveying landraces of pea (Pisum sativum) and faba bean (Vicia faba) in Qinghai province of China. Pl. Genet. Res. News. 156: 1–10.
- Hendrych, R. 1988. Die ersten nomenklatorischen Erganzungen zur Trifolium—Monographie von Zohary und Heller (taxa supraspecifica). Preslia 60: 215–236.
- Herridge, D.F., Rupela, O.P., Serraj, R., and Beck, D.P. 1994. Screening techniques and improved biological nitrogen fixation in cool season food legumes. Euphytica 73: 95–108.
- Heuzé, V., Tran, G., and Baumont, R. 2013. Common vetch (Vicia sativa). Feedipedia.org. A programme by INRA, CIRAD, AFZ and FAO. . http://www.feedipedia.org/node/239
- Heyn, C.C. 1981. Trifolieae. In: Advances in Legume Systematics. pp. 383–385. Part 1., Polhill, R.M., and Raven, P.H., Eds., Royal Botanic Gardens, Kew, UK.
- Heywood, V.H. 2011. The role of botanic gardens as resource and introduction centres in the face of global change. Biodiv. Conserv. 20: 221–239.
- Heywood, V.H., Kell, S.P., and Maxted, N. 2008. Towards a global strategy for the conservation and use of crop wild relatives. In: Crop Wild Relative Conservation and Use. pp. 653–662. Maxted, N., Ford-Lloyd, B.V., Kell, S.P., Iriondo, J., Dulloo, E., and Turok, J., Eds., Wallingford: ABI Publishing.
- Hirata, T., Abe, J., and Shimamoto, Y. 1999. Genetic structure of the Japanese soybean population. Genet. Res. Crop Evol. 46: 441–453.
- Hobson, K., Armstrong, R., Nicolas, M., Connor, D., and Materne, M. 2006. Response of lentil (Lens culinaris) germplasm to high concentrations of soil boron. Euphytica 151: 371–382.
- Hoffman, D.L., Soltis, D.E., Muehlbauer, F.J., and Ladizinsky, G. 1986. Isozyme polymorphism in Lens (Leguminosae). Syst. Bot. 11: 392–402.
- Holbrook, C.C., and Dong, W. 2005. Development and evaluation of a mini core collection for the U.S. peanut germplasm collection. Crop. Sci. 45: 1540–1544.
- Holbrook, C.C., Anderson, W.F., and Pittman, R.N. 1993. Selection of a core collection from the United States germplasm collection of peanut. Crop. Sci. 33: 859–861.
- Holbrook, C.C., Stalker, H.T. 2003. Peanut Breeding and Genetic Resources. In: Plant Breeding Reviews, Volume 34, p. 297–356. Janick, J., Ed., John Wiley & Sons, Inc. Hoboken, NJ, USA.
- Hopf, M. 1986. Archaeological evidence of the spread and use of some members of the Leguminosae family. In: The original and domestication of cultivated plants. pp. 35–60. Barigozzi, C. Ed., Oxford, Elsevier.
- Hoque, M.E. and Hasan, M.M. 2012. Molecular diversity analysis of lentil (Lens culinaris Medik.) through RAPD markers. Plant Tissue Cult. Biotech. 22: 51–58.
- Hossain, M. 1961. A revision of Trifolium in the Nearer East. Not. R. Bot. Gard. Edinburg 23: 387–481.
- Hu, J., Landry, E.J., Mwengi, J.E., and Coyne, C.J. 2011. Natural outcrossing rate of faba bean under Pullman field conditions and its implication to germplasm management and enhancement. Pisum Genet. 43: 59–61.
- Hughes, C. and Eastwood, R. 2006. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA 103: 10334–10339.
- Humphry, M.E., Lambrides, C.J., Chapman, S.C., Aitken, E.A. B, Imrie, B.C., Lawn, R.J., McIntyre, C.L., and Liu, C.J. 2005. Relationships between hard-seededness and seed weight in mungbean (Vigna radiata(L.) Wilczek) assessed by QTL analysis. Plant Breed. 124: 292–298.
- Hymowitz, T. 1970. On the domestication of the soybean. Econ. Bot. 24: 408–421.
- Hyten, D., Song, Q., Zhu, Y., Choi, I., Nelson, R., Costa, J., Specht, J., Shoemaker, R., and Cregan, P. 2006. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 103: 16666.
- Inder, P., Materne, M., Taylor, P.W. J., and Ford, R. 2008. Genotyping elite genotypes within the Australian lentil breeding program with lentil-specific sequenced tagged microsatellite site (STMS) markers. Crop Pasture Sci. 59: 222–225.
- Infantino, A., Kharrat, M., Riccioni, L., Coyne, C.J., McPhee, K.E., and Grünwald, N.J. 2006. Screening techniques and sources of resistance to root diseases in cool season food legumes. Euphytica 147: 201–221.
- Iriondo, J.M., Maxted, N., and Dulloo, E. 2008. Conserving Plant Genetic Diversity in Protected Areas: Population Management of Crop Wild Relatives. CAB International, Wallingford.
- Iriondo, J.M., Maxted, N., Kell, S.P., Ford-Lloyd, B.V., Lara-Romero, C., Labokas, J., and Magos Brehm, J. 2012. Quality standards for genetic reserve conservation of crop wild relatives. In: Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces. pp. 72–77. Maxted, N., Dulloo, M.E., Ford-Lloyd, B.V., Frese, L., Iriondo, J.M., and Pinheiro de Carvalho, M.A. A., Eds., CAB International, Wallingford.
- Isemura, T., Kaga, A., Tabata, S., Somta, P., and Srinives, P. 2012. Construction of a Genetic Linkage Map and Genetic Analysis of Domestication Related Traits in Mungbean (Vigna radiata). PLoS ONE 7: e41304.
- Ishii, H.S. 2013. Community-dependent foraging habits of flower visitors: cascading indirect interactions among five bumble bee species. Ecol. Res. 28: 603–613.
- ISI. 2013. ISI Web of Knowledge. Available from: http://apps.isiknowledge.com/WOS. Accessed October 2013.
- Jackson, M.T. and Yunus, A.G. 1984. Variation in the grasspea (L. sativus L.) and wild species. Euphytica 33: 549–559.
- Jahufer, M.Z., Cooper, M., Ayres, J.F., and Bray, R.A. 2002. Identification of research to improve the efficiency of breeding strategies for white clover in Australia: a review. Aust. J. Agric. Res. 53: 239–257.
- Jain, H.K. and Mehra, K.L. 1980. Evaluation, adaptation, relationship and cases of the species of Vigna cultivation in Asia. In: Advances in Legume Science. pp. 459–468. Summerfield, R.J. and Butnting, A.H., Eds., Royal Botanical Gardens, Kew, UK.
- Jain, M., Misra, G., Patel, R.K., Priya, P., Jhanwar, S., Khan, A.W., Shah, N., Singh, V.K., Garg, R., Jeena, G., Yadav, M., Kant, C., Sharma, P., Yadav, G., Bhatia, S., Tyagi, A.K., and Chattopadhyay, D. (2013). A draft genome sequence of the pulse crop chickpea. Plant J. 74: 715–29.
- Jaiswal, H.K., Singh A.K., and Singh, R.M. 1986. Introgression of genes for yield and yield traits from C. reticulatum into C. arietinum. Int. Chickpea Newsl. 14: 5–8.
- Jakesova, H., Repkova, J., Hampel, D., Cechova, L., and Hofbauer, J. 2011. Variation of morphological and agronomic traits in hybrids of Trifolium pratense x T. medium and a comparison with the parental species. Czech J. Genet. Pl. Breed. 47: 28–36.
- Jambunathan R. 1991. Groundnut quality characteristics. In: Uses of Tropical Grain Legumes. pp. 267–275. Proceedings of a Consultants Meeting ICRISAT, Patancheru, India.
- Janila, P., Nigam, S.N., Pandey, M.K., Nagesh, P., and Varshney, R.K. 2013. Groundnut improvement: use of genetic and genomic tools. Front. Plant Sci. 4: 23.
- Jiang, H.F., Ren, X.P., Liao, B.S., Huang, J.Q., Lei, Y., Chen, B.Y., Guo, B.Z., Holbrook, C.C., and Upadhyaya, H.D. 2008. Peanut core collection established in china and compared with ICRISAT mini core collection. Acta. Agron. Sin. 34: 25–30.
- Jing, R., Johnson, R., Seres, A., Kiss, G., Ambrose, M.J., Knox, M.R., Ellis, T.H., and Flavell, A.J. 2007. Gene-based sequence diversity analysis of field pea (Pisum). Genetics 177: 2263–2275.
- Jing, R., Knox, M.R., Lee, J.M., Vershinin, A.V., Ambrose, M., Ellis, T.H. N., and Flavell, A.J. 2005. Insertional polymorphism and antiquity of PDR1 retrotransposon insertions in Pisum species. Genetics 171: 741–752.
- Jing, R., Vershinin, A., Grzebyta, J., Shaw, P., Smýkal, P., Marshall, D., Ambrose, M.J., Ellis, T.H. N., and Flavell, A.J. 2010. The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 10: 44.
- Jukanti, A.K., Gaur, P.M., Gowda, C.L. L., and Chibbar, R.N. 2012. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br. J. Nutr. 108(S1): S11–S26.
- Kaass, E. and Wink, M. 1997. Molecular phylogeny and phylogeograph of Lupinus (Leguminosae) inferred from nucleotide sequences of the rbcL gene and ITS regions of rDNA. Plant Syst. Evol. 208: 139–167.
- Kaewwongwal, A., Jetsadu, A., Somta, P., Chankaew, S., and Srinives, P. 2013. Genetic diversity and population structure of Vigna exilis and Vigna grandiflora (Phaseoleae, Fabaceae) from Thailand based on microsatellite variation. Bot. 91: 653–661.
- Kaga, A., Shimizu, T., Watanabe, S., Tsubokura, Y., Katayose, Y., Harada, K., Vaughan, D.A., and Tomooka, N. 2012. Evaluation of soybean germplasm conserved in NIAS genebank and development of mini core collections. Breeding Sci. 61: 566–592.
- Kahraman, A., Kusmenoglu, I., Aydin, N., Aydogan, A., Erskine, W., and Muehlbauer, F.J. 2004. QTL mapping of winter hardiness genes in lentil. Crop Sci. 44: 13–22.
- Kajonphol, T., Sangsiri, C., Somta, P., Toojinda, T., and Srinives, P. 2012. SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek). Sabrao J. Breeding and Genet. 44: 71–86.
- Kaljund, K. and Leht, L. 2013. Extensive introgressive hybridization between cultivated lucerne and the native sickle medic (Medicago sativa subsp. falcata) in Estonia. Ann. Bot. Fennici 50: 23–31.
- Kasettranan, W., Somta, P., and Srinives, P. 2010. Mapping of quantitative trait loci controlling powdery mildew resistance in Mungbean (Vigna radiata (L.) Wilczek). J. Crop Sci. Biotech. 13: 155–161.
- Kasprzak, A., Šafar, J., Janda, J., Dolezel, J., Wolko, B., and Naganowska, B. 2006. The bacterial artificial chromosome (BAC) library of the narrow-leafed lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Letters 11: 396–407.
- Kass, E. and Wink, M. 1997. Molecular phylogeny and phylogeograph of Lupinus (Leguminosae) inferred from nucleotide sequences of the rbcL gene and ITS regions of rDNA. Plant Syst. Evol. 208: 139–167.
- Kassa, M.T., Penmetsa, R.V., Carrasquilla-Garcia, N., Sarma, B.K., Datta, S., Upadhyaya, H., Varshney, R., von Wettberg E.J. B., and Cook, D.R. 2012. Genetic patterns of domestication in pigeonpea (Cajanus cajan (L.) Millsp.) and wild Cajanus relatives. PLoS ONE 7: e39563.
- Kaur, S., Cogan, N.O., Pembleton, L.W., Shinozuka, M., Savin, K.W., Materne, M., and Forster, J.W. 2011. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genomics 12: 265.
- Keiša A., Maxted, N., and Ford-Lloyd, B.V. 2008. The assessment of biodiversity loss over time: wild legumes in Syria. Genet. Res. Crop Evol. 55: 603–612.
- Kenicer, G.J., Kajita, T., Pennington, R.T., and Murata, J. 2005. Systematics and biogeography of Lathyrus (Leguminosae) based on internal transcribed spacer and cpDNA sequence data. Am. J. Botany 92: 1199–1209.
- Khan, H.R., Link, W., Hocking, T.J., and Stoddard, F.L. 2007. Evaluation of physiological traits for improving drought tolerance in faba bean (Vicia faba L.). Plant Soil. 292: 205–217.
- Khera, P., Upadhyaya, H.D., Pandey, M.K., Roorkiwal, M., Sriswathi, M., Janila, P., Guo, Y., McKain, M.R., Nagy, E.D., Knapp, S.J., Leebens-Mack, J., Conner, J.A., Ozias-Akins, P., and Varshney, R.K. 2013. Single nucleotide polymorphism–based genetic diversity in the reference set of peanut (Arachis spp.) by developing and applying cost-effective kompetitive allele specific polymerase chain reaction genotyping assays. The Plant Genome 6. doi: 10.3835/plantgenome2013
- Khodapanahi, E., Lefsrud, M., Orsat, V., Singh, J., and Warkentin, T.D. 2012. Study of pea accessions for development of an oilseed pea. Energies 5: 3788–3802.
- Khoury, C., Laliberté, B., and Guarino, L. 2010. Trends in ex situ conservation of plant genetic resources: a review of global crop and regional conservation strategies. Genet. Res. Crop Evol. 57: 625–639.
- Khu, D.M., Reyno, R., Han, Y., Zhao, P.X., Bouton, J.H., Brummer, E.C., and Monteros, M.J. 2013. Identification of aluminum tolerance quantitative trait loci in tetraploid alfalfa. Crop Sci. 53:148–163.
- Kim, M.Y., Lee, S., Van, K., Kim, T.H., Jeong, S.C., Choi, I.Y., Kim, D.S., Lee, Y.S., Park, D., Ma, J., Kim, W.Y., Kim, B.C., Park, S., Lee, K.A., Kim, D.H., Kim, K.H., Shin, J.H., Jang, Y.E., Kim, K.D., Liu, W.X., Chaisan, T., Kang, Y.J., Lee, Y.H., Kim, K.H., Moon, J.K., Schmutz, J., Jackson, S.A., Bhak, J., and Lee, S.H. 2010. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci. USA 107: 22032–22037.
- Kislev, M.E. and Bar-Yosef, O. 1988. The Legumes: The Earliest Domesticated Plants in the Near East? Curr. Anthropol. 29: 175–179.
- Kjaergaard, T. 2003. A plant that changed the world: Rise and fall of clover 1000–2000. Landsc. Res. 28: 41–49.
- Kloz, J. 1971. Serology of the Leguminosae. In: Chemotaxonomy of the Leguminosae. pp. 309–365. Harborne, J.B., Boulter, D., and Turner, B.L., Eds., Academic Press, London.
- Knights, E.J., Southwell, R.J., Schwinghamer, M.W., and Harden, S. 2008. Resistance to Phytophthora medicaginis Hansen and Maxwell in wild Cicer species and its use in breeding root rot resistant chickpea (Cicer arietinum L.). Aust. J. Agric. Res. 59: 383–387.
- Kochert, G., Stalker, H.T., Gimenes, M., Galgaro, L., and Moore, K. 1996. RFLP and cytogenetic evidence for the progenitor species of allotetraploid cultivated peanut (Arachis hypogaea L.). Am. J. Bot. 83: 1282–1291.
- Kollipara, K.P., Singh, R.J., and Hymowitz, T. 1997. Phylogenetic and genomic relationships in the genus Glycine Willd. based on sequences from the ITS region of nuclear rDNA. Genome 40: 57–68.
- Kongjaimun, A., Kaga, A., Tomooka, N., Somta, P., Vaughan, P.A., and Srinives, P. 2012.The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. subsp. unguiculata cv.-gr. sesquipedalis. Ann Bot. 109: 1185–1200.
- Koppolu, R., Hari, U., Sangam, D., David, H., and Varshney, R.K. 2010. Genetic relationships among seven sections of genus Arachis studied by using SSR markers. BMC Plant Biol. 10: 15.
- Kosterin, O.E., Zaytseva, O.O., Bogdanova, V.S., and Ambrose, M.J. 2010. New data on three molecular markers from different cellular genomes in Mediterranean accessions reveal new insights into phylogeography of Pisum sativum L. sbsp. elatius (Bieb.) Schmalh. Genet. Res. Crop Evol. 57: 733–739.
- Kosterin, O.E. and Bogdanova, V.S. 2008. Relationship of wild and cultivated forms of Pisum L. as inferred from an analysis of three markers, of the plastid, mitochondrial and nuclear genomes. Genet. Res. Crop Evol. 55: 735–755.
- Kouame, C.N. and Quesenberry, K.H. 1993. Cluster analysis of a world collection of red clover germplasm. Genet. Res. Crop Evol. 40: 39–47.
- Krapovickas, A., and Gregory, W.C. 1994. Taxonomia del genero Arachis (Leguminosae). Bonplandia 8: 1–186.
- Krishna, T.G., and Reddy, L.J. 1982. Species affinities between Cajanus cajan and some Atylosia species based on esterase isozymes. Euphytica 31: 709–713.
- Kroc, M., Koczyk, G., Święcicki, W., Kilian, A., Nelson, M.N. 2014. New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L. (narrow-leafed lupin). Theor. Appl. Genet. 127: 1237–1249.
- Kumar, S., Bejiga, G., Ahmed, S., Nakkoul, H., and Sarker, A. 2011. Genetic improvement of grass pea for low neurotoxin (β-ODAP) content. Food Chem. Toxicol. 49: 589–600.
- Kumar, S., Gupta, P., Barpete, S., Sarker, A., Amri, A., Mathur, P.N., and Baum, M. 2013. Grass pea. pp. 269–305. In: Genetic and Genomic Resources of Grain Legume Improvement. Singh, M., and Upadhya, H., Eds., Elsevier, Netherlands.
- Kumar, V., Dikshit, H.K., Jain, N., Kumari, J., Singh, D., Singh, A., Tak, R., and Sharma, T.R. 2012. Genetic diversity in mungbean [Vigna radiata (L.) Wilczek] and related Vigna spp. detected by ISSR, URP and SSR markers. Indian Genet. Plant Breed. 72: 318.
- Kupicha, F.K. 1976. The infrageneric structure of Vicia L. Not. R. Bot. Gard. Edinburg 34: 287–326.
- Kupicha, F.K. 1977. The delimitation of the tribe Vicieae (Leguminosae) and the relationships of Cicer L. Bot. J. Lin. Soc. 74: 131–162.
- Kupicha, F.K. 1981. Vicieae (Adans.) DC. (1825) nom. conserv. prop. In: Advances in Legume Systematics. Vol. 34, pp. 377–381. Polhill, R.M. and Raven, P.H., Eds., Kew, Royal Botanical Gardens.
- Kupicha F.K. 1983. The infrageneric structure of Lathyrus. Not. R. Bot. Gard. Edinburg 41: 209–244.
- Kwon, S.J., Brown, A.F., Hu, J., McGee, R.J., Watt, C.A., Kisha, T., Timmerman-Vaughan, G.M., and Coyne, C.J. 2012. Population genetic sub-structure within the USDA ARS Pisum core collection and its potential as a platform for association mapping. Genes and Genomics 34: 305–320.
- Kwon, S.J., Hu, J., and Coyne, C.J. 2010. Genetic diversity and relationship among faba bean (Vicia faba L.) germplasm entries as revealed by TRAP markers. Plant Genet. Resour. Charact.Util. 8: 204–213.
- Ladizinsky, G. 1975. On the origin of the broad bean Vicia faba L. Israel J. Bot. 24: 80–88.
- Ladizinsky, G. 1979a. The origin of lentil and its wild genepool. Euphytica 28: 179–187.
- Ladizinsky, G. 1979b. Species relationships in the genus Lens as indicated by seed-protein electrophoresis. Botan. Gazette 140: 449–451.
- Ladizinsky, G. 1985. The genetics of hard seed coat in the genus Lens. Euphytica 34: 539–543.
- Ladizinsky, G. 1986. A new Lens species from the Middle-East. Not. R. Bot. Gard. Edinburg 43: 489–492.
- Ladizinsky, G. 1997. A new species of Lens from southeast Turkey. Bot. J. Linn. Soc. 123: 257–260.
- Ladizinsky, G. 1998. Plant Evolution under Domestication. Kluwer Academic Publishers, Dortrecht.
- Ladizinsky, G. and Adler, A. 1976a. The origin of chickpea Cicer arietinum L. Euphytica 25: 211–217.
- Ladizinsky, G. and Adler, A. 1976b. Genetic relationships among annual species of Cicer L. Theor. Appl. Genet. 48: 197–203.
- Ladizinsky, G., and Sakar, D. 1982. Morphological and cytogenetical and characterization of Vicia montbretii Fisch and Mey (Synonym Lens montbretii (Fisch and Mey) Davis and Plitmann. Bot. J. Linn. Soc. 85: 209–212
- Ladizinsky, G., Braun, D., Goshen, D., and Muehlbauer, F.J. 1984. The biological species of the genus Lens L. (Lens nigricans). Botan. Gazette 145: 253–261.
- Ladizinsky, G., Cohen, D., and Muehlbauer, F.J. 1985. Hybridization in the genus Lens by means of embryo culture. Theor. Appl. Genet. 70: 97–101.
- Lam, H.M., Xu, X., Liu, X., Chen, W.B., Yang, G.H., Wong, F.L., Li, M.W., He, W.M., Qin, N., Wang, B., Li, J., Jian, M., Wang, J., Shao, G.H., Wang, J., Sun, S.S. M., and Zhang, G.Y. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42: 1053–1059.
- Lamarck, J.B. 1778. Flore Françoise. Paris.
- Lamont, E.J., Zoghlami A., Sackville-Hamilton, R., and Bennett, S.J. 2001. Clovers (Trifolium L.). In: Plant Genetic Resources of Legumes in the Mediterranean. pp. 76–98. Maxted, N. and Bennett, S.J., Eds., The Netherlands: Kluwer Academic Publishers.
- Lane, L.A., Ayres, J.F., and Lovett, J.V. 1997. A review of the introduction and use of white clover (Trifolium repens L.) in Australia – significance for breeding objectives. Aust. J. Exp. Agric. 37: 831–839.
- Laserna-Ruiz, I., De-Los-Mozos-Pascual, M., Santana-Méridas, O., Sánchez-Vioque, R., and Rodríguez-Conde, M.F. 2012. Screening and selection of lentil (Lens Miller) germplasm resistant to seed bruchids (Bruchus spp.). Euphytica 73: 1–10.
- Lavin, M., Herendeen, P., and Wojchiechowski, M. 2005. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 54: 575–594.
- Lawn, R.J. 1995. The Asian Vigna species. In: The Evolution of Crop Plants. pp. 321–326. Smartt, J. and Simmonds, N.W. Eds., ( 2nd ed.). Longman, Harlow, UK.
- Lawn, R.J. 2014. Case study in the collection and conservation of crop wild relatives: The Australian Vigna species, chapter 18. In: Yadav, S.S., Redden, R., Dulloo, E., and Maxted, N. (eds.), Crop Wild Relatives and Climate Change. Chichester, West Sussez, UK: Wiley-Blackwell – John Wiley & Sons.
- Lawn, R.J. and Cottrell, A. 1988. Wild mungbean and its relaties in Australia. Biologist 35: 267–273.
- Lee, G.A., Crawford, G.W., Liu, L., Sasaki, Y., and Chen, X. 2010. Archaeological soybean (Glycine max) in East Asia: does size matter? PLoS ONE 6: e26720.
- Lee, Y.P., Mori, T.A., Sipsas, S., Barden, A., Puddey, I.B., Burke, V., Hall, R.S., and Hodgson, J.M. 2006. Lupin-enriched bread increases satiety and reduces energy intake acutely. Amer. J. Clinical Nutrition 84: 975–980.
- Lejeune-Hénaut, I., Hanocq, E., Béthencourt, L., Fontaine, V., Delbreil, B., Morin, J., Petit, A., Devaux, R., Boilleau, M., Stempniak, J.J., Thomas, M., Lainé, A.L., Foucher, F., Baranger, A., Burstin, J., Rameau, C., Giauffret, C. 2008. The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor. Appl. Genet. 116: 1105–1116.
- Lesins, K. 1976. Alfalfa, Lucerne. In: Evolution of Crop Plants. pp. 165–168. Simmonds, N.W., Ed., Longman, London, UK.
- Lesins, K.A. and Lesins, I. 1979. Genus Medicago (Leguminosae). A Taxogenetic Study. Dr.W. Junk, The Hague, The Netherlands.
- Lewis, G., Schrire, B., Mackinder, B., and Lock, M. 2005. Legumes of the World. Royal Botanic Gardens, Kew, UK.
- Li, D., Pfeiffer, T.W., and Cornelius, P.L. 2008. Soybean QTL for yield and yield components associated with Glycine soja alleles. Crop Sci. 48: 571–581.
- Li, F.S. 1994. A study on origin and evolution of soybean. Soybean Sci. 13: 61–66.
- Li, L., Redden, R.J., Zong, X., Berger, J.A. D., Bennett, S.J. 2013. Ecogeographic analysis of pea collection sites from China to determine potential sites with abiotic stresses. Genet. Res. Crop Evol. 60: 1801–1815.
- Li, X.H., Wang, K.J., and Jia, J.Z. 2009. Genetic diversity and differentiation of Chinese wild soybean germplasm (G. soja Sieb. & Zucc.) in geographical scale revealed by SSR markers. Plant Breeding 128: 658–664.
- Li, X., Acharya, A., Farmer, A.D., Crow, J.A., Bharti, A.K., Kramer, R.S., Wei, Y., Han, Y., Gou, J., May, G.D., Monteros, M.J., and Brummer, E.C. 2012. Prevalence of single nucleotide polymorphism among 27 diverse alfalfa genotypes as assessed by transcriptome sequencing. BMC Genomics 13: 568.
- Li, Y.H., Guan, R.X., Liu, Z., Ma, Y., Wang, L., Li, L., Lin, F., Luan, W., Chen, P., Yan, Z., and Qiu, L.J. 2008. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 117: 857–871.
- Li, Y.H., Li, W., Zhang, C., Yang, L., Chang, R.Z., Gaut, B.S., and Qiu, L.J. 2010. Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single‐nucleotide polymorphism loci. New Phytol. 188: 242–253.
- Li, Y.H., Zhang, C., Smulders, M.J. M., Li, W., Ma, Y.S., Xu, Q., Chang, R.Z., and Qiu, L.J. 2013a. Analysis of average standardized SSR allele size supports domestication of soybean along the Yellow River. Genet. Res.. Crop. Evol. 60: 763–776.
- Li, Y.H., Zhao, S.C., Ma, J.X., Li, D., Yan, L., Li, J., Qi, X.T., Guo, X.S., Zhang, L., He, W.M., Chang, R.Z., Liang, Q.S., Guo, Y., Ye, C., Wang, X.B., Tao, Y., Guan, R.X., Wang, J.Y., Liu, Y.L., Jin, L.G., Zhang, X.Q., Liu, Z.X., Zhang, L.J., Chen, J., Wang, K.J., Nielsen, R., Li, R.Q., Chen, P.Y., Li, W.B., Reif, J., Purugganan, M., Wang, J., Zhang, M.C., Wang, J., and Qiu, L.J. 2013b. Molecular footprints of domestication and improvement in soybean revealed by whole genome re-sequencing. BMC Genomics 14: 579.
- Link, W., Balko, C., and Stoddard, F.L. 2010. Winter hardiness in faba bean: physiology and breeding. Field Crop Res. 115: 287–296.
- Linnaeus, C. 1753. Species Plantarum. vol. 34. Salvius, Stockholm, Sweden.
- Liu, J., Guan, J.P., Xu, D.X., Zhang, X.Y., Gu, J., and Zong, X.X. 2008. Genetic diversity and population structure in lentil (Lens culinaris Medik.) germplasm detected by SSR markers. Acta Agron. Sinica 34: 1901–1909.
- Lock, M. and Maxted, N. 2005. Tribe Fabeae. In: Legumes of the World. Lewis, G., Schrire, B., Mackinder, B., and Lock, M., Eds., Kew, Richmond, Royal Botanic Gardens.
- Loridon, K., McPhee, K.E., Morin, J., Dubreuil, P., Pilet-Nayel, M.L., Aubert, G., Rameau, C., Baranger, A., Coyne, C.J., Lejeune-Hénault, I., and Burstin, J. 2005. Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor. Appl. Genet. 111: 1022–1031.
- Lubenets, P.A. 1953. Species composition and breeding assessment of cultivated and wild alfalfas. Bull. Appl. Bot. Genet. Sel. Forage Crops. 47: 3–155.
- Ma, Y., Bao, S.Y., Yang, T., Hu, J., Guan, J.P., He, Y.H., Wang, X.J., Wan, Y.L., Sun, X.L., Jiang, J.Y., Gong, C.X., and Zong, X.X. 2013. Genetic linkage map of Chinese native variety faba bean (Vicia faba L.) based on simple sequence repeat markers. Plant Breeding 132: 397–400.
- Mahalakshmi, V., Ng, Q., Lawson, M., and Ortiz, R. 2007. Cowpea [Vigna unguiculata (L.) Walp.] core collection defined by geographical, agronomical and botanical descriptors. Plant Genet. Res. Charact. Util. 5: 113–119.
- Mahé, F., Pascual, H., Coriton, O., Huteau, V., Navarro Perris, A., Misset, M.T., and Aïnouche, A. 2011. New data and phylogenetic placement of the enigmatic Old World lupin: Lupinus mariae-josephi H. Pascual. Genet. Res. Crop Evol. 58: 101–114.
- Majeed, M., Safdar, W., Ali, B., Mohammad, A., Ahmad, I., and Mumtaz, A.S. 2012. Genetic assessment of the genus Pisum L. based on sequence specific amplification polymorphism data. J. Medic. Plants Res. 6: 959–967.
- Makasheva, R.K. 1979. Gorokh (pea). In: Kulturnaya flora SSR. pp. 1–324. Korovina, Ed., Leningrad, Kolos Publishers (In Russian).
- Mallikarjuna, N., Jadhav, D.R., Nagamani, V., Amudhavalli, C., and Hoisington, D.A. 2007. Progress in interspecific hybridization between Cicer arietinum and wild species C. bijugum. Journal of SAT Agricultural Research 5: 1–2.
- Mallikarjuna, N. 1999. Ovule and embryo culture to obtain hybrids from interspecific incompatible pollinations in chickpea. Euphytica 110: 1–6.
- Mallikarjuna, N., Coyne, C.J., Cho, S., Rynearson, S., Rajesh, P., Jadhav, D.R., and Muehlbauer, F. 2011. Cicer. In Wild Crop Relatives: Genomic and Breeding Resources. Legume Crops and Forages. pp. 63–82. Kole, C., Ed., Springer, New York.
- Mallikarjuna, N., Jadhav, D., Reddy, P. 2006. Introgression of Cajanus platycarpus genome into cultivated pigeonpea C. cajan. Euphytica 149: 161–167.
- Mallikarjuna, N., Senthilvel, S., and Hoisington, D. 2011. Development of new sources of tetraploid Arachis to broaden the genetic base of cultivated groundnut (Arachis hypogaea L.). Genetic Resour. Crop Evol. 58: 889–907.
- Maréchal, R., Mascherpa, J.M., and Stainier, F. 1978. Etude taxonomique d’un groupe complexe d’espéces des genres Phaseolus et Vigna (Papilionaceae) surla base de données morphologiques et polliniques, traitées par l’analyse informatique. Boissiera 28: 1–273.
- Maroof, M.A. S., Glover, N.T., Biyashev, R.M., Buss, G.R., and Grabau, E.A. 2009. Genetic basis of the low-phytate trait in the soybean line CX1834. Crop Sci. 49: 69–76.
- Materne, M. and Siddique, K.H. M. 2009. Agroecology and crop adaptation. In: The Lentil: Botany, Production and Uses. pp. 47–63. Erskine, W., Muehlbauer, F., Sarker, F., and Sharma, F., Eds., CABI, Wallingford, UK.
- Maureira-Butler, I.J., Pfeil B.E., Muangprom, A.C. Osborn, T.C., and Doyle, J.J. 2008. The reticulate history of Medicago (Fabaceae). Syst. Biol. 57: 466–482.
- Maxted, N. 1995. An ecogeographic study of Vicia subgenus Vicia. In: Systematic and Ecogeographic Studies in Crop Genepools. pp. 184. International Board of Plant Genetic Resources, Rome, Italy.
- Maxted, N., and Ambrose, M. 2001. Peas (Pisum L.). In: Maxted, N., Bennett, S.J., (eds.). Plant Genetic Resources of Legumes in the Mediterranean. Dordrecht, The Netherlands: Kluwer Academic Publishers, 181–190.
- Maxted, N. and Bennett, S. 2001. Plant Genetic Resources of Legumes in the Mediterranean. Kluwer Academic Publisher, Dordrecht, the Netherlands.
- Maxted, N. and Kell, S.P. 2009. Establishment of a Global Network for the In Situ Conservation of Crop Wild Relatives: Status and Needs. FAO Commission on Genetic Resources for Food and Agriculture. Background Study Paper No. 39. Commission on Genetic Resources for Food and Agriculture. Food and Agriculture Organization of the United Nations, Rome.
- Maxted, N., Dulloo, E., Ford-Lloyd, B.V., Iriondo, J., and Jarvis, A. 2008. Genetic gap analysis: a tool for more effective genetic conservation assessment. Diversity and Distributions 14: 1018–1030.
- Maxted, N., Erskine, W., Singh, D.P., Robertson, L.D, and Asthana, A.N. 2000. Are our germplasm collections museum items? In: Proceedings of Third International Food Legumes. pp. 589–602. Research Conference (IFLRC): Linking Research and Marketing Opportunities for Pulses in the 21st Century. Adelaide, Australia. Knight, R., Ed., Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Maxted, N., Ford-Lloyd, B.V., and Hawkes, J.G. 1997. Complementary conservation strategies. In: Plant Genetic Conservation: The In Situ Approach. pp. 20–55. Maxted, N., Ford-Lloyd, B.V. and Hawkes, J.G., Eds., Chapman and Hall, London.
- Maxted, N., Hargreaves, S., Kell, S.P., Amri, A., Street, K., Shehadeh, A., Piggin, J., and Konopka, J. 2010. Temperate forage and pulse legume genetic gap analysis. In: XIII OPTIMA Meeting in Antalya, Turkey. pp. 22–26.
- Maxted, N., Hargreaves, S., Kell, S.P., Amri, A., Street, K., Shehadeh, A., Piggin, J., and Konopka, J., 2012. Temperate forage and pulse legume genetic gap analysis. Bocconea 24: 5–36.
- Maxted, N., Mabuza-Dlamini, P., Moss, H., Padulosi, S., Jarvis, A., and Guarino, L. 2004. An Ecogeographic Survey: African Vigna. In: Systematic and Ecogeographic Studies of Crop Genepools 10. pp. 1–468. IPGRI, Rome.
- Mayer, M.S. and Bagga, S.K. 2002. The phylogeny of Lens (Leguminosae): New insight from ITS sequence analysis. Plant Syst. Evol. 232: 145–154.
- Mayer, M.S. and Soltis, P.S. 1994. Chloroplast DNA phylogeny of Lens (Leguminosae): Origin and diversity of the cultivated lentil. Theor. Appl. Genet. 87: 773–781.
- McCaslin, M. and Reisen, P. 2012. New Technology for Alfalfa. Proceedings California Alfalfa & Grains Symposium December 10-12, 2012 Sacramento, CA.
- McCouch, S., Baute, G.J., Bradeen, J., Bramel, P., Bretting, P.K., Buckler, E., Burke, J.M., Charest, D., Cloutier, S., Cole, G., Dempewolf, H., Dingkuhn, M., Feuillet, C., Gepts, P., Grattapaglia, D., Guarino, L., Jackson, S., Knapp, S., Langridge, P., Lawton-Rauh, A., Lijua, Q., Lusty, C., Michael, T., Myles, S., Naito, K., Nelson, R.L., Pontarollo, R., Richards, C.M., Rieseberg, L., Ross-Ibarra, J., Rounsley, S., Hamilton, R.S., Schurr, U., Stein, N., Tomooka, N., van der Knaap, E., van Tassel, D., Tol, J., Valls, J., Varshney, R.K., Ward, J., Waugh, R., Wenzl, P., and Zamir, D. 2013. Agriculture: Feeding the future. Nature 499: 23–24.
- McCoy, T.J., and Bingham, E.T. 1988. Cytology and cytogenetics of alfalfa. In: Alfalfa and Alfalfa Improvement. pp. 737–776. Hanson, A.A., Barnes, D.K., and Hill, R.R. Jr., Eds., American Society of Agronomy, Crop Science of America and Soil Science Society of America, Madison, WI.
- McPhee, K.E., Tullu, A., Kraft, J.M., and Muehlbauer, F.J. 1999. Resistance to Fusarium wilt race 2 in the Pisum core collection. J. Am. Soc. Hortic. Sci. 124: 28–31.
- Meilleur, B.A. and Hodgkin, T. 2004. In situ conservation of crop wild relatives. Biodiversity Conserv. 13: 663–684.
- Mendel, G. 1866. Versuche über Pflanzen-Hybriden. [Experiments on Plant Hybrids] Verhandlungen des naturforschenden Vereines in Brünn,der naturfoschung Vereins. 4: 3–47. (http://www.mendelweb.org/)
- Michaud, R., Lehman, W.F., and Rumbaugh, M.D. 1988. World distribution and historical development. In: Alfalfa and Alfalfa Improvement. pp. 25–91. Hanson, A.A., Barnes, D.K., Hill, R.R. Jr., Eds., American Society of Agronomy, Crop Science of America and Soil Science Society of America, Madison, WI.
- Mikić, A. 2012. Origin of the words denoting some of the most ancient old world pulse crops and their diversity in modern European languages. PLoS ONE 7: e44512
- Mikič, A., Smýkal P., Kenicer, G., Vishnyakova, M., Akopian, J., Sarukhanyan, N., Gabrielyan, I., Vanyan, A., Toker, C., Cupina, B., Ambrose, M., Mihailovic, V., and Ellis, N. 2009. A revival of the research on beautiful vavilovia (Vavilovia formosa syn. Pisum formosum). Pisum Genet. 41: 14–19.
- Mikič, A., Smýkal P., Kenicer, G., Vishnyakova, M., Sarukhanyan, N., Akopian, J., Vanyan, A., Gabrielyan, I., Smykalova, I., Sherbakova, Zoric, L., Atlagic, J., Zeremski-Skoric, T., Cupina, B., Krstic, D., Jacic, I., Antanasovic, S., Dordevic, V., Mihailovic, V., Ivanov, A., Ochatt, S., and Ambrose, M. 2013. The bicentenary of the research on ‘beautiful’ vavilovia (Vavilovia formosa), a legume crop wild relative with taxonomic and agronomic potential. Bot. J. Linn. Soc. 172: 524–531.
- Mimura, M., Yasua, K., and Yamaguchi, H. 2000. RAPD variationin wild, weedy and cultivated adzuki beans in Asia. Genet. Res. Crop Evol. 47: 603–610.
- Morgan, J.M., Rodriguez-Maribona, B., and Knights, E.J. 1991. Adaptation to water deficit in chickpea breeding lines by osmoregulation: relationship to grain yields in the field. Field Crops Res. 27: 61–70.
- Muchero, M., Diop, N.N., Bhat, P.R., Fenton, R.D., Wanamaker, S., and Pottorff, M. 2009. A consensus genetic map of cowpea [Vigna unguiculata (L.) Walp.] and synteny based on EST-derived SNPs. Proc. Natl. Acad. Sci. USA 106: 18159–18164.
- Muehlbauer, F.J. and Singh, K.B. 1987. Genetics of chickpea. In: The Chickpea. pp. 99–125. Saxena, M.C. and Singh, K.B., Eds., CAB International Publication, Wallingford.
- Muehlbauer, F.J., Weeden, N.F., and Hoffman, D.L. 1989. Inheritance and linkage relationships of morphological and isozyme loci in lentil (Lens Miller). J. Hered. 80: 298–303.
- Muench, D.G., Slinkard, A.E., and Scoles, G.J. 1991. Determination of genetic variation and taxonomy in lentil (Lens Miller) species by chloroplast DNA polymorphism. Euphytica 56: 213–218.
- Muñoz, C., Duque, M.C., Debouck, D., and Blair, M.W. 2006. Taxonomy of tepary bean (Phaseolus acutifolius) and wild relatives as determined by amplified fragment length polymorphism (AFLP) markers. Crop Sci. 46: 1744–1754.
- Muñoz, L.C., Blair, M.W., Duque, M.C., Tohme, J., and Roca, W. 2004. Introgression in common bean x tepary bean interspecific congruity-backcross lines as measured by AFLP markers. Crop Sci. 44: 637–645.
- Muratova, V.S. 1931. Common beans (Vicia faba L.). Bull. Appl. Bot. Genet. Pl. Breed. Suppl. 50: 1–298.
- Nadimpalli, R.G., Jarret, R.L., Phatak, S.C., and Kochart, G. 1994. Phylogenetic relationships of pigeonpea (Cajanus cajan) based on nuclear restriction fragment length polymorphism. Genome 36: 216–223.
- Naganowska, B., Wolko, B., Sliwinska, E., and Kaczmarek, Z. 2003. Nuclear DNA content variation and species relationships in the genus Lupinus (Fabaceae). Ann. Botany 92: 349–355.
- Naganowska, B., Wolko, B., Śliwińska, E., Kaczmarek, Z., and Schifino-Wittmann, M.T. 2006. 2C DNA variation and relationships among New World species of the genus Lupinus (Fabaceae). Plant Syst. Evol. 256: 147–157.
- Nagel, M., Vogel, H., Landjeva, S., Buck-Sorlin, G., Lohwasser, U., Scholz, U., and Boerner, A. 2009. Seed conservation in ex situ genebanks—genetic studies on longevity in barley. Euphytica 170: 5–14.
- Nasiri, J., Haghnazari, A., and Saba, J. 2010. Genetic diversity among cultivars and wild species accessions of pea (Pisum sativum L.) based on SSR markers. African J. Biotech. 15: 3405–3417.
- Nelson, M.N., Moolhuijzen, P.M., Boersma, J.G., Chudy, M., Lesniewska, K., Bellgard, M., Oliver, R.P., Swiecicki, W., Wolko, B., Cowling, W.A., and Ellwood, S.R. 2010. Aligning a new reference genetic map of Lupinus angustifolius with the genome sequence of the model legume, Lotus japonicus. DNA Res. 17: 73–83.
- Nelson, M., Buirchell, B., and Berger, J.D. 2011. Genetic diversity in Lupinus angustifolius investigated through DArT marker genotyping. In: 13th International Lupin Conference, “Lupin crops – an opportunity for today, a promise for the future.” pp. 69–71. Naganowska, B., Wolko, B., and Kachlicki, P., Eds., Poznań, Poland.
- Nelson, M., Phan, H., Ellwood, S., Moolhuijzen, P., Hane, J., Williams, A., O’Lone, C., Fosu-Nyarko, J., Scobie, M., Cakir, M., Jones, M., Bellgard, M., Ksiazkiewicz, M., Wolko, B., Barker, S., Oliver, R., and Cowling, W. 2006. The first gene-based map of Lupinus angustifolius L.- Location of domestication genes and conserved synteny with Medicago truncatula. Theor. Appl. Genet. 113: 225–238.
- Nelson, R.L. 2009. Collection, conservation, and evaluation of soybean germplasm. Proceedings of the World Soybean Research Conference VIII, Beijing, China.
- O’Rourke, J.A., Yang, S.S., Miller, S.S., Bucciarelli, B., Liu, J., Rydeen, A., Bozsoki, Z., Uhde-Stone, C., Tu, Z.J., Allan, D., Gronwald, J.W., and Vance, C.P. 2013. An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 161: 705–724.
- Oakley, R.A. and Garver, S. 1917. Medicago falcata, a yellow flowered alfalfa. U.S. Dept. of Agric. Bull. 428. Washington, DC.
- Ocampo, B., Robertson, L.D., and Singh, K.B. 1998. Variation in seed protein content in the annual wild Cicer species. J. Sci. Food Agric. 78: 220–224.
- Ochatt, S.J., Benabdelmouna, A., Marget, P., Aubert, G., Moussy, F., Pontécaille, C., and Jacas, L. 2004. Overcoming hybridization barriers between pea and some of its wild relatives. Euphytica 137: 353–359.
- Oliveira, M.F., Nelson, R.L., Geraldi, I.O., Cruz, C.D., and de Toledo, J.F. F. 2010. Establishing a soybean germplasm core collection. Field Crops Res. 119: 277–289.
- Oomah, B.D., Luc, G., Leprelle, C., Drover, J.C. G., Harrison, J.E. and Olson, M. 2011. Phenolics, phytic acid, and phytase in canadian-grown low-tannin faba bean (Vicia faba L.) genotypes. J. Agric. Food Chem. 59: 3763–3771.
- Oskoueiyan, R., Kazempour, O.S., Maassoumi, A.A., Nejadsattari, T., and Mozaffarian, V. 2010. Phylogenetic status of Vavilovia formosa (Fabaceae-Fabeae) based on nrDNA ITS and cpDNA sequences. Biochem. Syst. Ecol. 38: 313–319.
- Ozturk, M. 2011. Revision of the Genus Cicer L. in Turkey via morphological, palynological, cytotaxonomical, molecular phylogenetic methods and analyses of seed protein and element contents. PhD Thesis, . The Graduate School of Natural and Applied Science of Selcuk Universıty, Konya.
- Ozturk, M., Duran, A., and Hakki, E.E. 2013. Cladistic and phylogenetic analyses of the genus Cicer in Turkey. Plant Syst. Evol. Doi: 10.1007/s00606-013-0850-6
- Ozturk, M., Duran, A., and Hakki, E.E. 2011. Cicer floribundum var. amanicola (Fabaceae), a new variety from south Anatolia, Turkey. Biological Diversity and Conservation 4/344–51.
- Padulosi, S. and Ng, N.Q. 1997. Origin, taxonomy and morphology of Vigna unguiculata (L.) Walp. In: Advances in Cowpea Research. pp. 1–12. Singh, B.B., Mohan Raj, D.R., Dashiell, K.E. and Jackai, L.E. N., Eds., Ibadan, Nigeria, IITA.
- Pandian, A., Ford, R., and Taylor, P.W. 2000. Transferability of sequence tagged microsatellite site (STMS) primers across four major pulses. Plant Mol. Biol. Rep. 18: 395–395.
- Pandiyan, M., Ramamoorthi, N., Ganesh, S.K., Jebaraj, S., Nagarajan, P., and Balasubramanian, P. 2008. Broadening the genetic base and introgression of MYMV resistance and yield improvement through unexplored genes from wild relative in mungbean. Plant Mut. Rep. 2:33–38.
- Pasternak, R. 1998. Investigations of botanical remains from Nevali Cori PPNB, Turkey: short interim report. In: Origins of Agriculture and Crop Domestication. pp. 170–177. Damania, A.B., Valkoun, J., Wilcox, G., and Qualset, C.O., Eds., ICARDA, Aleppo.
- Pearce, S.R., Knox, M., Ellis, T.H., Flavell, A.J., and Kumar, A. 2000. Pea Ty1-copia group retrotransposons: Transpositional activity and use as markers to study genetic diversity in Pisum. Mol. Gen. Genetics 263: 898–907.
- Percy, D.M. and Cronk, Q.C. B. 2002. Different fates of island brooms: contrasting evolution in Adenocarpus, Genista, and Teline (Genisteae, Fabaceae) in the Canary Islands and Madeira. Amer. J. Bot. 89: 854–864.
- Pérez-de-Luque, A., Eizenberg, H., Grenz, J.H., Sillero, J.C., Ávila, C.M., Sauerborn, J., and Rubiales, D. 2010. Broomrape management in faba bean. Field Crops Res. 115: 319–328.
- Phan, H.T. T., Ellwood, S.R., Adhikari, K., Nelson, M.N., and Oliver, R.P. 2007a. The first genetic and comparative map of white lupin (Lupinus albus L.): Identification of QTLs for anthracnose resistance and flowering time, and a locus for alkaloid content. DNA Res. 14: 59–70.
- Phan, H.T., Ellwood, S.R., Hane, J.K., Ford, R., Materne, M., and Oliver, R.P. 2007b. Extensive macrosynteny between Medicago truncatula and Lens culinaris subsp. culinaris. Theor. Appl. Genet. 114: 549–558.
- Picard, J. 1976. Aperçu sur l’hérédité du caractère absence de tannins dans les graines de féverole (Vicia faba L.). Ann. Amelior. Plantes 26: 101–106.
- Pinkas, R., Zamir, D., and Ladizinsky, G. 1985. Allozyme divergence and evolution in the genus Lens. Plant Syst. Evol. 151: 131–140.
- Podder, R., Banniza, S., and Vandenberg, A. 2012. Screening of wild and cultivated lentil germplasm for resistance to stemphylium blight. Plant Genet. Resour. Charact. Util. 1: 1–10.
- Polhill, R.M. 1981. Papilionoideae. In: Advances in Legume Systematics. pp. 191–208. Part 1. Polhill, R.M. and Raven, P.H., Eds., Royal Botanic Gardens, Kew, UK.
- Porch, T.G., Beaver, J.S., Debouck, D.G., Jackson, S.A., Kelly, J.D., and Dempewolf, H. 2013. Use of wild relatives and closely related species to adapt common bean to climate change. Agronomy 3: 433–461.
- Porch, T.G., Blair, M.W., Lariguet, P., Galeano, C.H., Pankhurst, C., and Broughton, W. 2009. Mutagenesis of common bean genotype BAT93 for the generation of a mutant population for TILLING. J. Amer. Soc. Hort. Sci. 134: 348–355.
- Provvidenti, R. 1990. Inheritance of resistance to pea seedborne mosaic virus in Pisum sativum. J. Heredity 81: 143–145.
- Pundir, R.P. S., and Singh, R.B. 1985. Cytogenetics of F1 hybrids between Cajanus and Atylosia species and its phylogenetic implications. Theor. Appl. Genet. 71: 216–220.
- Punguluri, S.K., Janaiah, K., Govil J.N., Kumar, P.A., and Sharma, P.C. 2006. AFLP fingerprinting in pigeonpea (Cajanus cajan (L.)Millsp) and its wild relatives. Genet. Resour. Crop Evol. 53: 423–431.
- Putnam, D., Russelle, M., Orloff, S., Kuhn, J., Fitzhugh, L., Godfrey, G., Kiess, A., and Longy, R. 2001. Alfalfa, Wildlife, and the Environment: The Importance and Benefits of Alfalfa in the 21st Century. California Alfalfa & Forage Association.
- Qiu, L.J., Chang, R.Z., Singh, G., Shivakumar, B.G., Kumudini, S., Mishra, S.K., and Verma, V.D. 2010. The origin and history of soybean. Oxfordshire: CABI.
- Qiu, L.J., Chen, P.Y., Liu, Z.X., Li, Y.H., Guan, R.X., Wang, L.H., and Chang, R.Z. 2011. The worldwide utilization of the Chinese soybean germplasm collection. Plant Genet. Resour. Charact. Util. 9: 109–122.
- Qiu, L.J., Xing, L.L., Guo, Y., Wang, J., Jackson, S.A., and Chang, R.Z. 2013. A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol. Biol. 83: 41–50.
- Rajora, O.P. and Mahon, J.D. 1994. Inheritance of mitochondrial DNA in lentil (Lens culinaris Medik.). Theor. Appl. Genet. 89: 206–210.
- Raman, R., Luckett, D.J., and Raman, H. 2008. Estimation of genetic diversity in albus lupin (Lupinus albus L.) using DArT and genic markers. In: Proceedings of the 12th International Lupin Conference, ‘Lupins for Health and Wealth.’ pp. 236–241. Palta, J.A. and Berger, J.D., Eds., International Lupin Association, Fremantle, Western Australia.
- Ramírez-Villegas, J., Khoury, C., Jarvis, A., Debouck, D.G., and Guarino, L. 2010. A gap analysis methodology for collecting crop gene pools: A case study with Phaseolus beans. PLoS ONE 5: e13497.
- Ratnaparkhe, M.B., Gupta, V.S., Venmurthy, M.R., and Ranjekar, P.K. 1995. Genetic fingerprinting of pigeonpea (Cajanus cajan (L.) Millsp) and its wild relatives using RAPD markers. Theor. Appl. Genet. 91: 893–898.
- Ravagnani, A., Abberton, M.T., and Skøt, L. 2012. Development of genomic resources in the species of Trifolium L. and its application in forage legume breeding. Agronomy 2: 116–131.
- Raven, P.H. and Polhill, R.M. 1981. Biogeography of the Leguminosae. In: Advances in Legume Systematics. pp. 27–34. Part 1., Polhill, R.M. and Raven, P.H., Eds., Royal Botanic Gardens, Kew, UK.
- Redden, R.J., Furman, B., and Coyne, C. 2007. Lens Biodiversity. In: Lentil, an Ancient Crop for Modern Times. Ch 2: pp. 11–22. Yadav, S.S., McNeil, D., and Stevenson, P.C., Eds., Springer, Dordrecht, Netherlands.
- Redden, R.J., Furman, B.J., Upadhyaya, H.D., Pundir, R.P. S., Gowda, C.L. L., Coyne, C.J., and Enneking, D. 2007. Biodiversity management in chickpea. In: Chickpea Breeding & Management. pp. 355–368. Yadav, S.S., Redden, R.J., Chen, W., and Sharma, B., Eds., CABI Press Oxfordshire, UK.
- Redden, B., Leonforte, T., Ford, R., Croser, J., and Slattery, J. 2005. Pea (Pisum sativum L.). In: Genetic Resources, Chromosome Engineering, and Crop Improvement: Series II - Grain Legumes. Chapter 3, pp. 49–83. Singh, R., Ed., Department of Crop Sciences, University of Illinois, USA.
- Redden, R.J., Basford, K.E., Kroonenberg, P.M., Islam, F.M., Ellis, R., Wang, S.M., Cao, Y.S., Zong, X.X., and Wang, X.M. 2009. Variation in adzuki bean (Vigna angularis) germplasm grown in China. Crop Sci. 49: 771–782.
- Redden, R. and Berger, J.D. 2007. History and origin of chickpea. In: Chickpea Breeding and Management. pp. 1–13. Yadav, S.S., Redden, R., Chen, W., and Sharma, B., Eds., CABI, Wallingford, UK.
- Reddy, L.J., Upadhyaya, H.D., Gowda, C.L. L., and Singh, S. 2005. Development of core collection in pigeonpea [Cajanus cajan (L.) Millsp.] using geographical and qualitative morphological descriptors. Genet. Res. Crop Evol. 52: 1049–1056.
- Reddy, M.R. K., Rathour, R., Kumar, N., Katoch, P., and Sharma, T.R. 2010. Cross‐genera legume SSR markers for analysis of genetic diversity in Lens species. Plant Breeding 129: 514–518.
- Rehman, A.U., Shunmugam, A., Arganosa, G., Bett, K.E., and Warkentin, T.D. 2012. Inheritance of the low-phytate trait in pea. Crop Sci. 52: 1171–1175.
- Reich, J. 2012. Alfalfa's role in feeding a hungry world. Proceedings California Alfalfa & Grains Symposium December 10-12, 2012 Sacramento, CA.
- Renfrew, J.M. 1969. The archaeological evidence for the domestication of plants: methods and problems. In: The Domestication and Exploitation of Plants and Animals. pp. 149–172. Ucko, P.J., and Dimbleby, G.W., Eds., Duckworth, London.
- Riday, H. and Brummer, E.C. 2002b. Forage yield heterosis in alfalfa. Crop Sci. 42: 716–723.
- Riday, H. and Brummer, E.C. 2005. Crop breeding, genetics and cytology: Heterosis in a broad range of alfalfa germplasm. Crop Sci. 45: 8–17.
- Riday, H., Brummer, E.C., and Moore, K.J. 2002a. Heterosis of forage quality in alfalfa. Crop Sci. 42: 1088–1093.
- Riehl, S., Zeidi, M., and Conard, N.J. 2013. Emergence of agriculture in the foothills of the Zagros Mountains of Iran. Science 341: 65–67.
- Robertson, L.D., Singh, K.B., and Ocampo, B. 1995. A Catalog of Annual Cicer Species, ICARDA, Aleppo, Syria.
- Robertson, L.D., Singh, K.B., Erskine, W., and El Moneim, A.M. A. 1996. Useful genetic diversity in germplasm collections of food and forage legumes from West Asia and North Africa. Genet. Res. Crop Evol. 43: 447–460.
- Robertson, M. 2006. Lucerne Prospects: Drivers for Widespread Adaptation of Lucerne for Profit and Salinity Management. CRC for Plant-Based Management of dryland salinity, Perth, W. Australia. 64 pp.
- Robins, J.G., Bauchan, G.R., and Brummer, E.C. 2007b. Genetic mapping forage yield, plant height, and regrowth at multiple harvests in tetraploid Alfalfa (Medicago sativa L.). Crop Sci. 47: 11–18.
- Robins, J.G., Hansen, J.L., Viands, D.R., and Brummer, E.C. 2008. Genetic mapping of persistence in tetraploid alfalfa. Crop Sci. 48: 1780–1786.
- Robins, J.G., Luth, D., Campbell, T.A., Bauchan, G.R., He, C., Viands, D.R., Hansen, J.L., and Brummer, E.C. 2007a. Genetic mapping of biomass production in tetraploid alfalfa. Crop Sci. 47: 1–10.
- Roskov, J.R. 1989. Trends in the evolution and the main taxonomic subdivisions in the group Trifolium s. l. (Fabaceae). Bot. Ž. 74: 36–43. [Russian with English summary]
- Rubeena, Taylor, P.W. J., Andes, P.K. and Ford, R. 2006. QTL mapping of resistance in lentil (Lens culinaris subsp. culinaris) to ascochyta blight (Ascochyta lentis). Plant Breeding 125: 506–512.
- Rubiales, D., Alcantara, C. and Sillero, J.C. 2004. Variation in resistance to Orobanche crenata in species of Cicer. Weed Res. 44: 27–32.
- Rubiales, D., Fernández-Aparicio, M., Pérez-de-Luque, A., Prats, E., Castillejo, M.A., Sillero, J., Rispail, N., Fondevilla, S. 2009. Breeding approaches for crenate broomrape (Orobanche crenata Forsk.) management in pea (Pisum sativum L.). Pest. Manag. Sci. 65: 553–559.
- Rubiales, D., Moreno, M.T., and Sillero, J.C. 2005. Search for resistance to crenate broomrape (Orobanche crenata) in pea germplasm. Genet. Res. Crop Evol. 52: 853–861.
- Rubiales, D., Pérez-de-Luque, A., Fernández-Aparicio, M., Sillero, J.C., Román, B., Kharrat, M., Khalil, S., Joel, D.M., and Riches, Ch. 2006. Screening techniques and sources of resistance against parasitic weeds in grain legumes. Euphytica 147: 187–199.
- Rubiales, D., Rojas-Molina, M.M., and Sillero, J.C. 2013. Identification of pre and posthaustorial resistance to rust (Uromyces viciae-fabae) in lentil (Lens culinaris) germplasm. Plant Breeding 132: 676–680.
- Russelle, M.P., Lamb, J.F. S., Turyk, N.B., Shaw, B.H., and Pearson, B. 2007. Managing nitrogen contaminated soils: Benefits of N2-fixing alfalfa. Agronomy J. 99: 738–746.
- Sadiki, M., Duc, G., and Furman, B. 2006. Genetic resources of faba bean worldwide. Grain Legumes 48: 18–19.
- Sakiroglu, M. and Brummer, E.C. 2011. Clarifying the ploidy of some accessions in the USDA alfalfa germplasm collection. Turk. J. Botany 35: 509–519.
- Sakiroglu, M. and Brummer, E.C. 2013. Presence of phylogeographic structure among wild diploid alfalfa accessions (Medicago sativa L. subsp. microcarpa Urb.) with evidence of the center of origin. Genet. Res. Crop Evol. 60: 23–31.
- Sakiroglu, M. and Kaya, M.M. 2012. Estimating genome size and confirming ploidy levels of wild tetraploid alfalfa accessions (Medicago sativa subsp. varia) using flow cytometry. Turk. J. Field Crops 17: 151–156.
- Sakiroglu, M., Doyle, J.J., and Brummer, E.C. 2010. Inferring population structure and genetic diversity of broad range of wild diploid alfalfa (Medicago sativa L.) accessions using SSR markers. Theor. Appl. Genet. 121: 403–415.
- Sangiri, C., Kaga, A., Tomooka, N., Vaughan, D., and Srinives, P. 2007. Genetic diversty of the mungbean (Vigna radiata, Leguminosae) gene pool on the basis of microsatellite analysis. Austr. J. Bot. 55: 837–847.
- Sanz, A.M., Gonzales, S.G., Syed, N.H., Suso, M.J., Caminero, C., and Flavell, A.J. 2007. Genetic diversity analysis in Vicia faba species using retrotransposon-based SSAP markers. Mol. Genet. Genomics 278: 433–441.
- Saraswat, K.S. 1980. The ancient remains of the crop plants at Atranjikera. J. Ind. Bot. Soc. 59: 306–319.
- Sardana, S., Mahajan, R.K., Gautam, N.K., and Ram, B. 2007. Genetic variability in pea (Pisum sativum L.) germplasm for utilization. Sabrao J. Breeding and Genet. 39: 31–41.
- Sari, D., Mutlu, N., and Toker, C. 2013. Resistance gene analog polymorphism in Lens species. Curr. Opin. Biotech. 24: 127–S128.
- Sarıkamış, G., Yanmaz, R., Ermiş, S., Bakir M., and Yüksel, C. 2010. Genetic characterization of pea (Pisum sativum) germplasm from Turkey using morphological and SSR markers. Genet. Mol. Res. 9: 591–600.
- Sarker A. and Erskine, W. 2006. Recent progress in ancient lentil. J. Agri. Sci. 144: 19–29.
- Sarker, A., El Moneim, A.A., and Maxted, N. 2001. Grasspea and chicklings (Lathyrus L.). In: Plant Genetic Resources of Legumes in the Mediterranean. pp. 159–180. Maxted, N. and Bennett, S.J. Eds., Springer, Dordrecht, Netherlands.
- Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Kato, T., Nakao, M., Sasamoto, S., Watanabe, A., Ono, A., Kawashima, K., Fujishiro, T., Katoh, M., Kohara, M., Kishida, Y., Minami, C., Nakayama, S., Nakazaki, N., Shimizu, Y., Shinpo, S., Takahashi, C., Wada, T., Yamada, M., Ohmido, N., Hayashi, M., Fukui, K., Baba, T., Nakamichi, T., Mori, H., and Tabata, S. 2008. Genome structure of the legume, Lotus japonicus. DNA Res. 15: 227–239.
- Saxena, K.B. 2008. Genetic improvement of pigeonpea—a review. Trop. Plant Biol. 1: 159–178.
- Saxena, K.B., Sultana, R., Mallikarjuna, N., Saxena, R.K., Kumar, R.V., Sawargaonkar, S.L., and Varshney, R.K. 2010. Male‐sterility systems in pigeonpea and their role in enhancing yield. Plant Breeding 129: 125–134.
- Saxena, M.C., Abd El Moneim, A.M., and Raninam, M. 1993. Vetches (Vicia spp.) and chicklings (Lathyrus spp.) in the farming systems in West Asia and North Africa and improvement of these crops at ICARDA. In: Potential for Vicia and Lathyrus Species as New Grain and Fodder Legumes for Southern Australia. pp. 2–9, Garlinge,J. R. and Perry, M.W., Eds., CLIMA, Perth, Australia.
- Saxena, N.P., Krishnamurthy, L., and Johansen, C. 1994. Registration of a drought resistant chickpea germplasm. Crop Sci. 33: 1424.
- Sbabou, L., Brhada, F., Alami, I.T., and Maltouf, A.F. 2010. Genetic diversity of Moroccan Lupinus germplasm investigated using ISSR and AFLP markers. Inter. J. Agric. Biol. 12: 26–32.
- Schaefer, H., Hechenleitner, P., Santos-Guerra, A., de Sequeira, M.M., Pennington, R.T., Kenicer, G., and Carine, M.A. 2012. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol. Biol. 12: 250.
- Schmutz, J., Cannon, S.B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., Hyten, D.L., Song, Q., Thelen, J.J., Cheng, J., Xu, D., Hellsten, U., May, G.D., Yu, Y., Sakurai, T., Umezawa, T., Bhattacharyya, M.K., Sandhu, D., Valliyodan, B., Lindquist, E., Peto, M., Grant, D., Shu, S., Goodstein, D., Barry, K., Futrell-Griggs, M., Abernathy, B., Du, J., Tian, Z., Zhu, L., Gill, N., Joshi, T., Libault, M., Sethuraman, A., Zhang, X.C., Shinozaki, K., Nguyen, H.T., Wing, R.A., Cregan, P., Specht, J., Grimwood, J., Rokhsar, D., Stacey, G., Shoemaker, R.C., and Jackson, S.A. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183.
- Schwarzbach, E., Smýkal, P., Dostál, O., Jarkovská, M., and Valová, S. 2014. Gregor J. Mendel – genetics founding father. Czech. J. Genet. Plant Breed. 50: 43–51.
- Sekhon, H.S., Singh, G., and Ram, R. 2007. Lentil-based cropping systems, chapter 7, pp. 107–126. In: Yadav, S.S., McNeil, D., Stevenson, P.C. (eds.), Lentil, an ancient crop for modern times. Dordrecht, The Netherlands: Springer.
- Shaikh, R., Diederichsen, A., Harrington, M., Adam, J., Conner, R.L., and Buchwaldt, L. 2013. New sources of resistance to Colletotrichum truncatum race Ct0 and Ct1 in Lens culinaris Medikus subsp. culinaris obtained by single plant selection in germplasm accessions. Genet. Res. Crop Evol. 60: 193–201.
- Sharma, K.D., Chen, W., Muehlbauer, F.J. 2005. Genetics of chickpea resistance to five races of Fusarium wilt and a concise set of race differentials for Fusarium oxysporum f. sp. ciceris. Plant Dis. 89: 385–390.
- Sharma, P.N., Díaz, L.M., and Blair, M.W. 2013. Genetic diversity of Indian common beans elucidated with two germplasm collections and by morphological and microsatellite markers. Plant Genet. Resour. Charact. Util. 11: 121–130.
- Sharma, S.K., Dawson, I.K., and Waugh, R. 1995. Relationships among cultivated and wild lentils revealed by RAPD analysis. Theor. Appl. Genet. 91: 647–654.
- Sharma, S.K., Knox, M.R., and Ellis, T.H. 1996. AFLP analysis of the diversity and phylogeny of Lens and its comparison with RAPD analysis. Theor. Appl. Genet. 93: 751–758.
- Sharpe, A.G., Ramsay, L., Sanderson, L.A., Fedoruk, M.J., Clarke, W.E., Li, R., Kagale S., Vijayan P., Vandenberg A., and Bett, K.E. 2013. Ancient orphan crop joins modern era: gene-based SNP discovery and mapping in lentil. BMC Genomics 14: 192.
- Shehadeh, A., Amri, A., and Maxted, N. 2013. Ecogeographic survey and gap analysis of Lathyrus L. species. Genet. Res. Crop Evol. 60: 2101–2113.
- Shimamoto, Y., Fukushi, H., Abe, J., Kanazawa, A., Gai, J., Gao, Z., and Xu, D. 1998. RFLPs of chloroplast and mitochondrial DNA in wild soybean, Glycine soja, growing in China. Genet. Resour. Crop Evol. 45: 433–439.
- Shiran, B. and Mashayek, A.M. 2004. Evaluation of chloroplast DNA among Vicia faba L. germplasm using restriction-site analysis. Iran. J. Sci. Technol. Trans. A 28: 51–55.
- Shockley, F.W., Backus, E.A., Ellersieck, M.R., Johnson, D.W., and McCaslin, M. 2002. Glandular-haired alfalfa resistance to potato leafhopper (Homoptera: Cicadellidae) and hopperburn: development of resistance indices. J. Econ. Entomol. 95: 437–447.
- Sholihin, and Hautea, D.M. 2002. Molecular mapping of drought resistance in mungbean (Vigna radiata): 1. Linkage map in mungbean using AFLP markers. J. Biotek. Pertanian. 7:17–24.
- Si, P., Sweetingham, M.W., Buirchell, B.J., Bowran, D., and Piper, T. 2006. Genotypic variation in metribuzin tolerance in narrow-leafed lupin (Lupinus angustifolius L.). Aus. J. Exp. Agric. 46: 85–91.
- Siddique, K.H. M., Regan, K.L., Tennant, D., and Thomson, B.D. 2001. Water use and water use efficiency of cool season grain legumes in low rainfall Mediterranean-type environments. Eur. J. Agronomy 15: 267–280.
- Silim, S.N., and Saxena, M.C. 1993a. Adaptation of spring-sown chickpea to the Mediterranean basin. 1. Response to moisture supply. Field Crops Res. 34: 121–136.
- Silim, S.N., and Saxena, M.C. 1993b. Adaptation of spring-sown chickpea to the Mediterranean basin. 2. Factors influencing yield under drought. Field Crops Res. 34: 137–146.
- Sillero, J.C., Fondevilla, S., Davidson, J., Vaz Patto, M.C., Warkentin, T.D., Thomas, J., and Rubiales, D. 2006. Screening techniques and sources of resistance to rusts and mildews in grain legumes. Euphytica 147: 255–272.
- Sillero, J.C., Moreno-Alías, I., and Rubiales, D. 2012. Identification and characterization of resistance to rust (Uromyces ciceris-arietini (Grognot) Jacz. & Boyd) in a germplasm collection of Cicer spp. Euphytica 188: 229–238.
- Sillero, J.C., Villegas-Fernández, A.M., Thomas, J., Rojas-Molina, M.M., Emeran, A.A., Fernández-Aparicio, M., and Rubiales, D. 2010. Faba bean breeding for disease resistance. Field Crops Res. 115: 297–307.
- Simon, C.J. and Hannan, R.M. 1995. Development and use of core subsets of cool-season food legume germplasm collections. Hort. Science 30: 907–907.
- Simpson, B. and Ogorzaly, M. 2001. Economic botany: plants in our world. New York: McGraw-Hill, Inc.
- Singh, A.K., Mishra, A., and Shukla, A. 2009. Genetic assessment of traits and genetic relationship in blackgram (Vigna mungo) revealed by isoenzymes. Biochem. Genet. 47: 471–485.
- Singh, K.B. 1990. Winter chickpea: problems and potential in the Mediterranean region. In: Options Mediterraneennes. pp. 25–34. Serie, A., Seminaires Mediterraneens, Zaragoza, Spain.
- Singh, K.B. and Ocampo, B. 1993. Interspecific hybridization in annual Cicer species. J. Genet. Breed. 47: 199–204.
- Singh, K.B., Malhotra, R.S. and Saxena, M.C. 1990. Sources for tolerance to cold in Cicer species. Crop Sci. 30: 1136–1138.
- Singh, K.B., Malhotra, R.S., Saxena, M.C., and Bejiga, G. 1997. Superiority of winter sowing over traditional spring sowing of chickpea in the Mediterranean region. Agronomy J. 89: 112–118.
- Singh, K.B., Ocampo, B. and Robertson, L.D. 1998. Diversity for abiotic and biotic stress resistance in the wild annual Cicer species. Genet. Resour. Crop Evol. 45: 9–17.
- Singh, M., Bisht, I.S., Sardana, S., Gautam, N.K., Husain, Z., and Gupta, S. 2006. Asiatic Vigna. In: Plant Genetic Resources: Foodgrain Crops. pp. 275–301. Dhillon, B.S., Saxena, S., Agrawal, A., and Tyagi, R.K., Eds., New Delhi, Narosa Publishing House Pvt. Ltd.
- Singh, S.P. and Schwartz, H.F. 2010. Breeding Common Bean for Resistance to Diseases: A Review. Crop Sci. 50: 2199–2223.
- Singh, S.P. and Schwartz, H.F. 2011. Review: Breeding common bean for resistance to insect pests and nematodes. Can. J. Plant Sci. 91: 239–250.
- Singh, S.R., Jackai, L.E. N., Thottappilly, G., Cardwell, K.F., and Myers, G.O. 1992. Status of research on constraints to cowpea production. In: Biotechnology: Enhancing Research on Tropical Crops in Africa. pp. 21–26. Thottappilly, G., Monti, L.M., Mohan Raj, D.T., and Moore, A.W., Eds., Ibadan, Nigeria: IITA. CTA/IITA copublication.
- Singh, S., Gumber, R.K., Joshi, N., and Singh, K. 2005. Introgression from wild Cicer reticulatum to cultivated chickpea for productivity and disease resistance. Plant Breeding 124: 477–480.
- Small, E. 1986. Taxonomy of glandular wild alfalfa (Medicago sativa L.). Can. J. Bot. 64: 2125–2129.
- Small, E. 1989. The evolution of genera in the Leguminosae. In: Advances in Legume Biology. 34: 467–486. Stirton, C.H. and Zarucchi, J.L., Eds., Monographs in Systematic Botany, from the Missouri Botanical Garden.
- Small, E. 2011. Alfalfa and Relatives: Evolution and Classification of Medicago. NRC Research Press, Ottawa, Ontario.
- Small, E. and Brookes, B. 1986. Glandular trichomes on cotyledonary petioles of Leguminosae tribe Trifolieae. Can. J. Pl. Sci. 66: 1019–1023.
- Small, E. and Jomphe, M. 1989. A synopsis of the genus Medicago (Leguminosae). Can. J. Bot. 67: 3260–3294.
- Smartt J. 1990. Grain Legumes: Evolution and Genetic Resources. Cambridge University Press
- Smithson, J.B., Redden, and R.M. Rawal, K.M. 1980. Methods of crop improvement and genetic resources in Vigna unguicalata, 445–457. In: Summerfield, R.F., and Bunting, A.H. (eds.), Advances in Legume Science. Kew, UK: Royal Botanic Gardens.
- Smýkal, P. 2014. Pea (Pisum sativum L.) in biology prior and after Mendel's discovery. Czech J. Genet. Plant Breed. 50: 52–64.
- Smýkal, P., Hýbl, M., Corander, J., Jarkovský, J., Flavell, A., and Griga, M. 2008a. Genetic diversity and population structure of pea (Pisum sativum L.) cultivars derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor. Appl. Genet. 117: 413–424.
- Smýkal, P., Coyne, C.J., Ford, R., Redden, R., Flavell, A.J., Hybl, M., Warkentin, T., Burstin, J., Duc, G., Ambrose, M., and Ellis, T.H. N. 2008b. Effort towards a world pea (Pisum sativum L.) germplasm core collection: The case for common markers and data compatibility. Pisum Genet. 40: 11–14.
- Smýkal, P., and Kosterin, O. 2010. Towards introgression library carrying wild pea (Pisum fulvum) segments in cultivated pea (Pisum sativum) genome background. In: Book of Abstracts of Vth International Congress on Legume Genetics and Genomics. pp. 123. Asilomar, USA.
- Smýkal, P., Aubert, G., Burstin, J., Coyne, C., Ellis, N., Flavell, A., Ford, R., Hýbl, M., Macas, J., Neumann, P., McPhee, K., Redden, R., Rubiales, D., Weller, J., and Warkentin, T.D. 2012. Pea (Pisum sativum L.) in the genomic era. Agronomy 2: 74–115.
- Smýkal, P., Coyne, C., Redden, R., and Maxted, N. 2013. Peas. In: Genetic and Genomic Resources of Grain Legume Improvement. Singh, M. and Upadhyaya, H., Eds. Elsevier, Netherlands.
- Smýkal, P. and Konečná E. 2014. Advances in pea genomics. In: Legumes in the Omic Era. Chapter 15. Gupta, S., Nadarajan, N. and Gupta, D., Eds., Springer, Dordrecht, Netherlands.
- Smýkal, P., Kenicer, G., Flavell, A.J., Kosterin, O., Redden, R.J., Ford, R., Zong, X., Coyne, C.J., Maxted, N., Ambrose, M.J., and Ellis, T.H. N. 2011. Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genetic Resour. Charact.Util. 9: 4–18.
- Snook, L.K., Dulloo, M.E., Jarvis, A., Scheldeman, X., and Kneller, M. 2011. Crop germplasm diversity: The Role of gene bank collections in facilitating adaptation to climate change. In: Crop Adaptation to Climate Change. Ch 25, pp. 495–506. Yadav, S.S., Redden, R.J., Hatfield, J.L., Lotze-Campen, H., and Hall, A.E., Eds. John Wiley and Sons, Blackwell Publishing Ltd.
- Sobolev, N.A. and Bugrii, V.P. 1970. A spontaneous intergeneric hybrid in the vetch tribe and its characteristics. In: Otdalenaja gibridizatsiya rastenij. pp. 415–421. Moscow, USSR, Kolos. [In Russian]
- Sobolev, N.A., Agarkova, S.N., and Adamchuk, G.K. 1971a. Overcoming cross incompatibility between pea and broad beans. 1. The effect of X irradiation on incompatibility. Nauchnye trudy zernobobovych kultur. 3: 136–146. [In Russian]
- Sobolev, N.A., Agarkova, S.N., and Adamchuk, G.K. 197lb. Overcoming incompatibility between pea and broad bean. II. Characteristics of progeny raised from seeds in pea-bean crosses. Nauchnye trudy zernobobovych kultur. 3: 147–156. (In Russian)
- Somta, P., Sommanas, W., and Srinives, P. 2009. Molecular diversity assessment of AVRDC-the world vegetable centre elite-parental mungbeans. Breeding Sci. 59: 149–157.
- Sonnante, G., and Pignone, D. 2001. Assessment of genetic variation in a collection of lentil using molecular tools. Euphytica 120: 301–307.
- Sonnante, G., Galasso, I., and Pignone, D. 2003. ITS sequence analysis and phylogenetic inference in the genus Lens mill. Ann. Botany 91: 49–54.
- Sprent, J.I. 2001. Nodulation in Legumes. Royal Botanic Gardens, Kew, UK.
- Srivastava, S.P., Bhandari, T.M. S., Yadav, C.R., Joshi, M., and Erskine, W. 2000. Boron deficiency in lentil: yield loss and geographic distribution in a germplasm collection. Plant Soil 219: 147–151.
- Steele, K.P. and Wojciechowski, M.F. 2003. Phylogenetic systematics of tribes Trifolieae and Vicieae (Fabaceae). In: Advances in Legume Systematics, part 10, pp. 355–370, Klitgaard, B. and Bruneau, A., Eds., Royal Botanic Garden, Kew, UK.
- Steele, K.P., Ickert-Bond, S.M., Zarre, S., and Wojciechowski, M.F. 2010. Phylogeny and character evolution in Medicago (Leguminosae): Evidence from analyses of plastid trnK/matK and nuclear GA3ox1 sequences Am. J. Bot. 97: 1142–1155.
- Stefanova, K.T., and Buirchell, B. 2010. Multiplicative mixed models for genetic gain assessment in lupin breeding. Crop Sci. 50: 880–891.
- Steiner, J.J., Robinson, W.A, Liston, A., and Taylor, N.L. 1997. ITS and RAPD phylogenetic hypotheses and the ecological distributions of North American Trifolium L. (Fabaceae). Amer. J. Bot. 84 (Suppl.): 235–236.
- Stoddard, F.L., Balko, C., Erskine, W., Khan, H.R., Link, W., and Sarker, A. 2006. Screening techniques and sources of resistance to abiotic stresses in cool-season food legumes. Euphytica 147: 167–186.
- Stolton, S., Maxted, N., Ford-Lloyd, B., Kell, S.P., and Dudley, N. 2006. Food Stores: Using Protected Areas to Secure Crop Genetic Diversity. WWF Arguments for protection series. Gland, Switzerland.
- Strzyzewska, C. 1995. Amphidiploid hybrids of Trifolium pratense L. (2n equals 14 + 2) with T. diffusum Ehrh. (2n equals 16). J. Appl. Genet. 36: 147–154.
- Stupar, R.M. 2010. Into the wild: The soybean genome meets its undomesticated relative. Proc. Natl. Acad. Sci. USA. 107: 21947–21948.
- Sultana, T. and Ghafoor, A. 2008. Genetic diversity in ex‐situ conserved Lens culinaris for botanical descriptors, biochemical and molecular markers and identification of landraces from indigenous genetic resources of Pakistan. J. Integr. Plant Biol. 50: 484–490.
- Sultana, T., Ghafoor, A., and Ashraf, M. 2006. Geographic patterns of diversity of cultivated lentil germplasm collected from Pakistan, as assessed by seed protein assays. Acta Biol. Cracoviensia, Series Botanica 48: 77–84.
- Sumner, D. and Rosen-Molina, J.T. 2011. Alfalfa in the context of global crop price prospects. Proceedings, 2011 Western Alfalfa & Forage Conference, Las Vegas, Nevada.
- Suso, M.J., Gilsanz, S., Duc, G., Marget, P., and Moreno, M.T. 2006. Germplasm management of faba bean (Vicia faba L.): Monitoring intercrossing between accessions with inter-plot barriers. Genet. Res. Crop Evol. 53: 1427–1439.
- Suso, M.J., Pierre J., Moreno, M.T., Esnault, R., and Le Guen, J. 2001. Variation in outcrossing levels in faba bean cultivars: role of ecological factors. J. Agric. Sci. Camb. 136: 399–405.
- Tahir, M., Båga, M., Vandenberg, A., and Chibbar, R.N. 2012. An assessment of raffinose family oligosaccharides and sucrose concentration in genus Lens. Crop Sci. 52: 1713–1720.
- Talhinhas, P., Leitão, J., and Neves-Martins, J. 2006. Collection of Lupinus angustifolius L. germplasm and characterisation of morphological and molecular diversity. Genet. Res. Crop Evol. 53: 563–578.
- Talhinhas, P., Neves-Martins, J., and Leitao, J. 2003. AFLP, ISSR and RAPD markers reveal high levels of genetic diversity among Lupinus spp. Plant Breeding 122: 507–510.
- Tan, A. 1998. Current status of plant genetic resources conservation in Turkey. In: The Proceedings of International Symposium on In Situ Conservation of Plant Diversity. pp. 5–16. Zencirci, N., Kaya, Z., Anikster, Y., and Adams, W.T., Eds., Central Research Institute for Field Crops, Ankara, Turkey.
- Tan, A. and Tan, A.S. 2002. In situ conservation of wild species related to crop plants: the case of Turkey. In: Managing Plant Genetic Diversity. pp. 195–204. Engels, J.M. M., Ramantha Rao, V., Brown, A.H. D., and Jackson, M.T., Eds., CAB International, Wallingford, UK.
- Tanksley, S.D. and McCouch, S.R. 1997. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063–1066.
- Tanno, K. and Willcox, G. 2006. The origins of cultivation of Cicer arietinum L. and Vicia faba L.: early finds from Tell el-Kerkh, north-west Syria, late 10th millennium B.P. Veget. Hist. Archaeobotany 15: 197–204.
- Tar’an, B., Zhang, C., Warkentin, T., Tullu, A., and Vandenberg, A. 2005. Genetic diversity among cultivars and wild species accessions of pea (Pisum sativum L.) based on molecular markers, and morphological and physiological characters. Genome 48: 257–272.
- Tateishi, Y. 1985. A revision of the azuki bean group, the subgenus Ceratotropis of the genus Vigna (Leguminoseae). Ph.D. Thesis. Japan: Tohoku University, p. 292.
- Tateishi, Y. 1996. Systematics of the species of Vigna subgenus Ceratotropis. In: Mungbean Germplasm: Collection, Evaluation and Utilisation for Breeding Programs. pp. 9–24. Srinives, P., Kitbamroong, C., and Miyazaki, S. Eds., Tsukuba, Japan: Japan International Research Center for Agricultural Sciences.
- Taylor, N.L. 1985. Clovers around the World. Clover Science and Technology, Agronomy Monograph 34: 1–6.
- Taylor, N.L. 2008. A century of clover breeding developments in the United States. Crop Sci. 48: 1–13.
- Taylor, N.L. and Smith, R.R. 1995. Red clover. In: An Introduction to Grassland Agriculture. pp. 217–226. Forages vol 34. Barnes, R.F., Miller, D.A., and Nelson, C.J., Eds. Ames, Iowa State University Press.
- Taylor, N.L., and Quesenberry, K.H. 1996. Red Clover Science. Kluwer Academic, The Netherlands.
- Taylor, N.L., Quarles, R.F., and Anderson, M.K. 1980. Methods of overcoming interspecific barriers in Trifolium. Euphytica 33: 431–441.
- Terzopoulos, P.J. and Bebeli, P.J. 2008. Genetic diversity of Mediterranean faba bean (Vicia faba L.) swith ISSR markers. Field Crops Res. 108: 39–44.
- Tewari, K., Dikshit, H.K., Jain, N., Kumari, J., and Singh, D. 2012. Genetic differentiation of wild and cultivated Lens based on molecular markers. J. Plant Biochem. Biotech. 21: 198–204.
- Thavarajah, D., Thavarajah, P., Sarker, A., and Vandenberg, A. 2009. Lentils (Lens culinaris Medikus subspecies culinaris): a whole food for increased iron and zinc intake. J. Agric. Food Chem. 57: 5413–5419.
- The Legume Phylogeny Working Group. 2013. Legume phylogeny and classification in the 21st century: Progress, prospects and lessons for other species-rich clades. Taxon 62: 217–248.
- Thompson, J.P., Reen, T.G., Clewett, T.G., Sheedy, J.G., Kelly, A.M., Gogel, B.J., and Knights, E.J. 2012. Hybridisation of Australian chickpea cultivars with wild Cicer spp. increases resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus). Aust. Plant Pathol. 40: 601–611.
- Tivoli, B., Baranger, A., Avila, C.M., Banniza, S., Barbetti, M., Chen, W., Davidson, J., Lindeck, K., Kharrat, M., Rubiales, D., Sadiki, M., Sillero, J.C., Sweetingham, M., and Muehlbauer, F.J. 2006. Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 147: 223–253.
- Toker, C. 2005. Preliminary screening and selection for cold tolerance in annual wild Cicer species. Genet. Resour.Crop Evol. 52: 1–5.
- Toker, C. 2009. A note on the evolution of kabuli chickpeas as shown by induced mutations in Cicer reticulatum Ladizinsky. Genet. Res. Crop Evol. 56: 7–12.
- Toker, C., Canci, H., and Yildirim, T. 2007a. Evaluation of perennial wild Cicer species for drought resistance. Genet. Resour. Crop Evol. 54: 1781–1786.
- Toker, C., Canci, H., Inci, N.E., and Ceylan, F.O. 2012. Improvement in imidazolinone resistance in Cicer species by induced mutation. Plant Breeding 131: 535–539.
- Toker, C., Lluch, C., Tejera, N.A., Serraj, R., and Siddique, K.H. M. 2007b. Abiotic Stresses. In: Chickpea Breeding and Management. pp. 474–496. Yadav, S.S., Redden, R., Chen, W., and Sharma, B., Eds., Wallingford: CAB International Publication, UK.
- Toklu, F., Karaköy, T., Haklı, E., Bicer, T., Brandolini, A., Kilian, B., and Özkan, H. 2009. Genetic variation among lentil (Lens culinaris Medik) landraces from Southeast Turkey. Plant Breeding 128: 178–186.
- Tomooka, N. 2009. The origins of rice bean (Vigna umbellata) and azuki bean (V. angularis): The evolution of two lesser-known Asian beans. In: An Illustrated Eco-history of the Mekong River Basin. Akimichi. T., Ed., White Lotus Publisher, Bangkok,Thailand.
- Tomooka, N.N., Senthi, Pandiyan, M., Ramamoorthi, N., Kaga, A., and Vaughan, D.A. 2008. Collection and conservation of leguminous crop and their wild relatives in Tamil Nadu. J. Agrobio. Sci. Jpn. 24:113–125.
- Tomooka, N., Kaga, A., and Vaughan, D.A. 2006. The Asian Vigna (Vigna subgenus Ceratotropis) Biodiversity and evolution. In: Plant Genome: Biodiversity and Evolution. Vol. 1, Part C Phan-erogams (Angiosperms- Dicotyledons). pp. 87–126. Sharma, A.K. and Sharma, A., Eds., Science Publishers, Enfield, New Jersey
- Tomooka, N., Pandiyan, M., Senthil, N., Ramamoorthi, N., Kaga, A., and Vaughan, D.A. 2009. Collection and conservation of leguminous crops and their wild relatives in Tamil Nadu, India. Annual Report on Exploration and Introduction of Plant Genetic Resources (NIAS, Tsukuba, Japan) 34: 83–109.
- Tomooka, N., Vaughan, D.A., and Moss, H. 2002. The Asian Vigna: Genus Vigna Subgenus Ceratotropis Genetic Resources. pp. 277. Kluwer Academic Publishers, The Netherlands.
- Tomooka, N., Vaughan, D.A., Kaga, A. 2005. Mungbean. In: Genetic Resources, Chromosome Engineering and Crop Improvement. II Grain Legumes. pp. 319–339. Singh, R.J. and Jauhar, P.P., Eds., CRC Press, Boca Rogue, Florida.
- Tomooka, N., Vaughan, D., Moss, H., and Maxted, N. 2003. The Asian Vigna: Vigna Subgenus Ceratotropis Genetic Resources. pp. 1–270. Kluwer, Dordrecht.
- Torres, A.M., C.M. Avila, N. Gutierrez, C. Palomino, M.T. Moreno, and J.I. Cubero, 2010: Marker-assisted selection in faba bean (Vicia faba L.). Field Crops Res. 115: 243–252.
- Torres, A.M., Roman, B., Avila, C.M., Satovic, Z., Rubiales, D., Sillero, J.C., Cubero, J.I., and Moreno, M.T. 2006. Faba bean breeding for resistance against biotic stresses: towards application of marker technology. Euphytica 147: 67–80.
- Tullu, A., Banniza, S., Tar’an, B., Warkentin, T., and Vandenberg, A. 2010. Sources of resistance to ascochyta blight in wild species of lentil (Lens culinaris Medik.). Genet. Res. Crop Evol. 57: 1053–1063.
- Tullu, A., Buchwaldt, L., Lulsdorf, M., Banniza, S., Barlow, B., Slinkard, A.E., and Vandenberg, A. 2006. Sources of resistance to anthracnose (Colletotrichum truncatum) in wild Lens species. Genet. Res. Crop Evol. 53: 111–119.
- Tullu, A., Diederichsen, A., Suvorova, G., and Vandenberg, A. 2011. Genetic and genomic resources of lentil: Status, use and prospects. Plant Genet. Resour. 9: 19–29.
- Tullu, A., Kusmenoglu, I., McPhee, K.E., and Muehlbauer, F.J. 2001. Characterization of core collection of lentil germplasm for phenology, morphology, seed and straw yields. Genet. Res. Crop Evol. 48: 143–152.
- Turner, N.C., Abbo, S., Berger, J.D., Chaturvedi, S.K., French, R.J., Ludwig, C., Mannur, D.M., Singh, S.J., and Yadava, H.S. 2007. Osmotic adjustment in chickpea (Cicer arietinum L.) results in no yield benefit under terminal drought. J. Exp. Bot. 58: 187–194.
- Undal, V.S., Thakare, P.V., Chaudhari, U.S., Deshmukh, V.P., and Gawande, P.A. 2011. Estimation of genetic diversity among wild Vigna species revealed by RAPD markers. Ann. Bio. Res. 2: 348–354.
- Upadhyaya, H.D., Dwivedi, S.L., Ambrose, M., Ellis, N., Berger, J., Smýkal, P.D., Debouck, D., Duc, G., Dumet, D., Flavell, A., Sharma, S.K., Mallikarjuna. N., and Gowda, C.L. 2011. Legume genetic resources: management, diversity assessment, and utilization in crop improvement. Euphytica 180: 27–47.
- Vail, S., Strelioff, J.V., Tullu, A., and Vandenberg, A. 2012. Field evaluation of resistance to Colletotrichum truncatum in Lens culinaris, Lens ervoides, and Lens ervoides × Lens culinaris derivatives. Field Crops Res. 126: 145–151.
- Vaillancourt, R.E., Weeden, N.F., Bruneau, A., and Doyle, J.J. 1993. Chloroplast DNA phylogeny of Old World Vigna (Leguminosae). Syst. Bot. 18: 642–651.
- van der Maesen, L.J. G. 1987. Origin, history and taxonomy of chickpea. In: The Chickpea. pp 11–34. Saxena, M.C., Singh, K.B., Eds., Wallingford. CAB International Publication, UK.
- van der Maesen, L.J. G. 1990. Pigeonpea origin, history, evolution, and taxonomy. In: The Pigeonpea. pp. 15–46. Nene, Y.L., Hall, S.D., and Sheila, V.K., Eds., CAB International Publication, UK.
- van der Maesen, L.J. G., Maxted, N., Javadi, F., Coles, S., and Davies, A.M. R. 2007. Taxonomy of the genus Cicer revisited. In: Chickpea Breeding and Management. pp. 14–46. Yadav, S.S., Redden, R., Chen, W., and Sharma, B., Eds., CAB International Publication, Wallingford.
- van Zeist, W., and de Roller, G. 1991, 1992. The plant husbandry of aceramic Çayönü, SE Turkey. Palaeohistoria 33/34: 65–96.
- Varshney, R.K., Chen, W., Li, Y., Bharti, A.K., Saxena, R.K., Schlueter, J.A., Donoghue, M.T. A., Azam, S., Fan, G., Whaley, A.M., Farmer, A.D., Sheridan, J., Iwata, A., Tuteja, R., Penmetsa, R.V., Wu, W., Upadhyaya, H.D., Yang, S.-P., Shah, T., Saxena, K.B., Michael, T., McCombie, W.R., Yang, B., Zhang, G., Yang, H., Wang, J., Spillane, C., Cook, D.R., May, G.D., Xu, X., and Jackson, S.A. 2012. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotech. 30: 83–89.
- Varshney, R.K., Close, T.J., Singh, N.K., Hoisington, D.A., and Cook, D.R. 2009. Orphan legume crops enter the genomics era! Curr. Opin. Plant Biol. 12: 202–210.
- Varshney, R.K., Song, C., Saxena, R.K., Azam, S., Yu, S., Sharpe, A.G., Cannon, S., Baek, J., Rosen, B.D., Tar’an, B., Millan, T., Zhang, X., Ramsay, L.D., Iwata, A., Wang, Y., Nelson, W., Farmer, A.D., Gaur, P.M., Soderlund, C., Penmetsa, R.V., Xu, C., Bharti, A.K., He, W., Winter, P., Zhao, S., Hane, J.K., Carrasquilla-Garcia, N., Condie, J.A., Upadhyaya, H.D., Luo, M.-C., Thudi, M., Gowda, C.L. L., Singh, N.P., Lichtenzveig, J., Gali, K.K., Rubio, J., Nadarajan, N., Dolezel, J., Bansal, K.C., Xu, X., Edwards, D., Zhang, G., Kahl, G., Gil, J., Singh, K.B., Datta, S.K., Jackson, S.A., Wang, J., and Cook, D.R. 2013. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotech. 31: 240–246.
- Vavilov, N.I. 1950. The phytogeographic basis of plant breeding. In: The Origin, Variation, Immunity, and Breeding of Cultivated Plants. pp. 13–54. Chester, K.S. Trans., Waltham, MA, Chronica Botanica.
- Vavilov, N.I. 1951. The origin, variation, immunity and breeding of cultivated plants. Translated from the Russian by K. Starrchester. Chronica Botanica 13: 1–364.
- Vaz Patto, M.C., Skiba, B., Pang, E.C. K., Ochatt, S.J., Lambein, F., and Rubiales, D. 2006. Lathyrus improvement for resistance against biotic and abiotic stresses: from classical breeding to marker assisted selection. Euphytica 147: 133–147.
- Verdcourt, B. 1970. Studies in the Leguminosae - Papilionoideae for the ‘Flora of Tropical East Africa’ IV. Kew Bulletin 24: 507–569.
- Verma, M., Ravi, M., and Sandhu, J.S. 1995. Characterization of interspecific cross Cicer arietinum L. × C. judaicum (Bioss). Plant Breed. 114: 549–551.
- Verma, P., Shah, N., and Bhatia, S. 2013. Development of an expressed gene catalogue and molecular markers from the de novo assembly of short sequence reads of the lentil (Lens culinaris Medik.) transcriptome. Plant Biotech. J. 11: 894–905.
- Vershinin, A.V., Allnutt, T.R., Knox, M.R., Ambrose, M.J., and Ellis, N.T. H. 2003. Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Mol. Biol. Evol. 20: 2067–2075.
- Vijaykumar, A., Saini, A., and Jawali, N. 2010. Phylogenetic analysis of subgenus Vigna species using nuclear ribosomal RNA ITS: evidence of hybridization among Vigna unguiculata subspecies. J. Heredity 101: 177–188.
- Vincent, H., Wiersema, J., Kell, S.P., Dobbie, S., Fielder, H., Castañeda Alvarez, N.P., Guarino, L., Eastwood, R., León, B. and Maxted, N. 2013. A prioritised crop wild relative inventory as a first step to help underpin global food security. Biol. Conserv. 167: 265–275.
- Vymyslicky, T., Pelikan, J., Gottwaldova, P., and Nedelnik, J. 2010. The Czech core collection of Trifolium repens L. In: Sustainable Use of Genetic Diversity in Forage and Turf Breeding. pp. 167–172. Huyghe, C., Ed., Springer, Dordrecht, Netherlands.
- Vymyslicky, T., Smarda, P., Pelikan, J., Cholastova, T., Nedelnik, J., Moravcova, H., Pokorny, R., Soldanova, M., and Polakova, M. 2012. Evaluation of the Czech core collection of Trifolium pratense, including morphological, molecular and phytopathological data. African J. Biotech. 11: 3583–3595.
- Waines, J.G. 1975. The Biosystematics and Domestication of Peas (Pisum L.). Bulletin of the Torrey Botanical Club 102: 385–395.
- Wang, L.X., Guan, Y., Guan, R.X., Li, Y.H., Ma, Y.S., Dong, Z.M., Liu, X., Zhang, H.Y., Zhang, Y.Q., Liu, Z.X., Chang, R.Z., Xu, H.M., Li, L.H., Lin, F.Y., Luan, W.J., Ya, N.Z., Ning, X.C., Zhu, L., Cui, Y.H., Piao, R.H., Liu, Y., Chen, P.Y., and Qiu, L.J. 2006. Establishment of Chinese soybean (Glycine max) core collections with agronomic traits and SSR markers. Euphytica 151: 215–223.
- Wang, M.L., Barkley, N. A., Gillaspie, G. A., and Pederson, G. A. 2008. Phylogenetic relationships and genetic diversity of the USDA Vigna germplasm collection revealed by gene-derived markers and sequencing. Genet. Res. 90: 467–480.
- Watson, L.E., Sayed-Ahmed, H., and Badr, A. 2000. Molecular phylogeny of Old World Trifolium (Fabaceae), based on plastid and nuclear markers. Plant Syst. Evol. 224: 153–171.
- Weeden, N.F. 2007. Genetic changes accompanying the domestication of Pisum sativum: is there a common genetic basis to the ‘Domestication Syndrome’ for legumes? Ann. Bot. 100: 1017–1025.
- Weeden, N.F. and Wolko, B. 2001. Allozyme analysis of Pisum sativum spp. abyssinicum and the development of genotypic definition for this subspecies. Pisum Genet. 33: 21–25.
- Weller, J.L., Hecht, V., Liew, L.C., Sussmilch, F.C., Wenden, B., Knowles, C.L., and Vander Schoor, J.K. 2009. Update on the genetic control of flowering in garden pea. J. Exp. Bot. 60: 2493–2499.
- Weller, J.L., Liew, L.C., Hecht, V.F. G., Rajandran, V., Laurie, R.E., Ridge, S., Wenden, B., Vander Schoor, J.K., Jaminon, O., Blassiau, C., Dalmais, M., Rameau, C., Bendahmane, A., Macknight, R.C., and Lejeune-Hénaut, I. 2012. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 109: 21158–21163.
- Wen, Z., Ding, Y., Zhao, T., and Gai, J. 2009. Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor. Appl. Genet. 119: 371–381.
- Werker, E., Marbach, I., and Mayer, A.M. 1979. Relation between the anatomy of the testa, water permeability and the presence of phenolics in the genus Pisum. Ann. Bot. 43: 765–771.
- Westengen, O.T., Jeppson, S., and Guarino, L. 2013. Global ex-situ crop diversity conservation and the Svalbard Global Seed Vault: assessing the current status. PLoS ONE 8: e64146.
- Wexelsen, H. 1928. Chromosome numbers and morphology in Trifolium. University of California Publications in Agricultural Sciences 2: 355–376.
- Wiersema, J. and León, B. 2013. World Economic Plants: a Standard Reference, 2nd Edition. CRC Press, Boca Raton.
- Wilcox, J.R. 1983. Breeding Soybeans Resistant to Diseases. In: Plant Breeding Reviews, Volume 34 Janick, J. Ed., John Wiley & Sons, Inc., Hoboken, NJ.
- Williams, W.M., Easton, H.S., and Jones, C.S. 2007. Future options and targets for pasture plant breeding in New Zealand. New Zealand J. Agric. Res. 50: 223–248.
- Williams, W.M., Ellison, N.W., Ansari, H.A., Verry, I.M., and Hussain, S.W. 2012. Experimental evidence for the ancestry of allotetraploid Trifolium repens and creation of synthetic forms with value for plant breeding. BMC Plant Biol. 12: 55.
- Wilson, V.E. and A.G. 1972: Natural crossing in Lens esculenta Moench. J. Soc. Hortic. Sci. 97:142–143.
- Wink, M. and Mohamed, G.I. A. 2003. Evolution of chemical defense traits in the Leguminosae: mapping of distribution patterns of secondary metabolites on a molecular phylogeny inferred from nucleotide sequences of the rbcL gene. Biochem. Syst. Ecol. 31: 897–917.
- Wojciechowski, M.F. 2003. Reconstructing the phylogeny of legumes (Fabaceae): an early 21st century perspective. In: Advances in Legume Systematics. Part 10, Higher Level Systematics, B. pp. 5–35. Klitgaard, B. and Bruneau, A., Eds., Royal Botanic Garden, Kew.
- Wojciechowski, M.F., Lavin, M., and Sanderson, M.J. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Botany 91: 1846–1862.
- Wolko, B. and Weeden, N. 1989. Estimation of Lupinus genome polyploidy on the basis of isozymic loci number. Genetica Polonica 30: 165–171.
- Wolko, B., Clements, J.C., Naganowska, B., Nelson, M.N., and Yang, H. 2011. Lupinus. In: Wild Crop Relatives: Genomic and Breeding Resources. pp. 153–206. Kole, C., Ed., Springer, Heidelberg, Germany.
- Xu, B., Lu, Q., and Zhuang, B. 1987. Analysis of ecotypes of wild soybean (G. soja) in China. Sci. Agri. Sin. 20: 29–35.
- Xu, D., Abe, J., Gai, J., and Shimamoto, Y. 2002. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor. Appl. Genet. 105: 645–653.
- Xu, D., Gao, Z., Tian, Q., Gai, J., Fukushi, H., Kitajima, S., Abe, J., and Shimamoto, Y. 1999. Genetic diversity of the annual wild soybean (Glycine soja) in China. Chinese J. Appl. Environ. Bio. 5: 439–443.
- Xu, D.H. and Gai, J.Y. 2003. Genetic diversity of wild and cultivated soybeans growing in China revealed by RAPD analysis. Plant Breeding 122: 503–506.
- Xu, H.X., Jing, T., Tomooka, N., Kaga, A., Isemura, T., and Vaughan, D.A. 2008. Genetic diversity of the azuki bean (Vigna angularis (Willd.) Ohwi & Ohashi) gene pool as assessed by SSR markers. Genome 51: 728–738.
- Yadav, P.B. S. and Padmaja, V. 2002. Interspecific hybridization and evolutionary relationships of Cajanus cajan (L.) Millspaugh and four of the wild species. Cytologia 67: 67–73.
- Yaish, M.W., Saenz de Miera, L.E., and Perez De La Vega, M. 2004. Isolation of a family of resistance gene analogue sequences of the nucleotide binding site (NBS) type from Lens species. Genome 47: 650–659.
- Yaklich, R.W., Helm, R.M., Cockrell, G., and Herman, E.M. 1999. Analysis of the distribution of the major soybean seed allergens in a core collection of Glycine max accessions. Crop Sci. 39: 1444–1447.
- Yang, H., Tao, Y., Zheng, Z., Li, C., Sweetingham, M., and Howieson, J. 2012. Application of next-generation sequencing for rapid marker development in molecular plant breeding: a case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genomics 13: 318.
- Yang, H., Tao, Y., Zheng, Z., Shao, D., Li, Z., Sweetingham, M., Buirchell, B., and Li, C. 2013a. Rapid development of molecular markers by next-generation sequencing linked to a gene conferring phomopsis stem blight disease resistance for marker-assisted selection in lupin (Lupinus angustifolius L.) breeding. Theor. Appl. Genet. 126: 511–522.
- Yang, H., Tao, Y., Zheng, Z., Zhang, Q., Zhou, G., Sweetingham, M.W., Howieson, J.G., and Li, C. 2013b. Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius L. PLoS ONE 8: e64799.
- Yang, T., Bao, S.Y., Ford, R., Jia, T.J., Guan, J.P., He, Y.H., Sun, X.L., Jiang, J.Y., Hao, J.J., Zhang, X.Y., and Zong, X.X. 2012. Highthroughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genomics 13: 602.
- Yarnell, S.H. 1962. Cytogenetics of the vegetable crops III. Legumes. A. Garden peas, Pisum sativum L. Bot. Rev. 28: 465–573.
- Yoder, J.B., Briskine, R., Mudge, J., Farmer, A., Paape, T., Steele, K., Weiblen, G.D., Bharti, A.K., Zhou, P., May, G.D., Young, N.D., and Tiffin, P. 2013. Phylogenetic signal variation in the genomes of Medicago (Fabaceae) Syst. Biol. 62: 424–438.
- Yoon, M.S., Lee, J., Kim, C.Y., and Baek, H.J. 2007. Geneticrelationships among cultivated and wild Vigna angularis (Willd.) Ohwi & Ohashi and relatives fromKorea based on AFLP markers. Genet. Res. Crop Evol. 54: 875–883.
- Young, N.D., Debelle, F., Oldroyd, G.E., Geurts, R., Cannon, S.B., Udvardi, M.K., Benedito, V.A., Mayer, K.F., Gouzy, J., Schoof, H., Van de Peer, Y., Proost, S., Cook, D.R., Meyers, B.C., Spannagl, M., Cheung, F., De Mita, S., Krishnakumar, V., Gundlach, H., Zhou, S., Mudge, J., Bharti, A.K., Murray, J.D., Naoumkina, M.A., Rosen, B., Silverstein, K.A., Tang, H., Rombauts, S., Zhao, P.X., Zhou, P., Barbe, V., Bardou, P., Bechner, M., Bellec, A., Berger, A., Berges, H., Bidwell, S., Bisseling, T., Choisne, N., Couloux, A., Denny, R., Deshpande, S., Dai, X., Doyle, J.J., Dudez, A.M., Farmer, A.D., Fouteau, S., Franken, C., Gibelin, C., Gish, J., Goldstein, S., Gonzalez, A.J., Green, P.J., Hallab, A., Hartog, M., Hua, A., Humphray, S.J., Jeong, D.H., Jing, Y., Jocker, A., Kenton, S.M., Kim, D.J., Klee, K., Lai, H., Lang, C., Lin, S., Macmil, S.L., Magdelenat, G., Matthews, L., McCorrison, J., Monaghan, E.L., Mun, J.H., Najar, F.Z., Nicholson, C., Noirot, C., O’Bleness, M., Paule, C.R., Poulain, J., Prion, F., Qin, B., Qu, C., Retzel, E.F., Riddle, C., Sallet, E., Samain, S., Samson, N., Sanders, I., Saurat, O., Scarpelli, C., Schiex, T., Segurens, B., Severin, A.J., Sherrier, D.J., Shi, R., Sims, S., Singer, S.R., Sinharoy, S., Sterck, L., Viollet, A., Wang, B.B., Wang, K., Wang, M., Wang, X., Warfsmann, J., Weissenbach, J., White, D.D., White, J.D., Wiley, G.B., Wincker, P., Xing, Y., Yang, L., Yao, Z., Ying, F., Zhai, J., Zhou, L., Zuber, A., Denarie, J., Dixon, R.A., May, G.D., Schwartz, D.C., Rogers, J., Quetier, F., Town, C.D., and Roe, B.A. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524.
- Yunus, A.G. and Jackson, M.T. 1991. The gene pools of the grasspea (Lathyrus sativus L.). Plant Breeding 106: 319–328.
- Zaccardelli, M., Lupo, F., Piergiovanni, A.R., Laghetti, G., Sonnante, G., Daminati, M.G., Sparvoli, F., and Lioi, L. 2012. Characterization of Italian lentil (Lens culinaris Medik.) germplasm by agronomic traits, biochemical and molecular markers. Genet. Res. Crop Evol. 59: 727–738.
- Zamir, D. 2001. Improving plant breeding with exotic genetic libraries. Nature Rev. Genet. 2: 983–989.
- Zamir, D. and Ladizinsky, G. 1984. Genetics of allozyme variants and linkage groups in lentil. Euphytica 33: 329–336.
- Zavodna, M., Kraic, J., Paglia, G., Gregova, E., and Morgante, M. 2000. Differentiation between closely related lentil (Lens culinaris Medik.) cultivars using DNA markers. Seed Sci. Technol. 28: 217–219.
- Zeid, M., Schon, C.C., and Link, W. 2003. Genetic diversity in recent elite faba bean lines using AFLP markers. Theor. Appl. Genet. 107: 1304–1314.
- Zhang, H., Pala, M., Oweis, T., and Harris, H. 2000. Water use and water use efficiency of chickpea and lentil in a Mediterranean environment. Aus. J. Agric. Res. 51: 295–304.
- Zhang, X., Blair, M.W., and Wang, S. 2008. Genetic diversity of Chinese Common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeat (SSR) markers. Theor. Appl. Genet. 117: 629–640.
- Zhao, L., Dong, Y., Liu, B., Hao, S., Wang, K., and Li, X. 2005. Establishment of a core collection for the Chinese annual wild soybean (Glycine soja). Chin. Sci. Bul. 50: 989–996.
- Zhao, Q.S., Nian, H., and Yang, C.Y. 2009. Genetic diversity of natural wild soybean populations in Xintian County, Hunan province. Acta Bot. Boreali-Occident. Sinica 29: 2221–2227.
- Zhao, R., Cheng, Z.H., Lu, W.F. and Lu, B.R. 2006. Sampling strategy for wild soybean populations based on their genetic diversity and fine scale spatial genetic structure. Chin. Sci. Bul. 51: 1042–1048.
- Zohary, D. and Hopf, M. 1973. Domestication of pulses in the Old World. Science 182: 887–894.
- Zohary, D., and Hopf, M. 2000. Domestication of Plants in the Old World. Oxford University Press, Oxford.
- Zohary, M. 1972. Origins and evolution in the genus Trifolium. Bot. Notiser. 125: 501–511.
- Zohary, M. and Heller, D. 1984. The Genus Trifolium. The Israel Academy of Sciences and Humanities, pp. 606.
- Zong, X., Guan, J.P., Wang, S.M., Liu, Q., Redden, R., and Ford, R. 2008. Genetic diversity and core collection of alien Pisum sativum L. germplasm. Acta Agronom. Sinica 34: 1518–1528.
- Zong, X., Kaga, A., Tomooka, N., Wang, XW..W., Han, O.K., and Vaughan, D. 2003. The genetic diversity of the Vigna angularis complex in Asia. Genome 46: 647–658.
- Zong, X., Liu, X., Guan, J., Wang S., Liu, Q., Paull, J.G., and Redden, R. 2009a. Molecular variation among Chinese and global winter faba bean germplasm. Theor. Appl. Genet. 118: 971–978.
- Zong, X., Redden, R., Liu, Q., Wang, S., Guan, J., Liu, J., Xu, Y., Liu, X., Gu, J., Yan, L., Ades, P., and Ford, R. 2009b. Analysis of a diverse Pisum sp. collection and development of a Chinese P. sativum core collection based on microsatellite markers. Theor. Appl. Genet. 118: 193–204.
- Zou, Y., Chang, S.K., Gu, Y., and Qian, S.Y. 2011. Antioxidant activity and phenolic compositions of lentil (Lens culinaris var. Morton) extract and its fractions. J. Agric. Food Chem. 59: 2268–2276.
