ABSTRACT
Plant diseases, caused by microbes, threaten world food, feed, and bioproduct security. Plant resistance has not been effectively deployed to improve resistance in plants for lack of understanding of biochemical mechanisms and genetic bedrock of resistance. With the advent of genome sequencing, the forward and reverse genetic approaches have enabled deciphering the riddle of resistance. Invading pathogens produce elicitors and effectors that are recognized by the host membrane-localized receptors, which in turn induce a cascade of downstream regulatory and resistance metabolite and protein biosynthetic genes (R) to produce resistance metabolites and proteins, which reduce pathogen advancement through their antimicrobial and cell wall enforcement properties. The resistance in plants to pathogen attack is expressed as reduced susceptibility, ranging from high susceptibility to hypersensitive response, the shades of gray. The hypersensitive response or cell death is considered as qualitative resistance, while the remainder of the reduced susceptibility is considered as quantitative resistance. The resistance is due to additive effects of several resistance metabolites and proteins, which are produced through a network of several hierarchies of plant R genes. Plants recognize the pathogen elicitors or receptors and then induce downstream genes to eventually produce resistance metabolites and proteins that suppress the pathogen advancement in plant. These resistance genes (R), against qualitative and quantitative resistance, can be identified in germplasm collections and replaced in commercial cultivars, if nonfunctional, based on genome editing to improve plant resistance.
I. Introduction
Plant pathogens are of huge economic importance as they threaten our food, fiber, and bioproduct production under field conditions and further in storage. Genetic improvement of plants is the best way to manage these losses. Molecular biologists have identified hundreds of disease resistance quantitative trait loci (QTLs), in several plant-pathogen systems, but their use in breeding is challenging because they contain several genes, including undesirable ones (Aghnoum et al., Citation2010; Ashkani et al., Citation2014; Buerstmayr et al., Citation2009; St. Clair, Citation2010). With the advent of sequencing of several plant and pathogen genomes, novel technologies have evolved including metabolomics, proteomics, and transcriptomics in addition to genomics and epigenomics (OMICs), offering new opportunities to unravel the mechanisms of qualitative and quantitative resistance in plants (Kushalappa and Gunnaiah, Citation2013; Liu et al., Citation2013b). In this review, we focus on hierarchies of regulatory and resistance metabolite and protein biosynthetic genes induced in plants following pathogen perception, in addition to those constitutively present, to produce resistance metabolites and proteins that directly suppress pathogen development in plants, leading to reduced susceptibility. Also, their deployment in improving crop plant resistance against microbial stress is based on genome editing tools.
II. Plant-pathogen interaction
Plants, unlike mammals, lack adaptive immunity but they have innate immune system in each cell with systemic signaling capability from infection sites (Jones and Dangl, Citation2006). Pathogens produce elicitors called pathogen/microbe-associated molecular patterns (PAMP/MAMP), including peptides, metabolites, cell wall components, enzymes, and toxins to suppress plant defense (Boller and Felix, Citation2009; Dodds and Rathjen, Citation2010; Giraldo and Valent, Citation2013; Wirthmueller et al., Citation2013). Following pathogen attack, the damaged host produces damage-associated molecular patterns (DAMP), including plant signal molecules (Boller and Felix, Citation2009). These elicitors or PAMP/MAMP/DAMP are recognized by the pattern recognition receptors (PRRs) that are biosynthesized in endoplasmic reticulum and transported to plasma membrane (Frescatada-Rosa et al., Citation2015). As a first line of defense response, the PAMP/MAMP trigger downstream genes resulting in no symptoms or race-non-specific hypersensitive response, generally referred to as the PAMP/pattern-triggered immunity (PTI) or non-host resistance (Baxter et al., Citation2014; Boller and Felix, Citation2009; Dodds and Rathjen, Citation2010; Macho and Zipfel, Citation2015; Stael et al., Citation2015; Trdá et al., Citation2015; Uma et al., Citation2011). The genotypes rendered susceptible are considered to vary in basal resistance, partial resistance, or horizontal resistance (Jones and Dangl, Citation2006; Niks et al. Citation2015; Dodds and Rathjen, Citation2010).
Specialized pathogens produce race-specific intracellular elicitors called effectors, produced by specific avirulence (AVR) genes (Boller and Felix, Citation2009; Oliver and Solomon, Citation2010). Though these are considered to be specific to biotrophs, several necrotrophs also produce effectors (Boller and Felix, Citation2009). These effectors suppress other PAMPs and also the host resistance genes to become more virulent (Lo Presti et al., Citation2015). The effectors, depending on their domains, are recognized by plant-produced specific receptors (R proteins), encoded by R genes (Boller and Felix, Citation2009; Du et al., Citation2015; Jones and Dangl, Citation2006; Sarris et al., Citation2015). As a second line of defense response, the effectors trigger downstream genes resulting in race-specific hypersensitive response to contain the pathogen, generally referred to as the effector-triggered immunity (ETI), qualitative resistance, or vertical resistance (Boller and Felix, Citation2009; Giraldo and Valent, Citation2013). Such a resistance is considered to be monogenic and spawned the gene-for-gene hypothesis (Flor, Citation1971). However, these effector recognition receptor genes are just surveillance genes and the real resistance genes that induce hypersensitive response are NADPH oxidase, callose synthase, etc., genes, which are reviewed in the next section. The genotypes rendered susceptible are considered to vary in basal resistance, partial resistance, or horizontal resistance (Dangl and Jones, Citation2001; Niks et al. Citation2015; Dodds and Rathjen, Citation2010).
The hallmark of ETI is the hypersensitive response or cell death, where the pathogen advancement in plant is completely suppressed by the effector-triggered downstream genes, following a complete suppression of PTI by effectors. Similarly, in PTI, the pathogen advancement is completely suppressed by the PAMP/pattern-triggered downstream genes, without any symptom or with hypersensitive response, following the absence of PTI suppression by effectors, also called non-host resistance (Dodds and Rathjen, Citation2010; Jones and Dangl, Citation2006). An incomplete suppression of PTI by effectors enables pathogen to advance, leading to basal or quantitative resistance (Niks et al., Citation2015). In this review, we consider both ETI and PTI to be qualitative resistance, where the plant immune response is either a complete resistance with hypersensitive response or susceptibility. Whereas a weaker immune response, PTI and ETI response, with a lack of hypersensitive response, due to reduced or non-functionality of genes that produce effectors/R proteins, and PAMP/PRR proteins, and also due to the production of enzymes and toxins by pathogens, enabling the pathogen to advance further is considered incomplete resistance, partial resistance, basal resistance or quantitative resistance, the third line of defense (Boyd et al., Citation2013; Kim and Hwang, Citation2015; Niks et al., Citation2015; Uma et al., Citation2011; Waszczak et al., Citation2015). All the same, neither the distinction between qualitative and quantitative resistance nor between the PTI and ETI is always clear, rather they are shades of gray (Poland et al., Citation2009).
A. Setting up the stage: Unifying concept of resistance and the terminologies
Here we propose a unifying concept of resistance. The resistance in plants to pathogen attack is expressed as reduced susceptibility, ranging from complete susceptibility to hypersensitive response, the shades of gray. The hypersensitive response or cell death is considered as qualitative resistance, while the remainder of the reduced susceptibility is considered as quantitative resistance. The quantitative resistance can be quantified either based on monocyclic process under greenhouse conditions as infection efficiency, latent period, lesion expansion, and amount of sporulation or as polycyclic process under field conditions as apparent infection rate and area under the disease progress curve (Kushalappa and Gunnaiah, Citation2013). The resistance in plants against pathogen stress is controlled by a hierarchy of genes, designated here as R genes with subscripts based on their functions, which eventually produce resistance-related (RR) metabolites (RRMs) and proteins (RRPs) that directly suppress the pathogen based on their antimicrobial properties or enforce cell walls to contain them. Following invasion, the pathogens produce elicitors (former: PAMP/MAMP) which are recognized by the plant elicitor recognition receptors (ELRRs produced by RELRR genes; former: PRR), and the specialized pathogens produce more virulent elicitors called effectors that are recognized by the effector recognition receptors (ERR by RERR genes; former: receptor proteins by R genes). In turn, they induce a hierarchy of downstream genes such as phytohormones (PHR by RPHR genes), mitogen-activated protein kinases (MAPKs by RMAPK genes), and transcription factors (TFs by RTF genes), which regulate downstream genes that biosynthesize RRMs (by RRRM genes) and RRPs (by RRRP genes) () (Kushalappa and Gunnaiah Citation2013). Following recognition of elicitors the RELRR genes induce elicitor-triggered immunity (ELTI; former: PTI), whereas following recognition of effectors the RERR genes induce ETI (former: same), both produce hypersensitive response, thus considered qualitative resistance. A weaker or absence of ELTI or ETI, due to non-functionality of genes involved and also due to their suppression by other elicitors, such as enzymes and toxins, enable the pathogen to advance, but are then suppressed by RRMs and RRPs leading to reduced susceptibility or quantitative resistance. The hypersensitive response, however, is also controlled by several genes that produce different resistance metabolites (RRM; callose) and proteins (RRP; chitinases), and so is the quantitative resistance. The amount of resistance thus depends on the type and abundances of RRM and RRP produced, which in turn depends on the hierarchy of genes that regulate them; rather the resistance is a product of the network of a hierarchy(ies) of R genes (; ) (Boyd et al., Citation2013; Brosché et al., Citation2014; Kushalappa and Gunnaiah, Citation2013; Liu et al., Citation2013a; Yogendra et al., Citation2015a). The products of a network of hierarchies of R genes (italicized R with subscripts representing their functions) involved can be represented by a simplified model:
where i is the ith hierarchy and n is the total number of hierarchies in a given genotype. The quantitative resistance is mainly due to additive effects of several such hierarchies of R genes that produce several RRMs and RRPs, which can be constitutive RR metabolites or proteins (RRC) or induced following pathogen invasion, the induced RR metabolites and proteins (RRI), suppressing pathogen by antimicrobial properties of RRM and RRP or further enforcement of cell walls by producing structural barriers (Kushalappa and Gunnaiah, Citation2013). Thus, a RRM or RRP is produced by a hierarchy of R genes, which are rather a chain of genes, and a missing link with a nonfunctional R gene would lead to reduced abundance or absence of a given RRM or RRP, resulting in reduced or no resistance contribution from that hierarchy. The R genes in a hierarchy, however, may have complex interactions with each other; for example, a TF can regulate several RRM genes and likewise a RRM gene can be regulated by several TF genes, thus affecting the abundances of several RRMs. These R genes, however, may have either functional (R) or nonfunctional (r) alleles. These R genes are often associated with several single nucleotide polymorphisms (SNPs) but all SNPs do not have similar values for the quality of protein produced, some may have minor effects on protein expression, while others with SNPs in domain regions may have detrimental effect on protein or enzyme quality. The R genes may control either a major or a minor trait or phenotype. More comprehensive studies, based on systems biology, are needed to unravel several interactions among the R genes in a hierarchy and also the interactions among hierarchies of R genes, involved in plant innate immune responses, the qualitative and quantitative resistance.
Table 1. Resistance genes (R), including regulatory (ELLR, ERR, PHR, MAPK, TF) and resistance-related metabolite (RRM) and resistance-related protein (RRP) biosynthetic genes (R), induced in plants against biotic stress resistance.
Figure 1. Snapshot of key players involved in plant-pathogen interaction. Plants face continuous challenges from several biotrophs, hemibiotrophs, and necrotrophic pathogens, which often attack and propagate in apoplastic space of plant tissues. Pathogens in general produce elicitors (former: PAMP/MAMP), except for specialized pathogens which also produce effectors. Plants recognize these elicitors/effectors and mount an immune response, by triggering a hierarchy of R genes (elicitor recognition receptor (ELRR), effector recognition receptor (ERR), phytohormone (PHR), mitogen-activated protein kinase (MAPK), transcription factor (TF)), eventually to produce resistance-related (RR) metabolites (RRMs) and proteins (RRPs), that directly supress the pathogen advancement. The elicitors are recognized by host membrane-localized ELRR (former: PRR), while the effectors are recognized by ERR (former: PRR produced by R genes). For example, effectors produced by biotrophs are often recognized by NBS-LRR proteins, leading to hypersensitive response via MAPK/SA/NPR1 pathway. On the other hand, elicitors such as chitins produced mainly by hemibiotrophs and necrotrophs are perceived by receptor-like kinases (RLK) and LysM domain chitin elicitor receptor kinase (CERK1), respectively, to activate downstream defense response through MAPK kinase kinase (MAPKKK) pathway. Necrotrophs produce elicitors such as enzymes and toxins, which damage the plant cell walls accumulating cell wall fragments and contents (Plant elicitors plant signal molecules; former: DAMPs), which activate plant defense response through wall-associated receptor kinases (WAKs). Concurrently, several secondary messengers such as calcium ions, reactive oxygen species (ROS), and plant hormone-mediated defense pathways (Ethylene, SA, and JA) are activated following biotic stress, which also trigger downstream genes resulting in hypersensitive response or reduced susceptibility. Overall, the signal perceived by receptor kinases are transmitted efficiently through cytosolic protein kinases such as MAPKKK pathway, to activate an array of plant transcription factors (WRKY, MYB, NAC, and ERF), which regulate several R genes to produce RRPs and RRMs. These RMs are phytoanticipins and phytoalexins, or their conjugate products that are deposited to enforce the secondary cell wall, thus containing the pathogen to initial infection area.
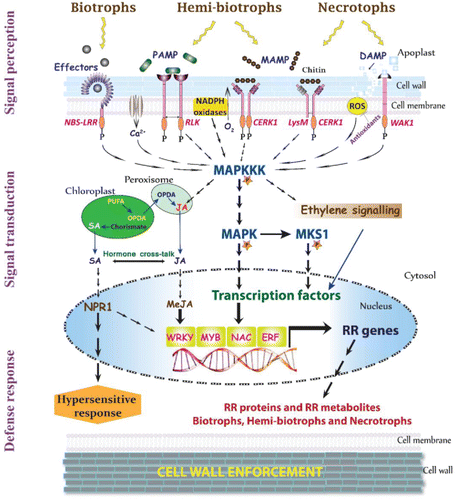
III. Hierarchies of Genes (R) Involved in Resistance
The resistance in plants against pathogen stress is mainly due to RRP and RRM, which are present only in, or in higher amounts, in resistant than in the susceptible genotype. These RRP and RRM are produced either constitutively (RRC) or induced (RRI) following pathogen invasion. The resistance is due to their antimicrobial properties or they are deposited to enforce cell walls, forming structural components, thus containing the pathogen (Kushalappa and Gunnaiah, Citation2013). These RRM and RRP are produced by R genes, which are regulated by a network of a hierarchy of R genes: receptors (ELRR/ERR), phytohormones (PHR), MAPKs (MAPK), and TFs (TF) genes, and they biosynthesize RRM and RRP that suppress pathogen advancement in plant leading to reduced susceptibility, including hypersensitive response. Any of these R genes can be replaced in a commercial cultivar, if its allele is nonfunctional (r gene), based on genome editing, to improve resistance against pathogen stress.
A. Plant receptor (ELRR and ERR) genes and regulation of downstream genes
The plant receptors localized at plasma membrane are receptor-like kinases (RLKs), which at C-terminal end bind to elicitors at the apoplast, and at N-terminal bind to kinases in the cytosol, or receptor-like proteins (RLPs), which have no intracellular kinase domains (Macho and Zipfel, Citation2015). The AVR genes produce hundreds of race-specific effectors that are recognized by specific receptors (ERR), belonging to the coiled-coil, nucleotide-binding, leucine-rich repeat (CC-NB-LRR) class of immune receptors, which in turn trigger downstream genes to induce hypersensitive response (Boller and Felix, Citation2009; Du et al. Citation2015). Broad-spectrum, race-non-specific and common to Phytophthora and Pythium species, elicitors called elicitins are recognized by RLPs (ELR) in potato triggering quantitative resistance (Du et al., Citation2015). Approximately 615 RLKs have been reported in Arabidopsis (Boller and Felix, Citation2009; Dodds and Rathjen, Citation2010). Extracellular ligand-binding domain activates the intracellular kinase domain leading to phosphorylation of substrates and signal transduction. Bacteria produce cell wall lipopolysaccharides which are recognized by a surface-localized lectin S-domain receptor kinase (Macho and Zipfel, 2015). Fungi produce cell wall chitins that are recognized by a lysine motif (lysM) domain containing the RLK1 gene, the chitin elicitor receptor kinase (CERK1) in Arabidopsis and rice, which in turn regulates TFs that regulate RRRM genes to biosynthesize RRMs that reinforce cell walls (Bashline et al., Citation2014; Macho and Zipfel, Citation2015; Miya et al., Citation2007) (). Necrotrophs and hemibiotrophs, and at advanced stages of infection the biotrophs, produce several enzymes (elicitors), such as cutinases, xylanase, cellulases, pectin lyases, and laccases to break down the host cell wall, which releases DAMP (plant signal molecules) (Boller and Felix, Citation2009; Pendleton et al., Citation2014). The host damage releases membrane lipids that activate a cascade of downstream genes prompted by phospholipases (PLs): PL-A activate jasmonic acid (JA), PL-C activate ion channels, and PL-D activate enzymes to produce phosphatidic acid, which activates MAPKs and oxidative bursts through NADPH oxidase controlled by RRRP gene (Ruelland et al., Citation2015). Oligogalacturonides, the plant signal molecules released from the breakdown of pectins in plant cell wall by Botrytis cinerea trigger phytohormones leading to the production of NADPH oxidase (RRP) and callose (RRM) in Arabidopsis (Galletti et al., Citation2008; Łaźniewska et al., Citation2012). Several necrotrophic and some hemibiotrophic pathogens produce toxins that not only act as effectors at low concentrations, activating downstream genes, but also inhibit plant defense mechanisms at high concentrations, facilitating pathogen advancement in the host (Boller and Felix, Citation2009; Gunnaiah and Kushalappa, Citation2014; Kabbage et al., Citation2015; Mengiste, Citation2012).
B. Phytohormone (PHR) genes and regulation of downstream genes
The plant receptor genes (ELRR/ERR), following perception of elicitors/effectors, activate phytohormones that may bind to nuclear proteins with specific domains, such as the F-box, to activate downstream regulatory and RRM genes (Lumba et al., Citation2010). Several phytohormones are activated by receptor proteins, the most common of which are salicylic acid (SA) and jasmonic acid (JA) (De Bruyne et al., Citation2014; Kazan and Lyons, Citation2014; Pieterse et al., Citation2012). While SA is mainly involved in resistance to biotrophs, methyl JA and ethylene (ET) play a significant role in resistance to necrotrophic pathogens. SA, after being sensed by the transcription cofactor NPR1, moves into the nucleus where it modulates expression of regulatory and RRM genes (Furniss and Spoel, Citation2015). JA, biosynthesized by allene oxide synthase (StAOS2), is associated with resistance in potato to late blight (Pajerowska-Mukhtar et al., Citation2008). ET influences the TF ORA59 in Arabidopsis and regulates the production of hydroxycinnamic acid amides (HCAAs), increasing cell wall thickness, which confers resistance to B. cinerea (Lloyd et al., Citation2011). ET in tomato regulates biosynthesis of tyramine hydroxycinnamoyl transferase (THT) to produce HCAAs acting against Cladosporium and Pseudomonas (Etalo et al., Citation2013).
C. Mitogen-activated protein kinase (MAPK) genes and regulation of downstream genes
The MAPKs, activated by receptor genes, activate other MAPKs (Cristina et al., Citation2010). In Arabidopsis infection by B. cinerea, receptors activate MAPKs that phosphorylate other MPK3/MPK6, which then phosphorylate WRKY33 leading to the biosynthesis of the phytoalexin camalexin (Mao et al., Citation2011). MPK4 phosphorylation upon Pseudomonas syringae infection of Arabidopsis, released MKS1 and WRKY33, which induced the biosynthesis of camalexin (Rushton et al., Citation2010). In rice, OsMKK4-OsMPK4/MPK6 play crucial roles in regulating the biosynthesis of diterpenoid- and phenylpropanoid-derived phytoalexins and also lignins in response to a fungal chitin elicitor of M. oryzae (Kishi‐Kaboshi et al., Citation2010). In maize following Fusarium verticillioides infection, brassinosteroid-insensitive-1-associated receptor kinase 1 (BAK1) and homologs of AtMKK4 were associated with ear rot resistance (Lanubile et al., Citation2014).
D. Transcription factor (TF) genes and regulation of resistance metabolite genes
TFs play a pivotal role in biotic stress resistance by directly regulating downstream RRM genes, and thus are excellent candidates in breeding for stress resistance (Alves et al., Citation2014; Kou and Wang, Citation2012; Rushton et al., Citation2010; Xu et al., Citation2011). More than 2000 TFs have been reported in Arabidopsis. Six major families of TFs involved in plant defense are the basic leucine zipper containing domain proteins (bZIP), amino-acid sequence WRKYGQK (WRKY), myelocytomatosis-related proteins (MYC), myeloblastosis-related proteins (MYB), Apetala2/ET-responsive element-binding factors (AP2/EREBP), and no apical meristem/Arabidopsis transcription activation factor/Cup-shaped cotyledon (NAC) (Alves et al., Citation2014). Phosphorylation of MAPK activates TFs by binding to the D site to then translocate into the nucleus (Whitmarsh, Citation2007). A TF can bind to itself, other TFs, or to several downstream RRM genes in a specific metabolic pathway; leading to enhanced production of specific resistance metabolites (RM) (Mao et al., Citation2011; Rushton et al., Citation2010; Shan et al., Citation2015). TF WRKY33 positively regulates resistance to necrotrophic pathogen B. cinerea through activation of tryptophan-derived camalexin biosynthesis in Arabidopsis (Ahuja et al., Citation2012; Mao et al., Citation2011). The overexpression of VvWRKY1 induced a cinnamoyl alcohol dehydrogenase and a caffeic acid O-methyl transferase gene in grape vines, providing higher resistance to downy mildew (Marchive et al., Citation2013) and the transcriptional activation of StWRKY1 by heat shock protein (sHSP17.8)-induced phenylpropanoid genes in potato conferring resistance to late blight (Yogendra et al., Citation2015a). In plants, a battery of NAC and MYB TFs activate secondary cell wall formation based on lignin monomers (Nakano et al., Citation2015). WRKY29, a JA-responsive TF, was expressed only in F. graminearum-resistant genotype, Wangshuibai, but not when the QTL-Fhb1 region was deleted (Xiao et al., Citation2013).
E. Resistance-related protein (RRP) and resistance-related metabolite (RRM) biosynthetic genes
The RRP and RRM directly suppress pathogens and are biosynthesized by resistance (RRRP, RRRM) genes (). These RRP and RRM are either constitutive (RRC), present before the pathogen invasion, or induced (RRI), induced following pathogen invasion. The amount of proteins and metabolites produced depends not only on the functional RRP and RRM genes, but also on the hierarchy of functional regulatory R genes that regulate them. The resistance related proteins (RRP) are the pathogenesis-related (PR) proteins that suppress pathogens, detoxify toxins or virulence factors produced by pathogens and prevent pathogen advancement by enforcing cell walls (Egorov and Odintsova, Citation2012; Łazniewska et al., Citation2012). The resistance related metabolites (RRM) may be antimicrobial, such as phytoanticipins and phytoalexins, or may form complex conjugates that are deposited to reinforce cell walls and suppress pathogen progress (Boller and Felix, Citation2009; Nawrot et al., Citation2014; Yogendra et al., Citation2014). Polymorphisms in introns, exons, and promoter regions, especially in the domain sequence, of these hierarchy of R genes determine the functionality of the proteins/enzymes produced, and thus the amount of proteins (RRP) and metabolites (RRM) biosynthesized (Eudes et al., Citation2014; Krattinger et al., Citation2009; Pushpa et al., Citation2014; Yogendra et al., Citation2015b; Yogendra et al., Citation2014). Thus, the amount of RRP or RRM biosynthesized by an R gene is significantly controlled by a hierarchy of regulatory R genes, especially the TFs, activated following the perception of elicitors/effectors. The resistance, both qualitative and quantitative, in plants is due to the cumulative effects of RRP and RRM biosynthesized by RRRP and RRRM genes, respectively, which in turn are affected by regulatory R genes that can regulate several RRRP and RRRM genes. These defense compounds are generally delivered to the site of infection in vesicles by several transporters, such as ABC and Arabidopsis PEN3 transporters (R genes) (Frescatada-Rosa et al., Citation2015).
IV. Resistance-related (RR) proteins and metabolites
A. Resistance-related proteins (RRPs) and the mechanisms of pathogen suppression
Following pathogen invasion plants induce several proteins (IRP), which are commonly known as PR proteins and about 17 of their families have been reported (Golshani et al., Citation2015; Van Loon et al., Citation2006). The PR proteins that are RRP include those with antimicrobial, toxin-degrading, and cell wall enforcing properties (Nawrot et al., Citation2014). PR proteins, β-1-3-endoglucanases (PR-2) and endochitinases (PR-3, 4, 8, and 11), break down pathogen cell walls (Jach et al., Citation1995). Chitinases breakdown chitin cell wall of fungi, β-1-3-glucanases break down cell wall glucans of bacteria and Chromista (Van Loon et al., Citation2006). PR peptides (molecular weight of <10 kDa) family more specifically proteinase inhibitors (PR-6), defensins (PR-12), thionins (PR-13), and lipid transfer proteins (PR-14) have broad antibacterial and antifungal activities (Sels et al., Citation2008). Members of PR-1 and thaumatin-like PR-5 families have resistance against Oomycetes (Van Loon et al., Citation2006). PR-7 is an endoproteinase, which helps in microbial cell wall cessation in tomato (Jordá et al., Citation2000).
Several hemibiotrophic and necrotrophic pathogens produce toxins that are peptides or metabolites, which act as elicitors inducing hypersensitive response at low concentrations and necrosis at high concentrations (Boller and Felix, Citation2009; Karlovsky, Citation1999). The toxic metabolites such as deoxynivalenol (DON) produced by F. graminearum in wheat and barley is a pathogen virulence factor, which is glycosylated by DON-3-glucosyl transferase gene, converting toxic DON to less toxic DON-3-glycosides, thus reducing further spread of pathogen from initial infection (Schweiger et al., Citation2010). Some toxins produced by pathogens are transported by ABC transporter proteins, such as pleotropic drug resistance transporters, to store them in vacuoles (Kang et al., Citation2011; Walter et al., Citation2015). Hydroxyproline-rich glycoproteins, called extensins, are deposited along with lignin monomers, to thicken the cell walls of Arabidopsis against P. syringae (Łaźniewska et al., Citation2012).
B. Resistance-related metabolites (RRM) and the mechanisms of pathogen suppression
RRMs called phytoanticipins are constitutively (RRC) produced in growing plants and are generally stored in trichomes, oil glands, and epidermal cell layers as nontoxic glycosides, while the toxic forms are released following simple hydrolysis. Plants produce thousands of phenols, flavonoids, terpenes, fatty acids, and alkaloids that are antimicrobial. Resistance metabolites may also be biosynthesized de novo following pathogen invasion; these are commonly known as phytoalexins (RRI) () (Ahuja et al., Citation2012; Pedras and To, Citation2015; Piasecka et al., Citation2015). Resistance depends not only on the amount of metabolite biosynthesized by the plant, but also on the antimicrobial property of a given metabolite (Piasecka et al., Citation2015). These phytoalexins, in hundreds, are biosynthesized by RRRM genes in various specific metabolic pathways () (Ahuja et al., Citation2012). Phenylpropanoids and flavonoids: isoflavonoids, isoflavones, pterocarpans, isoflavans, coumestans, arylbenzofurans, and stilbenes; terpenes: monoterpene, sesquiterpene, carboxylic sesquiterpene, and diterpene families; indole: camalexin (Jeandet et al., Citation2014; Kang et al., Citation2014; Yamamura et al., Citation2015). The pathogens also try to neutralize the effect of these phytoalexins by converting them to less toxic oxidized forms or conjugate with glycosides through production of enzymes (Jeandet et al., Citation2014).
Figure 2. Satellite metabolic pathways, involved in the biosynthesis of RRMs by plants, in response to biotic stress. These resistance metabolites are biosynthesized by the catalytic proteins that are coded by the plant RRRM genes. The biosynthesis of RRMs in a plant is controlled by a hierarchy or several hierarchies of R genes, which may have regulatory or RRM production roles.
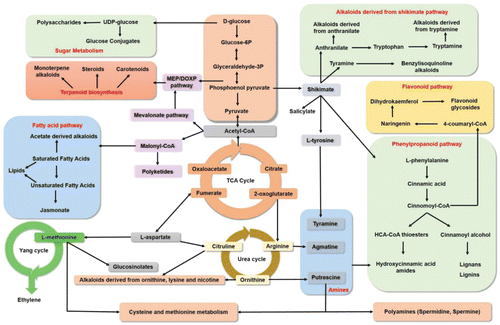
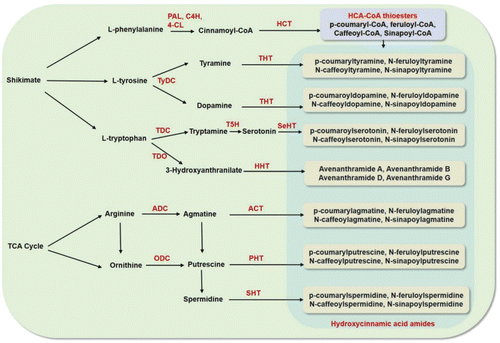
Primary cell walls produced by cellulose and pectins are constitutively enforced by the deposition of secondary metabolites (RRC) (Bashline et al., Citation2014; Eudes et al., Citation2014; Nakano et al., Citation2015). In addition, following pathogen invasion, more secondary metabolites are induced (RRI) and these form complex polymers that are deposited in xylem vessels to enforce cell walls (Gunnaiah et al., Citation2012; Wang et al., Citation2014b; Yogendra et al., Citation2015a). The conjugated complex metabolites, not degraded by the enzymes produced by most pathogens, are produced in the phenylpropanoid, flavonoid, fatty acid, and alkaloid metabolic pathways () (Nakano et al., Citation2015; Yogendra et al., Citation2015b). These play a significant role in plant resistance; Lignin: These are three-dimensional phenolic heteropolymers resulting from the oxidative coupling of three p-hydroxycinnamoyl (p-coumaryl, coniferyl, and sinapyl) alcohols. The cross-coupling reaction of monolignols forms p-hydroxyphenyl, guaiacyl, and syringyl lignins. These, and also lignans, are deposited to enforce secondary cell walls, thus preventing pathogen spread mainly through stem and rachis of inflorescence (Fernández‐Pérez et al., Citation2015; Niks et al., Citation2015). Hydroxycinnamic acid amides (HCAAs): Hydroxycinnamic acid amides are produced as polymers of amines and hydroxyphenols, in several combinations () (Dong et al., Citation2015; Kusano et al., Citation2015; Wen et al., Citation2014; Yogendra et al., Citation2015a). They are deposited as secondary cell walls to prevent the advance of pathogens as proved in wheat against F. graminearum (Gunnaiah et al., Citation2012), in tomato against P. syringae (López-Gresa et al., Citation2011), in Arabidopsis against B. cinerea (Lloyd et al., Citation2011), and in potato against late blight (Yogendra et al., Citation2015a). Callose: A β-1,3-glucan polymer is deposited around the invading hyphae of the biotroph Blumeria graminis in barley and wheat to form papillae leading to hypersensitive response or resistance against powdery mildew; this is quite common against several other biotrophs in other crops (Łaźniewska et al., Citation2012; Nedukha, Citation2015). Kaempferol and quercetin glycosides: Flavonol glycosides form a hard, crystalline structure as a physical barrier against pathogens. Kaempferol, quercetins, and their glycosylated forms prevent the spread of F. graminearum in barley (Bollina et al., Citation2010; Kumaraswamy et al., Citation2012) and P. infestans in potato (Fellenberg and Vogt, Citation2015; Pushpa et al., Citation2014; Yogendra et al., Citation2014). Pectin-alkaloid: Pectins are primary cell wall components and following pathogen invasion they conjugate with alkaloids and deposit to enforce secondary cell walls (Didi et al., Citation2015). Cutin and wax: These are polymers of fatty acids deposited on the cuticle and peridermal layers to prevent pathogen invasion (Andersen et al., Citation2015; Łaźniewska et al., Citation2012; Shi et al., Citation2013). Suberin: Suberins are polymers of polyaromatic, such as hydroxycinnamic acid, and polyaliphatic, such as ω-hydroxy acid, metabolites. They are deposited not only in periderm but also in xylem (Gunnaiah and Kushalappa, Citation2014; Vishwanath et al., Citation2015; Yogendra et al., Citation2014).
Figure 3. Proposed pathway of hydroxycinnamic acid amides biosynthesis by R genes. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4-CL, 4-coumarate: CoA ligase; HCT, hydroxycinnamoyl transferase; TyDC, tyrosine decarboxylase; THT, tyramine hydroxycinnamoyl transferase; TDC, tryptophan decarboxylase; TDO, tryptophan 2,3-dioxygenase; T5H, tryptamine 5-hydroxylase; SeHT, Serotonin hydroxycinnamoyl transferase; HHT, hydroxyanthranilate hydroxycinnamoyl transferase; ADC, arginine decarboxylase; ACT, agmatine coumaroyl transferase; ODC, ornithine decarboxylase; PHT, putrescine hydroxycinnamoyl transferase; SHT, spermidine hydroxycinnamoyl transferase.
C. Metabolic pathways of resistance metabolite (RRM) biosynthesis
Plants produce more than 200 000 secondary metabolites. These are biosynthesized from glucose in specific metabolic pathways (). Thus, the production of a metabolite and its amount depends on a network of functional genes (R), including receptors (ELRR, ERR), phytohormones (PHR), MAPKs (MAPK), TFs (TF), and resistance metabolite (RRM) genes. A nonfunctional gene in this chain, a missing link, can lead to reduced amount or complete absence of a metabolite. The amount of metabolites biosynthesized by RRM gene thus can be significantly influenced by the TFs that regulate them. A TF can bind to several RRM genes thus affecting the abundances of several resistance metabolites. Similarly, a RRRM gene can be regulated by more than one TF. TFs also can bind and regulate each other (Shan et al., Citation2015). Thus, the metabolic pathway regulation is quite complex, requiring further research.
The resistance metabolites (RRM) identified in different pathosystems are () Shikimic acid pathway: The biosynthesis of phenylalanine, tyrosine, and tryptophan leads to the production of phenylpropanoids, flavonoids, and some alkaloids. The production of cinnamic acid from phenylalanine and cinnamoyl-CoA leads to the production of phenylpropanoid metabolites, and 4-coumaroyl CoA leads to the production of flavonoid metabolites (Eudes et al., Citation2014). The production of anthranilate leads to the production of some alkaloids. Mevalonate pathway: Acetyl-CoA leads to the production of terpenes, consisting of isoprene units forming mono- di-, tri-, and sesquiterpenes (Tholl, Citation2015). Fatty acid pathway: Acetyl-CoA produces malonyl CoA which leads to the production of fatty acids and lipids (Ahuja et al., Citation2012; Baetz and Martinoia, Citation2014; Grosjean et al., Citation2015). Polyamine pathway: The biosynthesis of arginine leads to the production of putrescine, spermidine, and spermine (Dong et al., Citation2015). Glucocinolate pathway: The biosynthesis of aspartate and methionine leads to the production of sulfur-containing compounds (Ahuja et al., Citation2012; Bednarek, Citation2012). These also produce, in the Yang cycle, ET; which is a signaling molecule mainly involved in response to necrotrophic infection (Mengiste, Citation2012).
V. Gene (R) pyramiding based on genome editing
Hundreds of cultivars have been bred for each staple food crop such as wheat, rice, and potato in order to meet the demands for growing in niche climatic regions, food, feed, and bioproducts. For lack of disease resistance, some of the old cultivars, in spite of several good traits, are going out of production. Thus the cultivated crops in general lack genetic diversity. Natural as well as intentional hybridization and mutations caused the gain or loss of thousands of gene functions in cultivars and germplasm collections, thus a plethora of biotic stress resistance genes (R) are available for plant resistance improvement (Kage et al., Citation2015; Palmgren et al., Citation2015).
Several RERR genes have been pyramided in potato against late blight but the resistance is not durable, and the pathogen overcomes resistance by producing new effectors (Jacobsen, Citation2013). In these cultivars, the presence of functional RRM and/or RRP genes must be confirmed to guarantee production of resistance metabolites and/or resistance proteins, the end products that give resistance. A few quantitative resistance genes, such as Lr34 in wheat against leaf rust (Puccinia triticina), Yr36 in wheat against stripe rust (Puccinia striiformis), and Pi21 in rice against blast (Magnaporthe oryzae), have been cloned and used successfully in breeding, though their mechanism of resistance are not yet completely understood (Niks et al., Citation2015). Though the resistance in plants against pathogen stress is due to several plausible plant-pathogen gene interactions (Niks et al., Citation2015), the resistance metabolites and resistance proteins that are biosynthesized by R genes, which in turn are activated by regulatory genes such as ELRR, ERR, PHR, MAPK, and TF, as reviewed here, play a major role in plant resistance. It is crucial to make sure that any candidate R gene selected for breeding has rest of the R gene team to ensure production of a given resistance biochemical that can directly suppress the pathogen. A cultivar may have a complete hierarchy of these functional genes except for a missing link that is nonfunctional. Candidate genes, or missing links, with significant resistance effects, can be identified in germplasms based on several approaches (Chen et al., Citation2014; Wen et al., Citation2014) and these candidate genes can be DNA sequenced in commercial cultivars, and if nonfunctional, they can be replaced with functional candidate genes to improve resistance. Some of the regulatory genes, especially TFs, are excellent candidates, if found to be polymorphic in commercial cultivars: TaWRKY45 in wheat QTL 2D, which regulates agmatine coumaroyl transferase (TaACT) and other genes to produce coumaroyl agmatine that are deposited to enforce cell walls, significantly suppressed the biomass of F. graminearum (Kage and Kushalappa, Citation2015). In potato, the StWRKY1 gene regulates THT to produce N-feruloyltyramine and N-feruloyloctopamine (Yogendra et al., Citation2015a). When these genes in the resistant plants were silenced not only the candidate metabolite abundances were significantly reduced but also the pathogen biomass and disease severity significantly increased, proving the resistance function of these genes. Based on our ongoing research, on candidate genes in the QTLs conferring resistance, these TF genes are excellent candidates to improve resistance in commercial cultivars.
Cisgenic transformation is the transfer or replacement of genes between sexually compatible genotypes. These genes must contain their own promoter, coding region, introns, and terminators (Jacobsen, Citation2013). A paradigm shift is now needed, from conventional and molecular breeding to genome editing, in order to replace these nonfunctional gene sequences in commercial cultivars with functional sequences. The cisgenic and intergenic gene pool can be explored to improve plants (Holme et al., Citation2013; Zhan et al., Citation2015). Among several DNA sequence replacement technologies available, the regularly interspaced short palindromic repeats (CRISPR-Cas9 system) are the most preferred for their simplicity to replace either based on agro-transformation or direct introduction to the protoplast (Shan et al., Citation2014). The background DNA can be removed based on self-crossing. Preassembled CRISCR-Cas9 ribonucleoproteins have now enabled DNA-free genome editing in plants (Woo et al., Citation2015). Cisgenics, unlike transgenics, face less regulatory challenges, as such transformations are expected to be similar to conventional breeding (Clasen et al., Citation2015; Jones, Citation2015).
VI. Conclusion and future research
Resistance in plants against microbial stress is either qualitative or quantitative, but the mechanisms of resistance are quite common, controlled by oligo- or polygenes. Plant receptors (ELRR, ERR), following perception of pathogen (elicitor, effector), trigger a hierarchy of regulatory genes (PHR, MAPK, and TF) that control expression of RRM and RRP genes, which biosynthesize resistance metabolites and resistance proteins that directly suppress pathogens. The hierarchy of these R genes with significant trait/resistance effect can be identified in world germplasm collections and used to replace the nonfunctional genes, the missing link, in commercial cultivars to improve resistance. The future research should focus on revealing the hierarchical link of genes involved in pathogen perception and further triggering of downstream regulatory, and RRRM and/or RRRP, genes that are responsible in plant to biosynthesize resistance metabolites and resistance proteins, both constitutive and induced, and biochemical and structural, which directly suppress pathogen, leading to resistance.
Acknowledgments
This project was funded by the International Development Research Center (IDRC), with the financial support of the Department of Foreign Affairs, Trade and Development Canada (DFATD), Ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec (MAPAQ), Québec, Canada, and Natural Sciences and Engineering Research Council of Canada (NSERC). We apologize to all colleagues whose work has not been mentioned due to space limitations. The authors declare that there is no conflict of interest.
References
- Aghnoum, R., Marcel, T.C., Johrde, A., Pecchioni, N., Schweizer, P., and Niks, R.E. 2010. Basal host resistance of barley to powdery mildew: connecting quantitative trait loci and candidate genes. Mol. Plant Microbe Interact. 23: 91–102.
- Ahmad, S., Veyrat, N., Gordon-Weeks, R., Zhang, Y., Martin, J., and Smart, L. 2011. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 157: 317–327.
- Ahuja, I., Kissen, R., and Bones, A.M. 2012. Phytoalexins in defense against pathogens. Trends Plant Sci. 17:73–90.
- Al-Attala, M. N., Wang, X., Abou-Attia, M., Duan, X., and Kang, Z. 2014. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol. Biol. 84: 589–603.
- Alves, M.S., Dadalto, S.P., Gonçalves, A.B., de Souza, G.B., Barros, V.A., and Fietto, L.G. 2014. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes. 2: 85–106.
- Anand, A., Zhou, T., Trick, H. N., Gill, B. S., Bockus, W. W., and Muthukrishnan, S. 2003. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin‐like protein, chitinase and glucanase against Fusarium graminearum. J. Exp. Bot. 54: 1101–1111.
- Andersen, T.G., Barberon, M., and Geldner, N. 2015. Suberization—the second life of an endodermal cell. Curr. Opin. Plant Biol. 28: 9–15.
- Ashkani, S., Rafii, M., Shabanimofrad, M., Ghasemzadeh, A., Ravanfar, S., and Latif, M. 2014. Molecular progress on the mapping and cloning of functional genes for blast disease in rice (Oryza sativa L.): current status and future considerations. Crit. Rev. Biotechnol. 8: 1–15.
- Baetz, U., and Martinoia, E. 2014. Root exudates: the hidden part of plant defense. Trends Plant Sci. 19: 90–98.
- Bashline, L., Lei, L., Li, S., and Gu, Y. 2014. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol Plant. 7: 586–600.
- Baxter, A., Mittler, R., and Suzuki, N. 2014. ROS as key players in plant stress signalling. J Exp. Bot. 65: 1229–1240.
- Bednarek, P. 2012. Sulfur‐Containing Secondary Metabolites from Arabidopsis thaliana and other Brassicaceae with Function in Plant Immunity. ChemBioChem. 13: 1846–1859.
- Bellés, J.M., López-Gresa, M.P., Fayos, J., Pallás, V., Rodrigo, I., and Conejero, V. 2008. Induction of cinnamate 4-hydroxylase and phenylpropanoids in virus-infected cucumber and melon plants. Plant Sci. 174: 524–533.
- Blümke, A., Somerville, S. C., and Voigt, C. A. 2013. Transient expression of the Arabidopsis thaliana callose synthase PMR4 increases penetration resistance to powdery mildew in barley. Adv. Biosci. Biotechnol. 4: 810–813.
- Boller, T., and Felix, G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406.
- Bollina, V., Kumaraswamy, G.K., Kushalappa, A.C., Choo, T.M., Dion, Y., Rioux, S., Faubert, D., and Hamzehzarghani, H. 2010. Mass spectrometry‐based metabolomics application to identify quantitative resistance‐related metabolites in barley against Fusarium head blight. Mol. Plant Pathol. 11: 769–782.
- Boyd, L.A., Ridout, C., O'Sullivan, D.M., Leach, J.E., and Leung, H. 2013. Plant–pathogen interactions: disease resistance in modern agriculture. Trends Genet. 29: 233–240.
- Brosché, M., Blomster, T., Salojärvi, J., Cui, F., Sipari, N., Leppälä, J., Lamminmäki, A., Tomai, G., Narayanasamy, S., and Reddy, R.A. 2014. Transcriptomics and functional genomics of ROS-induced cell death regulation by radical-induced cell death1. PLoS Genet. 10: e1004112.
- Buerstmayr, H., Ban, T., and Anderson, J.A. 2009. QTL mapping and marker‐assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed. 128: 1–26.
- Chen, W., Gao, Y., Xie, W., Gong, L., Lu, K., Wang, W., Li, Y., Liu, X., Zhang, H., and Dong, H. 2014. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46: 714–721.
- Chen, L., Zhang, L., Li, D., Wang, F., and Yu, D. 2013. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 110: E1963–E1971.
- Chen, L., Zhang, Z., Liang, H., Liu, H., Du, L., and Xu, H. 2008. Overexpression of TiERF1 enhances resistance to sharp eyespot in transgenic wheat. J. Exp. Bot. 59: 4195–4204.
- Christensen, S.A., Nemchenko, A., Park, Y.-S., Borrego, E., Huang, P.-C., Schmelz, E.A., Kunze, S., Feussner, I., Yalpani, N., and Meeley, R. 2014. The novel monocot-specific 9-lipoxygenase ZmLOX12 is required to mount an effective jasmonate-mediated defense against fusarium verticillioides in Maize. Mol. Plant Microbe Interact. 27: 1263–1276.
- Clasen, B.M., Stoddard, T.J., Luo, S., Demorest, Z.L., Li, J., Cedrone, F., Tibebu, R., Davison, S., Ray, E.E., and Daulhac, A. 2015. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14: 169–76.
- Cristina, M.S., Petersen, M., and Mundy, J. 2010. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649.
- Dangl J.L, Jones J.D.G. 2001. Plant pathogens and integrated defence responses to infection. Nature. 411: 826–33.
- De Bruyne, L., Höfte, M., and De Vleesschauwer, D. 2014. Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant. 7: 943–959.
- Didi, V., Jackson, P., and Hejátko, J. 2015. Hormonal regulation of secondary cell wall formation. J. Exp. Bot. 66: 5015–27.
- Dodds, P.N., and Rathjen, J.P. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11: 539–548.
- Dong, X., Gao, Y., Chen, W., Wang, W., Gong, L., Liu, X., and Luo, J. 2015. Spatiotemporal Distribution of Phenolamides and the Genetics of Natural Variation of Hydroxycinnamoyl Spermidine in Rice. Mol. Plant. 8: 111–121.
- Dong, N., Liu, X., Lu, Y., Du, L., Xu, H., and Liu, H. 2010. Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana. Funct. Integr. Genomics. 10: 215–226.
- Dracatos, P. M., Weerden, N. L., Carroll, K. T., Johnson, E. D., Plummer, K. M., and Anderson, M. A. 2014. Inhibition of cereal rust fungi by both class I and II defensins derived from the flowers of Nicotiana alata. Mol. Plant Pathol. 15: 67–79.
- Du, J., Verzaux, E., Chaparro-Garcia, A., Bijsterbosch, G., Keizer, L.P., Zhou, J., Liebrand, T.W., Xie, C., Govers, F., and Robatzek, S. 2015. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants. 1: 15034.
- Duan, Y., Guo, J., Shi, X., Guan, X., Liu, F., and Bai, P., 2013. Wheat hypersensitive-induced reaction genes TaHIR1 and TaHIR3 are involved in response to stripe rust fungus infection and abiotic stresses. Plant Cell Rep. 32: 273–283.
- Egorov, T.A., and Odintsova, T. 2012. Defense peptides of plant immunity. Russ. J. Bioorg. Chem. 38: 1–9.
- Etalo, D.W., Stulemeijer, I.J., van Esse, H.P., de Vos, R.C., Bouwmeester, H.J., and Joosten, M.H. 2013. System-wide hypersensitive response-associated transcriptome and metabolome reprogramming in tomato. Plant Physiol. 162: 1599–1617.
- Eudes, A., Liang, Y., Mitra, P., and Loque, D. 2014. Lignin bioengineering. Curr. Opin. Biotechnol. 26: 189–198.
- Fellenberg, C., and Vogt, T. 2015. Evolutionarily conserved phenylpropanoid pattern on angiosperm pollen. Trends Plant Sci. 20: 212–218.
- Fernández‐Pérez, M., Villafranca‐Sánchez, M., Flores‐Céspedes, F., and Daza‐Fernández, I. 2015. Lignin‐polyethylene glycol matrices and ethylcellulose to encapsulate highly soluble herbicides. J. Appl. Polym. Sci. 132: 41422.
- Flor, H.H. 1971. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9: 275–296.
- Frescatada-Rosa, M., Robatzek, S., and Kuhn, H. 2015. Should I stay or should I go? Traffic control for plant pattern recognition receptors. Curr. Opin. Plant Biol. 28: 23–29.
- Furniss, J.J., and Spoel, S.H. 2015. Cullin-RING ubiquitin ligases in salicylic acid-mediated plant immune signaling. Front Plant Sci. 6: 154.
- Gallego‐Giraldo, L., Jikumaru, Y., Kamiya, Y., Tang, Y., and Dixon, R.A. 2011. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New. Phytol. 190: 627–639.
- Galletti, R., Denoux, C., Gambetta, S., Dewdney, J., Ausubel, F.M., De Lorenzo, G., and Ferrari, S. 2008. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 148: 1695–1706.
- Geng, S., Li, A., Tang, L., Yin, L., Wu, L., and Lei, C., 2013. TaCPK2-A, a calcium-dependent protein kinase gene that is required for wheat powdery mildew resistance enhances bacterial blight resistance in transgenic rice. J. Exp. Bot. 64: 3125–3136.
- Giberti, S., Bertea, C.M., Narayana, R., Maffei, M.E., and Forlani, G. 2012. Two phenylalanine ammonia lyase isoforms are involved in the elicitor-induced response of rice to the fungal pathogen Magnaporthe oryzae. J. Plant Physiol. 169: 249–254.
- Giraldo, M.C., and Valent, B. 2013. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11: 800–814.
- Golshani, F., Fakheri, B.A., Behshad, E., and Vashvaei, R.M. 2015. PRs proteins and their Mechanism in Plants. In: Biological Forum: Research Trend. 7: 477–495.
- Grosjean, K., Mongrand, S., Beney, L., Simon-Plas, F., and Gerbeau-Pissot, P. 2015. Differential Effect of Plant Lipids on Membrane Organization specificities of phytosphingolipids and phytosterols. J. Biol. Chem. 290: 5810–5825.
- Gunnaiah, R., and Kushalappa, A.C. 2014. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 83: 40–50.
- Gunnaiah, R., Kushalappa, A.C., Duggavathi, R., Fox, S., and Somers, D.J. 2012. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS One. 7: e40695.
- Han, J., Lakshman, D. K., Galvez, L. C., Mitra, S., Baenziger, P. S., and Mitra, A. 2012. Transgenic expression of lactoferrin imparts enhanced resistance to head blight of wheat caused by Fusarium graminearum. BMC Plant Biol. 12: 33.
- Holme, I.B., Wendt, T., and Holm, P.B. 2013. Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotech. J. 11:395–407.
- Jach, G., Görnhardt, B., Mundy, J., Logemann, J., Pinsdorf, E., Leah, R., Schell, J., and Maas, C. 1995. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8: 97–109.
- Jacobsen, E. 2013. Cisgenesis: a modern way of domesticating traits of the breeders' gene pool. CAB Reviews. 8: 56.
- Jeandet, P., Hébrard, C., Deville, M.-A., Cordelier, S., Dorey, S., Aziz, A., and Crouzet, J. 2014. Deciphering the role of phytoalexins in plant-microorganism interactions and human health. Molecules 19:18033–18056.
- Jha, S., Tank, H. G., Prasad, B. D., and Chattoo, B. B. 2009. Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res. 18: 59–69.
- Jones, H.D. 2015. Regulatory uncertainty over genome editing. Nat. Plants. 1.
- Jones, J.D., and Dangl, J.L. 2006. The plant immune system. Nature 444: 323–329.
- Jordá, L., Conejero, V., and Vera, P. 2000. Characterization of P69E and P69F, two differentially regulated genes encoding new members of the subtilisin-like proteinase family from tomato plants. Plant Physiol. 122: 67–74.
- Kabbage, M., Yarden, O., and Dickman, M.B. 2015. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233: 53–60.
- Kage, U., and Kushalappa, A. C. 2015. Identification of Candidate Genes in QTL 2DL and Deciphering Mechanisms of Resistance Against Fusarium Head Blight Based on Metabolo-Genomics in Wheat. In: Plant and Animal Genome XXIII Conference: Plant and Animal Genome.
- Kage, U., Kumar, A., Dhokane, D., Karre, S., and Kushalappa, A., 2015. Functional molecular markers for crop improvement. Crit. Rev. Biotechnol. 16: 1–14.
- Kang, J.-H., McRoberts, J., Shi, F., Moreno, J.E., Jones, A.D., and Howe, G.A. 2014. The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164: 1161–1174.
- Kang, J., Park, J., Choi, H., Burla, B., Kretzschmar, T., Lee, Y., and Martinoia, E. 2011. Plant ABC transporters. The Arabidopsis book/American Society of Plant Biologists. 9: e0153.
- Karlovsky, P. 1999. Biological detoxification of fungal toxins and its use in plant breeding, feed and food production. Nat. Toxins. 7: 1–23.
- Kazan, K., and Lyons, R. 2014. Intervention of phytohormone pathways by pathogen effectors. Plant Cell. 26: 2285–2309.
- Kim, N.H., and Hwang, B.K. 2015. Pepper pathogenesis‐related protein 4c is a plasma membrane‐localized cysteine protease inhibitor that is required for plant cell death and defense signaling. Plant J. 81: 81–94.
- Kishi‐Kaboshi, M., Okada, K., Kurimoto, L., Murakami, S., Umezawa, T., Shibuya, N., Yamane, H., Miyao, A., Takatsuji, H., and Takahashi, A. 2010. A rice fungal MAMP‐responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63: 599–612.
- Kou, Y., and Wang, S. 2012. Toward an understanding of the molecular basis of quantitative disease resistance in rice. J. Biotechnol. 159: 283–290.
- Krattinger, S.G., Lagudah, E.S., Spielmeyer, W., Singh, R.P., Huerta-Espino, J., McFadden, H., Bossolini, E., Selter, L.L., and Keller, B. 2009. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 323: 1360–1363.
- Krishnamurthy, K., Balconi, C., Sherwood, J. E., and Giroux, M. J. 2001. Wheat puroindolines enhance fungal disease resistance in transgenic rice. Mol. Plant Microbe Interact. 14: 1255–1260.
- Kumaraswamy, G., Kushalappa, A., Choo, T., Dion, Y., and Rioux, S. 2012. Differential metabolic response of barley genotypes, varying in resistance, to trichothecene‐producing and‐nonproducing (tri5−) isolates of Fusarium graminearum. Plant Pathol. 61: 509–521.
- Kusano, T., Kim, D.W., Liu, T., and Berberich, T. 2015. Polyamine Catabolism in Plants. In: Polyamine: A Universal Molecular Nexus for Growth, Survival and Specialized Metabolism, pp. 77–88. T. Kusano, & H. Suzuki (Eds.). Tokyo, Japan: Springer.
- Kushalappa, A.C., and Gunnaiah, R. 2013. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 18: 522–531.
- Lanubile, A., Ferrarini, A., Maschietto, V., Delledonne, M., Marocco, A., and Bellin, D. 2014. Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC genomics. 15: 710.
- Lauvergeat, V., Lacomme, C., Lacombe, E., Lasserre, E., Roby, D., and Grima-Pettenati, J. 2001. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry. 57: 1187–1195.
- Łaźniewska, J., Macioszek, V.K., and Kononowicz, A.K. 2012. Plant-fungus interface: the role of surface structures in plant resistance and susceptibility to pathogenic fungi. Physiol. Mol. Plant Pathol. 78: 24–30.
- Leba, L. J., Cheval, C., Ortiz‐Martín, I., Ranty, B., Beuzón, C. R., and Galaud, J. P., 2012. CML9, an Arabidopsis calmodulin‐like protein, contributes to plant innate immunity through a flagellin‐dependent signalling pathway. Plant J. 71: 976–989.
- Lee, H., Ko, Y. J., Cha, J.-Y., Park, S. R., Ahn, I., and Hwang, D.-J. 2013. The C-terminal region of OsWRKY30 is sufficient to confer enhanced resistance to pathogen and activate the expression of defense-related genes. Plant Biotechnol. Rep. 7: 221–230.
- Li, J., Wang, J., Wang, N., Guo, X., and Gao, Z. 2015a. GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tissue Organ Cult. 121: 127–140.
- Li, W., Wang, F., Wang, J., Fan, F., Zhu, J., and Yang, J. 2015b. Overexpressing CYP71Z2 Enhances Resistance to Bacterial Blight by Suppressing Auxin Biosynthesis in Rice. Plos one. 10: e0119867.
- Li, X., Zhang, Y., Huang, L., Ouyang, Z., Hong, Y., and Zhang, H. 2014. Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea. BMC Plant Biol. 14: 166.
- Liu, B., Hong, Y.-B., Zhang, Y.-F., Li, X.-H., Huang, L., and Zhang, H.-J. 2014a. Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227: 145–156.
- Liu, B., Ouyang, Z., Zhang, Y., Li, X., Hong, Y., and Huang, L. 2014b. Tomato NAC Transcription Factor SlSRN1 Positively Regulates Defense Response against Biotic Stress but Negatively Regulates Abiotic Stress Response. Plos One. 9: e102067.
- Liu, W., Liu, J., Ning, Y., Ding, B., Wang, X., Wang, Z., and Wang, G.-L. 2013. Recent progress in understanding PAMP-and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol. Plant. 6: 605–620.
- Lloyd, A.J., William Allwood, J., Winder, C.L., Dunn, W.B., Heald, J.K., Cristescu, S.M., Sivakumaran, A., Harren, F.J., Mulema, J., and Denby, K. 2011. Metabolomic approaches reveal that cell wall modifications play a major role in ethylene‐mediated resistance against Botrytis cinerea. Plant J. 67: 852–868.
- López-Gresa, M.P., Torres, C., Campos, L., Lisón, P., Rodrigo, I., Bellés, J.M., and Conejero, V. 2011. Identification of defence metabolites in tomato plants infected by the bacterial pathogen Pseudomonas syringae. Environ. Exp. Bot. 74: 216–228.
- Lumba, S., Cutler, S., and McCourt, P. 2010. Plant nuclear hormone receptors: a role for small molecules in protein-protein interactions. Annu. Rev. Cell Dev. Biol. 26: 445–469.
- Macho, A.P., and Zipfel, C. 2015. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 23: 14–22.
- Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. 2011. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 23: 1639–1653.
- Marchive, C., Léon, C., Kappel, C., Coutos-Thévenot, P., Corio-Costet, M.-F., Delrot, S., and Lauvergeat, V. 2013. Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS One. 8: e54185.
- Maury, S., Geoffroy, P., and Legrand, M. 1999. Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 121: 215–224.
- Mei, C., Qi, M., Sheng, G., and Yang, Y. 2006. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19: 1127–1137.
- Mengiste, T. 2012. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50: 267–294.
- Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. 2003. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 15: 2551–2565.
- Miao, W., Wang, X., Li, M., Song, C., Wang, Y., and Hu, D. 2010. Genetic transformation of cotton with a harpin-encoding gene hpaXoo confers an enhanced defense response against different pathogens through a priming mechanism. BMC Plant Biol. 10: 67.
- Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and Shibuya, N. 2007. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104: 19613–19618.
- Muroi, A., Ishihara, A., Tanaka, C., Ishizuka, A., Takabayashi, J., Miyoshi, H., and Nishioka, T. 2009. Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta. 230: 517–527.
- Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N.A., and Ohtani, M. 2015. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci. 6: 288.
- Nawrot, R., Barylski, J., Nowicki, G., Broniarczyk, J., Buchwald, W., and Goździcka-Józefiak, A. 2014. Plant antimicrobial peptides. Folia microbiologica. 59: 181–196.
- Nedukha, O. 2015. Callose: Localization, functions, and synthesis in plant cells. Cytol. Genet. 49: 49–57.
- Niks, R., Li, X., and Marcel, T.C. 2015. Quantitative Resistance to Biotrophic Filamentous Plant Pathogens: Concepts, Misconceptions, and Mechanisms. Annu. Rev. Phytopathol. 53: 445–470.
- Ogata, T., Kida, Y., Arai, T., Kishi, Y., Manago, Y., and Murai, M. 2012. Overexpression of tobacco ethylene response factor NtERF3 gene and its homologues from tobacco and rice induces hypersensitive response-like cell death in tobacco. J. General Plant Pathol. 78: 8–17.
- Oliver, R.P., and Solomon, P.S. 2010. New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 13: 415–419.
- Pajerowska-Mukhtar, K.M., Mukhtar, M.S., Guex, N., Halim, V.A., Rosahl, S., Somssich, I.E., and Gebhardt, C. 2008. Natural variation of potato allene oxide synthase 2 causes differential levels of jasmonates and pathogen resistance in Arabidopsis. Planta. 228: 293–306.
- Palmgren, M.G., Edenbrandt, A.K., Vedel, S.E., Andersen, M.M., Landes, X., Østerberg, J.T., Falhof, J., Olsen, L.I., Christensen, S.B., and Sandøe, P. 2015. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 20: 155–164.
- Petersen, L. N., Ingle, R. A., Knight, M. R., and Denby, K. J. 2009. OXI1 protein kinase is required for plant immunity against Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 60: 3727–3735.
- Pedras, M.S.C., and To, Q.H. 2015. Non-indolyl cruciferous phytoalexins: Nasturlexins and tridentatols, a striking convergent evolution of defenses in terrestrial plants and marine animals? Phytochemistry. 113: 57–63.
- Pendleton, A.L., Smith, K.E., Feau, N., Martin, F.M., Grigoriev, I.V., Hamelin, R., Nelson, C.D., Burleigh, J.G., and Davis, J.M. 2014. Duplications and losses in gene families of rust pathogens highlight putative effectors. Front Plant Sci. 5: 299.
- Piasecka, A., Jedrzejczak‐Rey, N., and Bednarek, P. 2015. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206: 948–964.
- Pieterse, C.M., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S.C. 2012. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521.
- Poland, J.A., Balint-Kurti, P.J., Wisser, R.J., Pratt, R.C., and Nelson, R.J. 2009. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14: 21–29.
- Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., Zuccaro, A., Reissmann, S., and Kahmann, R. 2015. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66: 513–545.
- Pushpa, D., Yogendra, K.N., Gunnaiah, R., Kushalappa, A.C., and Murphy, A. 2014. Identification of late blight resistance-related metabolites and genes in potato through nontargeted metabolomics. Plant Mol. Biol. Rep. 32: 584–595.
- Raffaele, S., Vailleau, F., Léger, A., Joubès, J., Miersch, O., and Huard, C., 2008. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 20: 752–767.
- Rong, W., Qi, L., Wang, J., Du, L., Xu, H., and Wang, A. 2013. Expression of a potato antimicrobial peptide SN1 increases resistance to take-all pathogen Gaeumannomyces graminis var. tritici in transgenic wheat. Funct. Integr. Genomics. 13: 403–409.
- Ruelland, E., Kravets, V., Derevyanchuk, M., Martinec, J., Zachowski, A., and Pokotylo, I. 2015. Role of phospholipid signalling in plant environmental responses. Environ. Exp. Bot. 114: 129–143.
- Rushton, P.J., Somssich, I.E., Ringler, P., and Shen, Q.J. 2010. WRKY transcription factors. Trends Plant Sci. 15: 247–258.
- Sarowar, S., Kim, Y. J., Kim, E. N., Kim, K. D., Hwang, B. K., and Islam, R. 2005. Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant cell Rep. 24: 216–224.
- Sarowar, S., Kim, Y. J., Kim, K. D., Hwang, B. K., Ok, S. H., and Shin, J. S. 2009. Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant cell Rep. 28: 419–427.
- Sarris, P.F., Duxbury, Z., Huh, S.U., Ma, Y., Segonzac, C., Sklenar, J., Derbyshire, P., Cevik, V., Rallapalli, G., and Saucet, S.B. 2015. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell. 161: 1089–1100.
- Schweiger, W., Boddu, J., Shin, S., Poppenberger, B., Berthiller, F., Lemmens, M., Muehlbauer, G.J., and Adam, G. 2010. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Mol. Plant Microbe Interact. 23: 977–986.
- Sels, J., Mathys, J., De Coninck, B.M., Cammue, B.P., and De Bolle, M.F. 2008. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46: 941–950.
- Serazetdinova, L., Oldach, K. H., and Lörz, H. 2005. Expression of transgenic stilbene synthases in wheat causes the accumulation of unknown stilbene derivatives with antifungal activity. J. Plant Physiol. 162: 985–1002.
- Shan, Q., Wang, Y., Li, J., and Gao, C. 2014. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9: 2395–2410.
- Shan, W., Chen, J.Y., Kuang, J.F., and Lu, W.J. 2015. Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance expressions of Pathogenesis‐Related genes against Colletotrichum musae. Mol. Plant. Pathol. 1–9.
- Sharma, P., Ito, A., Shimizu, T., Terauchi, R., Kamoun, S., and Saitoh, H. 2003. Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol. Genet. Genomics. 269: 583–591.
- Shi, J., Zhang, L., An, H., Wu, C., and Guo, X. 2011. GhMPK16, a novel stress responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12: 1.
- Shi, J.X., Adato, A., Alkan, N., He, Y., Lashbrooke, J., Matas, A.J., Meir, S., Malitsky, S., Isaacson, T., and Prusky, D. 2013. The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol. 197: 468–480.
- Singh, A.K., Dwivedi, V., Rai, A., Pal, S., Reddy, S.G.E., Rao, D.K.V., Shasany, A.K., and Nagegowda, D.A. 2015. Virus‐induced gene silencing of Withania somnifera squalene synthase negatively regulates sterol and defence‐related genes resulting in reduced withanolides and biotic stress tolerance. Plant Biotech. J. 13:1287–99.
- St. Clair, D.A. 2010. Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol. 48:247–268.
- Stael, S., Kmiecik, P., Willems, P., Van Der Kelen, K., Coll, N.S., Teige, M., and Van Breusegem, F. 2015. Plant innate immunity–sunny side up? Trends Plant Sci. 20: 3–11.
- Taniguchi, S., Hosokawa‐Shinonaga, Y., Tamaoki, D., Yamada, S., Akimitsu, K., and Gomi, K. 2014. Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 37: 451–461.
- Tholl, D. 2015. Biosynthesis and Biological Functions of Terpenoids in Plants. Adv. Biochem. Eng. Biotechnol. 148: 63–106.
- Trdá, L., Boutrot, F., Claverie, J., Brulé, D., Dorey, S., and Poinssot, B. 2015. Perception of pathogenic or beneficial bacteria and their evasion of host immunity: pattern recognition receptors in the frontline. Front Plant Sci. 6: 219.
- Tronchet, M., Balague, C., Kroj, T., Jouanin, L., and Roby, D. 2010. Cinnamyl alcohol dehydrogenases‐C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant. Pathol. 11: 83–92.
- Uma, B., Rani, T.S., and Podile, A.R. 2011. Warriors at the gate that never sleep: non-host resistance in plants. J. Plant Physiol. 168: 2141–2152.
- Van Loon, L.C., Rep, M., and Pieterse, C. 2006. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162.
- Vishwanath, S.J., Delude, C., Domergue, F., and Rowland, O. 2015. Suberin: biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Rep. 34: 573–586.
- Walter, S., Kahla, A., Arunachalam, C., Perochon, A., Khan, M.R., Scofield, S.R., and Doohan, F.M. 2015. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. erv048.
- Wan, J., Zhang, X.-C., Neece, D., Ramonell, K. M., Clough, S., and Kim, S.Y. 2008. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 20: 471–481.
- Wang, H., Hao, J., Chen, X., Hao, Z., Wang, X., and Lou, Y. 2007. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65: 799–815.
- Wang, X., Tang, C., Huang, X., Li, F., Chen, X., and Zhang, G., 2012. Wheat BAX inhibitor-1 contributes to wheat resistance to Puccinia striiformis. J. Exp. Bot. 3:4571–84.
- Wang, X., Wang, X., Deng, L., Chang, H., Dubcovsky, J., and Feng, H., 2014a. Wheat TaNPSN SNARE homologues are involved in vesicle-mediated resistance to stripe rust (Puccinia striiformis f. sp. tritici). J. Exp. Bot. 65: 4807–20.
- Wang, S., Li, E., Porth, I., Chen, J.-G., Mansfield, S.D., and Douglas, C.J. 2014b. Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 4: 5054.
- Waszczak, C., Akter, S., Jacques, S., Huang, J., Messens, J., and Van Breusegem, F. 2015. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 66: 2923–2934.
- Wen, W., Li, D., Li, X., Gao, Y., Li, W., Li, H., Liu, J., Liu, H., Chen, W., and Luo, J. 2014. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5: 3438.
- Whitmarsh, A.J. 2007. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. BBA-Mol. Cell. Res. 1773: 1285–1298.
- Wirthmueller, L., Maqbool, A., and Banfield, M.J. 2013. On the front line: structural insights into plant-pathogen interactions. Nat. Rev. Microbiol. 11: 761–776.
- Woo, J.W., Kim, J., Kwon, S.I., Corvalán, C., Cho, S.W., Kim, H., Kim, S.-G., Kim, S.-T., Choe, S., and Kim, J.-S. 2015. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33: 1162–4.
- Xia, N., Zhang, G., Liu, X.-Y., Deng, L., Cai, G. L., and Zhang, Y. 2010. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 37: 3703–3712.
- Xiao, J., Jin, X., Jia, X., Wang, H., Cao, A., Zhao, W., Pei, H., Xue, Z., He, L., and Chen, Q. 2013. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC genomics 14: 197.
- Xu, Z.S., Chen, M., Li, L.C., and Ma, Y.Z. 2011. Functions and application of the AP2/ERF transcription factor family in crop improvementF. J. Integr. Plant. Biol. 53: 570–585.
- Xu, Y.-H., Wang, J.-W., Wang, S., Wang, J.-Y., and Chen, X.-Y. 2004. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 135: 507–515.
- Yamamura, C., Mizutani, E., Okada, K., Nakagawa, H., Fukushima, S., Tanaka, A., Maeda, S., Kamakura, T., Yamane, H., and Takatsuji, H. 2015. Diterpenoid Phytoalexin Factor, a bHLH Transcription Factor, Plays a Central Role in the Biosynthesis of Diterpenoid Phytoalexins in Rice. Plant J. 84: 1100–13.
- Yeh, Y.-H., Chang, Y.-H., Huang, P.-Y., Huang, J.-B., and Zimmerli, L. 2015. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6: 322.
- Yogendra, K.N., Kumar, A., Sarkar, K., Li, Y., Pushpa, D., Mosa, K.A., Duggavathi, R., and Kushalappa, A.C. 2015a. Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J. Exp. Bot. 66: 7377–7389.
- Yogendra, K.N., Kushalappa, A.C., Sarmiento, F., Rodriguez, E., and Mosquera, T. 2015b. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Funct. Plant Biol. 42: 284–298.
- Yogendra, K.N., Pushpa, D., Mosa, K.A., Kushalappa, A.C., Murphy, A., and Mosquera, T. 2014. Quantitative resistance in potato leaves to late blight associated with induced hydroxycinnamic acid amides. Funct. Integr. Genomics. 14: 285–298.
- Zeng, W., and He, S. Y. 2010. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 153: 1188–1198.
- Zhan, J., Thrall, P.H., Papaïx, J., Xie, L., and Burdon, J.J. 2015. Playing on a Pathogen's Weakness: Using Evolution to Guide Sustainable Plant Disease Control Strategies. Annu. Rev. Phytopathol. 53: 19–43.
- Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., and Li, H., 2012. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. Plos One. 7: e51091.
- Zhang, F., Huang, L.-Y., Ali, J., Cruz, C. V., Zhuo, D.-L., and Du, Z.-L. 2015. Comparative transcriptome profiling of a rice line carrying Xa39 and its parents triggered by Xanthomonas oryzae pv. oryzae provides novel insights into the broad-spectrum hypersensitive response. BMC genomics. 16: 111.
- Zhang, H., Hu, Y., Yang, B., Xue, F., Wang, C., and Kang, Z. 2013. Isolation and characterization of a wheat IF2 homolog required for innate immunity to stripe rust. Plant Cell Rep. 32: 591–600.
- Zhang, J., Peng, Y., and Guo, Z. 2008. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 18: 508–521.
- Zhu, X., Qi, L., Liu, X., Cai, S., Xu, H., and Huang, R. 2014. The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164: 1499–1514.
- Zuo, W., Chao, Q., Zhang, N., Ye, J., Tan, G., and Li, B. 2015. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47: 151–157.
