Abstract
For thousands of years, humans have been improving crops to better suit their needs. These enhancements are driven by changes in the genetic makeup of the plant. While this was initially unintentional, there has been a steady push to increase the pace and precision of crop breeding, something that has occurred alongside a growing understanding of genetics and an escalating capacity to thoroughly assess genomes at the molecular level. With the advent and rapid uptake of molecular breeding techniques, such as transgenics and genome editing over the past few decades, there has been much trepidation regarding the possibility of off-target effects derived from unanticipated mutations at loci other than those intended for alteration, and the unintended risks that this might confer. These concerns persist regardless of the fact that a growing number of studies indicate that the occurrence of off-target mutations derived from newer biotechnological breeding techniques are negligible compared to what is observed with many conventional breeding approaches, and even spontaneously from one generation to the next. Given the impending food security crisis that we are facing in the short-term, there is a critical need to implement a wide range of breeding tools as a means of meeting growing demand, withstanding climate change-related pressures, increasing nutrition, and providing environmental benefits. While food safety is clearly of the utmost importance, now is certainly not the time to prevent the use of particular breeding technologies based on unfounded doubts. Therefore, in this review, we attempt to shed light on these apprehensions by putting purported “risks” into the context of plant breeding as a whole by comparing frequencies of spontaneous mutations with those (both anticipated and unanticipated) that occur through various conventional and biotechnological breeding approaches, including transgenics and genome editing. We then consider how these changes may, or may not, translate into unanticipated risk, and discuss the current global regulatory asynchrony surrounding genome edited crops.
I. Introduction
It is predicted that our global population will increase from approximately 7.7 billion to 9.7 billion by the year 2050 (United Nations Department of Economic and Social Affairs, Citation2017). Together with a rising need for plant-based renewable resources and our ever-growing per capita calorific consumption (Tilman and Clark, Citation2015), this will translate into a substantial elevation in demand for crop-derived products. With more than 800 million people already chronically hungry and 2 billion suffering micronutrient deficiencies (FAO, Citation2017), it is clear that without substantial increases in crop production, this deficit in food security will continue to expand. Although reducing food waste and equalizing global food distribution would go a long way toward meeting demand in the future, increasing crop yields is also critical since there is little, if any, room for expansions in arable land area (Bruinsma, Citation2009; Jansson et al., Citation2018). Indeed, it is proposed that we will need to achieve an approximately 50% increase in crop productivity by 2050, compared with 2010, just to support human food-related needs (Gouel and Guimbard, Citation2019; Matthews, Citation2019). Such a feat will be challenging in and of itself; however, factors such as water scarcity, climbing fertilizer costs and additional regulatory restrictions on their use will impede these efforts even further. Moreover, climate change-related environmental factors, such as increased temperatures, augmentation in weather variability and the intensity of extreme weather events, and shifting disease/pest outbreaks are known to instigate both yield and nutritional penalties in many crop species (Deryng et al., Citation2014; Scheelbeek et al., Citation2018; Schleussner et al., Citation2018; Asseng et al., Citation2019). These negative effects are already evident in parts of the world (e.g., Ali et al., Citation2017; Ray et al., Citation2019), and will only worsen as climate change escalates. In addition to the clear food security crisis that we are facing, we are also seeing an erosion in traditional farming knowledge with an increasing number of farmers moving away from this occupation due to diminishing returns. Therefore, in order to ensure a sufficient supply of food during a period when considerable environmental changes are predicted to occur, there is an urgent need for the development of high-yielding, climate change-tolerant, nutrient-rich and environmentally-friendly crop cultivars that are profitable to farmers.
Plant breeding in a broad sense involves a human-led shift in plant evolution to improve traits that are beneficial specifically to us, typically in the context of either cultivation or our preferences. It could be argued that the first human-led crop improvement endeavors began approximately 10,000 to 13,000 years ago during the initiation of domestication (reviewed by Purugganan and Fuller, Citation2009; ). Prior to this time, plant genomes, like those of all other living organisms, had been evolving through the occurrence of spontaneous genetic alterations for better survivability. Those genetic changes that conferred traits that enhanced the plant’s ability to survive and adapt were maintained by the species, while those that did not either accumulated through genetic drift or were eliminated through natural selection (reviewed by Saini et al., Citation2020). During the conversion from a nomadic lifestyle to one that was more sedentary, humans harnessed this natural evolution of plant species, and altered its course by selecting, and then propagating, plants with traits that facilitated agriculture, such as a reduction in the dispersal of seeds and fruits, alterations in plant phenology and morphology, and palatability (e.g., Meyer and Purugganan, Citation2013; Berger et al., Citation2017). In practice, traits selected during domestication sometimes opposed those favored during natural selection (Hillman and Davies, Citation1990), and in 1859, Charles Darwin coined the term “artificial selection” to make the distinction between selection that occurred “in nature” and that which derived from human activities (Darwin, Citation1859). Domestication has led to profound changes in the crops that we grow currently compared to their wild progenitors; changes that would have almost certainly never have taken place without the intervention of humans (reviewed by Custers et al., Citation2018). While this process was essential to agriculture as we know it today, it unfortunately also led to a reduction in genomic diversity (Cowling et al., Citation2009; Gross and Olsen, Citation2010; Olsen and Wendel, Citation2013), as well as a narrowing of food crop species, which have contributed to agriculture’s vulnerability to climate change and disease/pest outbreaks (reviewed by Smýkal et al., Citation2018).
Figure 1. Timeline of major events in crop breeding. BCE, Before the Common Era; CRISPR/Cas, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein; DNA, deoxyribonucleic acid; US, United States. References: 1Robbins, Citation1922; 2Laibach, Citation1925; 3Stadler, 1928 b; 4Blakeslee and Avery, Citation1937; 5Watson and Crick, Citation1953; 6Carlson et al., Citation1972; 7Caplan et al., Citation1983; 8Burr et al., Citation1983. Image sources: corn, John Doebley; hand pollination, tissue culture, wheat field, DNA fingerprinting, DNA sequence, and gene editing obtained from iStock; Gregor Mendel, wikipedia; wheat embryos, Andriy Bilichak, Agriculture and Agri-Food Canada; radiation, colchicine and double helix obtained from pngegg.com; protoplasts, Mnolf; petri dish, Udaya Subedi, Agriculture and Agri-Food Canada – Dr. Stacy Singer; tomato, pngfuel.com.
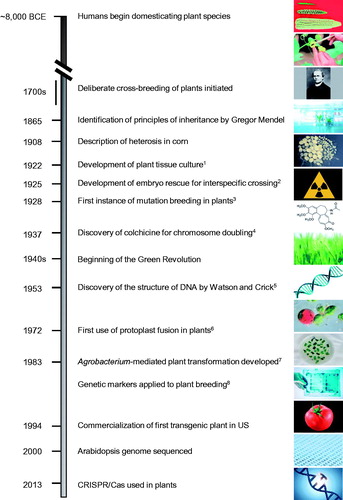
Domestication and other early breeding endeavors were achieved without any theoretical knowledge of genetics. However, the discoveries of Gregor Mendel in relation to how traits (phenotypes) were transferred from parents to offspring through genes transformed plant breeding in the 20th century (; Hoßfeld et al., Citation2017), and it has been strongly influenced by scientific progress ever since. Due to the slow rate at which spontaneous mutations occur, the unpredictable nature of their manifestation, and the crucial need for genetic variability for breeding success, all breeding innovations since then have centered on a desire to increase the pace and/or precision of such efforts, along with an impetus to broaden genetic diversity in crop species (). For example, while traditional breeding has maintained genetic gains in most cereal crops where grain quality holds importance, in other cereals such as corn, the development of hybrids has brought higher genetic gains. George Harrison Shull coined the term “heterosis” in the early 1900s to describe the vigor displayed by F1 hybrids of genetically diverse corn lines (Shull, Citation1908), resulting in an almost complete shift to the cultivation of hybrids in the US Midwest by the late 1930s. This practice resulted in large yield gains at the time, and continues to this day in crops that are capable of forming hybrids as a means of obtaining vigorous crops that are more stress tolerant, synchronized in flowering and higher yielding. Similarly, the discovery of mutation breeding using ionizing radiation in the late 1920s (Stadler, Citation1928a, Citation1928b), and later using chemical mutagens such as ethyl methanesulfonate (EMS; e.g., Neuffer and Ficsor, Citation1963), allowed an extremely large number of mutations to be induced in plants in a short period of time. This conventional breeding technology provided an important component of the subsequent Green Revolution during the 1960s, largely led by Norman Borlaug, whereby sizeable increases in yield were achieved primarily in wheat, rice and maize through the development of germplasm with a high harvest index that responded to increased rates of irrigation and fertilizers (reviewed by Llewellyn, Citation2018; Vietmeyer, Citation2010).
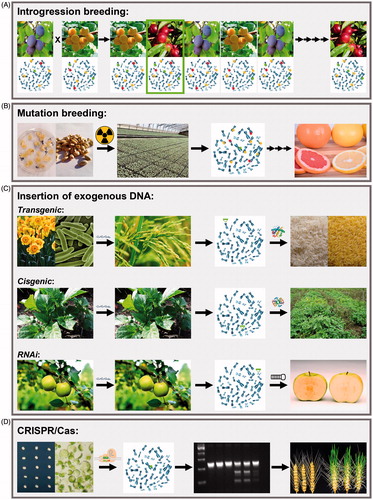
Figure 2. Schematic representation of common plant breeding techniques. A) Introgression breeding involves the crossing of two plant genotypes, the selection of offspring with a desirable trait, and multiple rounds of backcrossing. B) Mutation breeding involves the treatment of plant tissue with physical or chemical mutagens, selection of a genotype with a desirable trait from a large mutagenized population, and multiple rounds of backcrossing. C) Transgenic, cisgenic and RNAi approaches involve the insertion of an exogenous sequence of DNA into the plant genome, which results in the production of either a protein or double-stranded RNA molecule, and consequently the improved trait. D) Simple NHEJ-based CRISPR/Cas gene editing involves the introduction of Cas and sgRNA into plant cells, resulting in a double stranded DNA break at the target site and the subsequent generation of an indel at a precise genomic location, which leads to trait improvement. CRISPR/Cas, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein; DNA, deoxyribonucleic acid; RNAi, ribonucleic acid interference. Image sources: plums, apricots, grapefruit, daffodils, bacteria, rice plant, potato plant, and apple tree obtained from Pixabay; radiation, scissors, proteins, double helix and Cas obtained from pngegg.com; pluots, dreamstime; chromosomes, iStock; callus on media, Udaya Subedi, Agriculture and Agri-Food Canada – Dr. Stacy Singer; citrus seeds, pixnio.com; greenhouse, Government of Newfoundland and Labrador, Department of Fisheries, Forestry and Agriculture; Golden Rice, Golden Rice Project (www.goldenrice.org.); potato blight, Ronald Hutten, Laboratory of Plant Breeding, Wageningen University – Dr. Henk Schouten; nonbrowning apple, webcomicms.net; wheat immature embryos, derived from Hayta et al., Citation2019; protoplasts, Mnolf; gel image, Agriculture and Agri-Food Canada – Dr. Stacy Singer; edited wheat, National Agriculture and Food Research Organization and Okayama University (for more information, please refer to the following article: https://doi.org/10.1016/j.celrep.2019.06.090).
Although rates of yield gain for cereal crops (both hybrid and nonhybrid) were relatively high between the 1960s and 1990s, they are increasingly difficult to sustain under sub-optimal farming conditions (Brisson et al., Citation2010; Ray et al., Citation2012; Hall and Richards, Citation2013) and targeted crosses between elite germplasm to maintain quality often comes at the cost of performance stability across different environments. The advent of genomics- and molecular-assisted selection methods has enhanced breeding efficiency (Parry and Hawkesford, Citation2012); however, the targeted improvement of many traits, especially in crops with large polyploid genomes, will require biotechnological breeding tools to improve elite breeding germplasm. Such contemporary breeding techniques have tended to involve the over-expression of native genes, the heterologous expression of foreign genes, or the down-regulation of endogenous gene expression and are considered “genetically modified” (“GM”) in most countries (reviewed by Kamthan et al., Citation2016). However, despite the promise of these strategies to improve a vast number of traits in a wide selection of crop species (e.g., Xiao et al., Citation2007; Driever et al., Citation2017; Paul et al., Citation2017), and the exceptionally rapid uptake of commercially available cultivars to date, only a relatively small number of traits have made it to the market thus far (reviewed by Qaim, Citation2016). This hindrance in market acceptability has occurred as a result of misconceptions concerning the breeding technologies themselves, as well as the rigorous regulatory processes currently required prior to the commercialization of such cultivars (e.g., Qaim, Citation2016; Macnaghten and Habets, Citation2020).
To further increase the speed and precision of improvement, and with the added benefit of potentially minimizing concerns that surround the use of transgenic plants, highly specific genome editing platforms (including clustered regularly interspaced short palindromic repeats/CRISPR-associated protein [CRISPR/Cas]) have made substantial headway over the past decade due to their capacity to elicit mutations at specific, predefined genomic loci (reviewed by Zhang et al., Citation2017; Songstad et al., Citation2017). A growing number of countries are either in the process of updating, or have already modernized, their regulatory or policy frameworks to clarify the requirements for products developed through genome editing. While the vast majority have indicated that at least certain genetic outcomes derived from genome editing will not be considered “GM”, there remains global asynchrony regarding the regulation of such plants, which will almost certainly impact biotechnological innovation and complicate trade in the future (Parrott et al., Citation2020; Schmidt et al., Citation2020). In light of the short timeframe before food demand is projected to exceed supply in the midst of a climate change crisis, it is clear that we are in urgent need of a new revolution in crop biotechnology as a critical component of ensuring food security in the future. In order to achieve this, it is of the utmost importance that crop breeders have access to a wide range of complementary breeding technologies; a prerequisite for which will be regulatory policies that are based on scientific findings rather than political or emotional sentiments, as well as an understanding that all of these methods are simply different means of achieving the same thing – genetic variations. In this review, we will compare the types and propensity for mutations derived spontaneously with those that occur through traditional, induced mutagenesis, transgenic and genome editing breeding approaches. Furthermore, we will put this into the perspective of how these changes might translate into unanticipated risk, which may help to inform regulatory and policy decisions related to the commercialization of new crop cultivars in the future.
II. Spontaneous genetic variation in plants
As environments change, organisms must adapt to the new conditions or face the possibility of extinction. For species to adapt, they require genetic variation. Simply put, mutations are the ultimate source of all genetic variation, and can include a wide variety of changes, including single base pair substitutions and small insertions/deletions (indels), as well as larger changes such as whole genome duplications, large insertions or deletions, chromosomal rearrangements and inversions, horizontal gene transfer from unrelated organisms, and the replication/movement of transposable elements (TEs), for example. If these genetic alterations occur in the meristematic/germ cells, they can then be shuffled into offspring via recombination, providing the cornerstone for plant evolution and adaptation ().
Figure 3. Means by which DNA mutations are incurred spontaneously in plants. Although many naturally occurring mutations have no observable effect on plant growth and appearance, some do, and it is these that are important for plant adaptation and the breeding of new crop cultivars using traditional techniques. DNA, deoxyribonucleic acid; TE, transposable element.
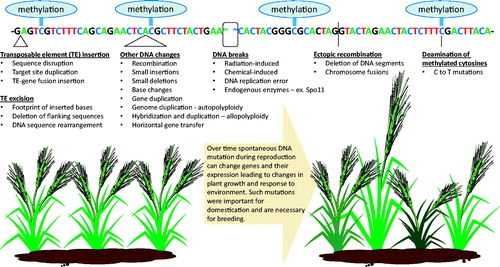
A. Main mechanisms driving nucleotide level mutations in plants
DNA can be damaged through endogenous processes, the production of genotoxic metabolites, and exposure to certain environmental factors, for example, and this is a constant threat to organisms, including plants (reviewed by Spampinato, Citation2017). As a result, plants utilize various DNA repair mechanisms to preserve genomic integrity and minimize potentially deleterious effects; however, some of these repair processes are themselves error-prone, and lead to mutations (Gorbunova and Levy, Citation1999). For example, reactive oxygen species, which often accumulate in response to various types of stress, can cause various types of DNA damage, including base alkylation or oxidation, which when repaired leads to the production of a single nucleotide variation (SNV; Gill and Tuteja, Citation2010). Another means through which SNVs can occur is through C to T transitions that arise from the spontaneous deamination of methylated cytosines (Zhou et al., Citation2020), which are very common in plant genomes and play a substantial role in epigenetic gene silencing (Pikaard and Scheid, Citation2014).
Double-strand breaks (DSBs), which can be triggered following exposure to certain chemicals, radiation, and during DNA replication, are another type of lesion that are highly mutagenic and often lead to mutations following repair. The cellular enzyme Spo11, which generates cuts along chromosomes to facilitate recombination during meiosis (sexual reproduction), is a well-known cause of DSBs in plant cells. While a small number of these DSBs lead to recombination, the remainder are repaired using error-prone mechanisms (Sanchez-Moran et al., Citation2007), and therefore every passage through reproduction results in a number of DSB-induced mutations that exist within progeny along with their recombined chromosomes.
Two main strategies are used by plant cells to repair DSBs, and include the following: error-free homology-dependent repair process (HDR) that is active predominantly during the S/G2 phase of the cell cycle in somatic cells and also during meiosis, as well as the far more prevalent error-prone nonhomologous end-joining (NHEJ) pathways, which incorporate canonical NHEJ (C-NHEJ; active in all cell phases) and microhomology-mediated end joining (MMEJ; maximal activity in S phase of the cell cycle), for example (Sfeir and Symington, Citation2015; Schmidt et al., Citation2019). Unlike high fidelity HDR, which requires the presence of a homologous template (typically a sister chromatid) for repair, C-NHEJ requires no homologous sequences for repair and MMEJ only necessitates small microhomologies of approximately 1–20 nt flanking the break site. In the case of C-NHEJ-mediated repair of DSBs, small indels are typically generated at the break site in the process, while MMEJ generally yields deletions (Sfeir and Symington, Citation2015).
B. Major factors driving larger scale genetic variation in plants
Larger structural variations in the form of polyploidization, TE re-location and abundance, horizontal gene transfer, and alterations in gene copy number or presence/absence are also important sources of genetic variation in plants (Gabur et al., Citation2019). Much of the success of flowering plants is believed to have been achieved through genome expansion, typically through polyploidy and massive expansions in TE families, followed by contraction (Puttick et al., Citation2015; Pellicer et al., Citation2018). Polyploidy involves a whole genome duplication, and can occur within a species spontaneously or following hybridization between two different species. The end result of this is two or more complete genomes within a single organism. Although this process is rare, all flowering plants show signatures in their genomes of several past polyploidy events (Wood et al., Citation2009) and most of our important crops are polyploid (Renny-Byfield and Wendel, Citation2014). Wheat, for example, is a recent polyploid with three distinct subgenomes, while maize is the result of a more ancient event where genome fractionation and subgenome dominance has greatly reduced the overall genome content from its initial polyploid state (Schnable et al., Citation2011; Renny-Byfield et al., Citation2017).
TEs make up a large proportion of plant genomes, with maize and wheat genomes consisting of approximately 85% TE-derived DNA (Schnable et al., Citation2009; Wicker et al., Citation2018), while rice and soybean genomes comprise 47% and 59% TE-derived DNA, respectively (Schmutz et al., Citation2010; Jiang and Panaud, Citation2013). These TEs are very active in terms of their propagation over time, “jumping” to and integrating in different locations within the genome. Although in many cases they do not insert within genic sequences, there are nonetheless many examples of gene disruption and influence of nearby TE insertions on gene expression (Hirsch and Springer, Citation2017). Some examples of important traits caused by TEs are the single stock growth habit in maize, fruit flesh color in citrus, fruit color in grape, tolerance to preharvest sprouting in wheat and barley, and fruit shape in tomato (Singh and Singh, Citation2012; Lisch, Citation2013; Singh et al., Citation2013). In addition to TE-mediated gene disruption and changes in gene expression, TEs are also known to mediate gene duplications and fusions. This can occur through the copy and reinsertion of genes alongside TEs, and from the fusion of copied genes with other copied DNA. During the replication process of TEs, genes can also be multiplied in the form of extra-chromosomal circular DNA, which can then reinsert into the genome resulting in increased gene copy number. Recently, these events have been linked to the amplification of a 400-kb region containing the 5-ENOYLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) gene and 58 other genes leading to herbicide resistance in Amaranthus palmeri (Molin et al., Citation2020). Above and beyond all of these rather large scale genetic modifications, TE excision is imprecise, leaving behind small SNVs or indels, and TE insertion is usually associated with a target site duplication (Singh et al., Citation2006; Singh et al., Citation2012; Wicker et al., Citation2016).
Although genome expansion through polyploidization and TE bursts is easy to comprehend, genome contraction through fractionation is less well-studied. However, it appears that the main driver for this process is most likely recombination, where ectopic events lead to DNA deletion and shuffling (Li et al., Citationin press). One possible consequence of genome fractionation is that when genes are deleted in certain lineages, presence-absence variation (PAV) among lineages ensues. The extent of genome rearrangement through genome expansion and contraction, as well as TE activity, has been shown to be profound in plants with genes changing positions along chromosomes over time (Zhao and Schranz, Citation2019). Indeed, while gene order along chromosomes (synteny) is highly conserved in mammals, considerable disruptions are evident in flowering plants, suggesting extensive DNA perturbation through TE-mediated gene movement and ectopic recombination, which all require DNA breakage and repair.
In addition to genome expansion and contraction, the spontaneous and stable introduction of exogenous DNA through horizontal gene transfer from unrelated organisms can also contribute to the evolution of plant genomes (e.g., Fang et al., Citation2017). For example, Agrobacterium tumefaciens is a naturally occurring soil bacteria that normally induces abnormal root growths through the delivery, genomic integration and expression of specialized oncogenes present on a tumor-inducing plasmid (Joos et al., Citation1983). Scientists have leveraged this ability to transfer DNA into plant cells by removing the oncogenes and replacing them with genetic elements encoding agronomic traits of interest (reviewed by Lemay and Moineau, Citation2020). However, this transfer of DNA does not inherently require human intervention, and spontaneous horizontal gene transfer from A. tumefaciens to an ancestor of sweet potato has led to the presence of several “transgenes” in this crop that are still intact and expressed today. Indeed, there is even evidence that these “transgenes” may have played a role during the domestication of this crop (Kyndt et al., Citation2015).
C. Prevalence of spontaneous mutations in plants
Prior to the late 1960s, it was argued that there was a wild type genome for each species, where a single genome existed without any mutations. This is now known to be incorrect, and in actuality, each single species is composed of many different genomes. The human HapMap project highlighted this concept, whereby genome-wide genetic variation in the form of mutations can be seen in representative humans from across the globe, outlining our long history of global movement and migration (International HapMap Consortium, Citation2005; International HapMap 3 Consortium, Citation2010). This is also evidenced by the genetic variation observed within individual plants of the same cultivar (e.g., Haun et al., Citation2011; Yates et al., Citation2012; Mercati et al., Citation2016), which derives from de novo spontaneous mutations and the segregation of heterozygous loci during reproduction. In two individuals of the soybean “Williams 82” cultivar, for example, 1,838 SNVs were identified between exomes, while 25 genes were found to be differentially present/absent (Haun et al., Citation2011). However, the prevalence of such intra-cultivar genetic heterogeneity can differ substantially from species to species depending on their mating strategy (for example selfing, outcrossing or asexual) and level of homozygosity (Haun et al., Citation2011), with the lowest levels of heterogeneity expected to occur in crops that are propagated asexually.
For species to adapt, an intermediate spontaneous mutation rate is thought to be optimal since high mutational loads have the potential to erode important information. In humans (genome size of ∼3,100 Mbp), spontaneous mutations lead each of us to possess around 60 mutations that were not present in our parents (Pennisi, Citation2018). In plants, the number of spontaneous mutations that occur per generation varies depending on several factors, including genome size/ploidy level; however, SNVs tend to occur at rates of ∼10−8 to 10−9 mutations per site per generation (reviewed by Graham et al., Citation2020), which is similar to estimates in humans (Roach et al., Citation2010). For example, in the model species, Arabidopsis thaliana (genome size of ∼135 Mbp), it has been estimated that 1-5 de novo spontaneous mutations occur per generation per diploid plant (; Ossowski et al., Citation2010; Willing et al., Citation2016; Weng et al., Citation2019). This number tends to be higher in plants with larger genomes, and in rice for instance (genome size ∼430 Mbp), an average of 23 SNVs and 18 indels have been estimated to arise from parents to progeny (; Tang et al., Citation2018).
Table 1. Examples of approximate ranges in genetic variability derived spontaneously and from various breeding methods as determined by whole genome sequencing in rice and Arabidopsis.
However, these estimates are likely often underrepresentations overall since highly repetitive regions of the genome are difficult to assess, which means that only a proportion of mutations within a genome are identified in cases where de novo assemblies of whole genome sequencing data are not carried out. In addition, these studies were carried out under controlled growth conditions, and therefore do not take into account the substantial acceleration of de novo genetic variation from one generation to the next that occurs under stress conditions, such as elevated temperatures or salinity, that are typically encountered in the field (DeBolt, Citation2010; Jiang et al., Citation2014). Regardless of the exact numbers of both small and large scale genetic alterations that take place from generation to generation in plants, an understanding of the fact that mutations occur naturally and create the genetic variation that has been of utmost importance for the adaptation of all species is crucial when considering crop genomics.
III. Genetic alterations in the context of “conventional” breeding
A. Introgression through crossing
Many traditional plant breeding platforms rely upon the presence of existing genetic variation, which has occurred spontaneously in plants over many years, to elicit improvements in traits using methods such as crossing and selection. The most commonly used traditional breeding method involves the introgression of a gene(s) responsible for a desired trait into a crop cultivar from another plant of the same species via intraspecific crossing, or from another species or even genera through interspecific and intergeneric hybridization, respectively (e.g., reviewed by Schnell et al., Citation2015; Katche et al., Citation2019). While intraspecific crossing is a rather straightforward process, the more distant the phylogenetic relationship between two parent plants, the more difficult their hybridization becomes due to various barriers. As such, an assortment of tissue culture-dependent technologies, including somatic hybridization (often through protoplast fusion; Tiwari et al., Citation2018), embryo rescue (Sharma et al., Citation1996) and chromosome doubling (Van Tuyl and De Jeu, Citation1997), are commonly used to successfully achieve these outcomes. Such crosses have also been prevalent in the domestication of many crop species, and the domestication of bread wheat, for example, has involved not only a whole genome duplication event and an intergeneric cross between wild goatgrass (Aegilops tauschii) and domesticated emmer wheat (Triticum dicoccon; Gornicki and Faris, Citation2014), but also introgressions from wild related and unrelated species (e.g., Ali et al. Citation2016; Rahmatov et al., Citation2016; Fedak et al., Citation2017).
Introgression through crossing or hybridization is inherently random and not only the gene(s) of interest will be introgressed into progeny, but also many other genes, including those associated with large regions of linked DNA that surrounds the desired gene. Unfortunately, such genes can encode mildly unfavorable traits, resulting in a phenomenon termed “linkage drag” (e.g., Vikram et al., Citation2015). Multiple rounds of extensive backcrossing with the original parental cultivar followed by selection can eliminate introgressed DNA associated with detrimental traits that is unlinked to the gene of interest. This process can also slowly reduce the size of linked introgressions; however, linked regions can still be hundreds of kilobases to tens of megabases even in elite cultivars (e.g., Ballini et al., Citation2007; Lin et al., Citation2014; Calafiore et al., Citation2016; Li, Chitwood, et al., Citation2018; Lobaton et al., Citation2018). For example, in cultivated rice, an introgression bearing a gene that provides resistance to blast disease also includes at least 185 other putative genes with more than 100 different potential functions (Ballini et al., Citation2007). Similarly, modern wheat varieties contain disease resistance genes from dozens of species belonging to six distinct genera, and in some cases entire chromosomes or chromosome arms made up of megabase pairs of DNA were introgressed along with the resistance gene (Jones et al., Citation1995). In any case, despite the fact that the required backcrossing is a very lengthy process, and the substantial challenge that linkage drag presents, introgression has played a critical role in the history of today’s crop cultivars (e.g., Hajjar and Hodgkin, Citation2007).
In order to accelerate the pace of traditional breeding, marker- and genomics-assisted breeding platforms have been developed in recent years (Jiang, Citation2015; Leng et al., Citation2017), which has allowed the large-scale mapping of agronomically-important quantitative trait loci (QTL)/genes, mining for elite alleles, and high-throughput genotyping. These approaches do not change the breeding outcome in terms of genetic consequences because the method of achieving the desired trait (typically introgression) remains the same. Instead, these strategies are simply used to increase the efficiency of the selection process by enabling the selection of a specific genetic variation that is known to be associated with a desired trait, rather than selecting for the trait itself. This means that the initial screening of large populations can be carried out in a high-throughput manner without any need to assess a specific growth stage or response to environmental conditions, or to carry out technically challenging physiological/biochemical assessments. This can be particularly useful in the context of backcrossing (Jiang, Citation2013), and allows for selection of the allele eliciting the desirable trait, selection for other desirable traits in the recurrent backcross genotype, or selection against the undesirable genome of the donor parent to hasten the elimination of undesirable genes (e.g., Soto-Cerda et al. Citation2013; Pratap et al., Citation2017). Similarly, the development of other technologies has also led to accelerations in traditional breeding. For example, the production of doubled haploids has become an important tissue culture-requiring component of many traditional crop breeding programs that allows for the immediate production of completely homozygous lines (reviewed by Lenaerts et al., Citation2019).
While there is a relative paucity of information regarding the precise genetic changes that have occurred as a result of traditional breeding approaches, there is growing evidence to suggest that they have been incredibly widespread (e.g., Ballini et al., Citation2007; Lin et al., Citation2014; Anderson et al., Citation2016; Sun et al., Citation2018). An accumulation of spontaneous mutations over many generations, along with introgressions into elite cultivars via traditional breeding, has led to a remarkable amount of genetic diversity within any given plant species, including SNVs leading to amino acid changes, protein truncations, aberrations in splicing and changes in transcriptional regulation, as well as transposon insertions, large deletions, and the introduction of entire genes (Olsen and Wendel, Citation2013). Wide crosses can lead to even more substantial genetic consequences than intraspecific crosses, including genome rearrangements, translocations, recombination and chromosomal elimination (Liu and Li, Citation2007; Du et al., Citation2008; Xu et al., Citation2011).
Furthermore, since tissue culture is utilized as an essential step for plant regeneration across a wide range of traditional breeding methods, additional mutations resulting from somaclonal variation, which derives from genetic changes incurred during plant growth in tissue culture (Evans, Citation1989), can also be evident. Somaclonal variation can lead to genome-wide SNVs, indels, variations in chromosome number, chromosome rearrangements, the activation of transposable elements, and epigenetic alterations (Karp and Maddock, Citation1984; Lee and Phillips, Citation1987; Jiang et al., Citation2011; Miguel and Marum, Citation2011). In rice, mutations in the range of tens to tens of thousands of SNVs and indels per individual plant have been observed following tissue culture (Miyao et al., Citation2012; Zhang, Wang, et al., Citation2014; Wei et al., Citation2016; Qin et al., Citation2018; Tang et al., Citation2018). Similarly, between 9 and 65 novel homozygous SNVs and 2–6 homozygous indels have been discerned in the R1 generation compared to its progenitor plant in Arabidopsis (Jiang et al., Citation2011). In line with this, mutation rates following tissue culture tend to be 1–2 orders of magnitude higher than spontaneous mutation rates across plant species in general (e.g., Jiang et al., Citation2011; Tang et al., Citation2018; Park et al., Citation2020); however, rates can differ quite dramatically depending on various factors related to in vitro culture conditions and the length of culturing itself ().
Given that traditional breeding can elicit substantial genetic changes during the development of new crop cultivars, and spontaneously-derived genetic variation is prevalent in all species, it is not surprising that genetic variation between traditionally-bred cultivars of the same species can be immense. For example, between approximately 25,000 to 9.8 million SNVs and 1,400 to 1.4 million indels have been observed between individuals of different cultivars/accessions of soybean (Lam et al., Citation2010; Yadav et al., Citation2015; Anderson et al., Citation2016), cotton (Li, Manghwar, et al., Citation2019), sweet cherry (Xanthopoulou et al., Citation2020), maize (Sun et al., Citation2018), rice (e.g., Subbaiyan et al., Citation2012; Kawakatsu et al., Citation2013; Fu et al., Citation2016; Qin et al., Citation2018; Wang, Mauleon, et al., Citation2018), and tomato (Causse et al., Citation2013), respectively (). Indeed, SNVs have been found to be present on average every 61–540 bp in maize, wheat, soybean, B. oleracea and rice cultivars (Ching et al., Citation2002; Somers et al., Citation2003; Van et al., Citation2005; Subbaiyan et al., Citation2012; Agnieska et al., Citation2016; Fu et al., Citation2016; Golicz et al., Citation2016; Sun et al., Citation2018), while indels have been estimated to occur every 126–900 bp in maize lines (Ching et al., Citation2002; Sun et al., 2018). Furthermore, larger structural rearrangements, including differences in the presence/absence of certain genes among individuals within species, are also typically widespread, with tens to tens of thousands of such variations apparent between cultivars depending on the species (e.g., McHale et al., Citation2012; Causse et al., Citation2013; Agnieska et al., Citation2016; Anderson et al., Citation2016; Du et al., Citation2017; Sun et al., Citation2018; Wang, Mauleon, et al., Citation2018; Xanthopoulou et al., Citation2020; Zhang, Sun, et al., Citation2020). When comparing two inbred lines of maize, for instance, roughly 180 single copy genes were present in one inbred line but not the other (Springer et al., Citation2009), while approximately 49 genes on average have been found to be unique to individual wheat cultivars (Montenegro et al., Citation2017).
B. Random mutagenesis
Although introgression through crossing has led to the generation of a multitude of improvements in crop species thus far, it has its limitations. These include the relatively slow rate of occurrence for new spontaneous mutations and a dependence upon hybridization compatibility between species. Furthermore, genetic variation does not always exist for certain traits of interest in available germplasm (Meyer and Purugganan, Citation2013; Bedő and Láng, Citation2015). As such, many of the newer breeding technologies developed over the past 100 years have been aimed at mitigating these issues by randomly re-introducing new genetic variability in a rapid and random manner, including somaclonal variation and chemical/physical mutagenesis, which are all considered conventional breeding approaches (reviewed by Songstad et al., Citation2017).
As discussed previously, somaclonal variation elicits increased mutation rates during in vitro plant tissue culture. Above and beyond the use of tissue culture in many traditional breeding platforms, this process has also been used directly as a means of stimulating improvements in agronomic traits, and at least 22 food crop cultivars, including rice, strawberry and potato, have been derived from such an approach and registered to date (FAO/IAEA mutant variety database, https://mvd.iaea.org/). Similarly, for almost a century, genetic variability within species has also been rapidly increased through the treatment of plant tissue with physical (e.g., ionizing radiation), chemical (e.g., alkylating agents) or biological (e.g., transposon-mediated) mutagens (reviewed by Jankowicz-Cieslak and Till, Citation2015), followed by selection. As is the case with traditional breeding methods, these platforms can be based solely on phenotypic selection, or can make use of molecular-assisted selection through a process termed Targeting Induced Local Lesions in Genomes (TILLING), whereby large mutant populations can be directly screened for mutations in a desired gene(s) (Kumar et al., Citation2017).
Ionizing radiation, such as X-rays, gamma-rays, fast-neutrons and heavy ion beams, can damage DNA directly or indirectly through the production of hydroxyl radicals, which then target DNA molecules (Jo and Kim, Citation2019). The spectrum and frequency of mutations observed with irradiation is highly dependent on many factors, including the type, dose and linear energy transfer (LET) of the radiation, as well as the plant tissue type used and treatment conditions (Jo and Kim, Citation2019). High-LET forms of radiation, such as fast neutrons and heavy ion beams, tend to lead mainly to the production of DSBs, although single-strand breaks (SSBs) and other DNA lesions can also be evident (Hada and Georgakilas, Citation2008). Conversely, low-LET forms of radiation such as X-rays and gamma-rays typically result in a higher proportion of SSBs and base or sugar lesions (Wallace, Citation1998). The repair of this DNA damage results in a range of mutations, including deletions ranging in size from tens to millions of base pairs, SNVs, and rearrangements at break sites (e.g., Shirley et al., Citation1992; Ashikari et al., Citation2002; Morita et al., Citation2009), leading to potentially novel alleles that may not have been available previously in the breeding population. Due to the prevalence of DSBs, which are repaired mainly through the error-prone NHEJ, high-LET irradiation tends to elicit a greater proportion of indels than SNVs compared to low-LET irradiation (Jo and Kim, Citation2019). Furthermore, high-LET irradiation also typically leads to larger deletions and more complex DNA rearrangements than low-LET irradiation, which may derive, at least in part, through the action of an alternative end-joining repair pathway in these instances (Hirano et al., Citation2015). In rice, tens up to hundreds of thousands of SNVs and indels, along with 0 to approximately 11,000 structural variations (SVs), have been observed per mutant plant following treatment with ionizing radiation (; e.g., Cheng et al., Citation2014; Zhenga et al., Citation2017; Li, Shimizu, et al., Citation2019; Yang et al., Citation2019; Hwang et al., Citation2020).
The most commonly used chemical mutagen in plant breeding is EMS, which selectively alkylates guanine bases, resulting in mispairing with thymine and primarily inducing G/C to A/T transitions (Greene et al., Citation2003). Indeed, chemical mutagens in general predominantly provoke single base substitutions (Olsen et al., Citation1993; Xin et al., Citation2008; Satoh et al., Citation2010) and often lead to mutation rates that are higher than those observed following irradiation. While EMS-mutagenized diploid plants often exhibit thousands of SNVs per mutant genotype (; e.g., Mohd-Yusoff et al., Citation2015; Shirasawa et al., Citation2016; Sevanthi et al., 2018), polyploid species can tolerate higher numbers of mutations as a result of the presence of homologous gene copies, which serve to buffer mutations in essential genes. While this means that the populations required for mutation saturation can be much smaller than with diploid species, recessive mutations in single homologs are less likely to exhibit a phenotype in early generations (Parry et al., Citation2009). In wheat and B. napus, for example, EMS-induced mutations occur every 23–51 kb (Slade et al., Citation2005; Wang et al., Citation2008; Dong et al., Citation2009; Uauy et al., Citation2009; Chen et al., Citation2012), which can lead to tens of thousands to hundreds of thousands of mutations throughout the genome of each mutant plant (e.g., Wang et al., Citation2008; Hussain et al., Citation2018).
Differences in chemically/physically-induced mutational frequencies noted among studies are likely due to species-specific disparities in DSB repair (Kirik et al., Citation2000), variations in irradiation/treatment conditions (Li, Shimizu, et al., Citation2019) and/or evaluation method used. In any case, these approaches invariably result in large numbers of unpredictable mutations, most of which will be neutral, and some of which will lead to positive and negative characteristics, respectively. Therefore, as is the case with traditional crosses, laborious and time-consuming backcrossing and selection steps are required to segregate out as many of the unknown deleterious mutations as possible during cultivar development (reviewed by Troadec and Pagès, Citation2019). In addition, this mutational load poses a serious challenge in terms of identifying causal mutations resulting in particular phenotypes, and therefore the genetic alterations that have been elicited in even commercially released varieties derived from mutation breeding frequently remain unknown. Despite the widespread genetic changes that result from chemical and physical mutagenesis, these breeding strategies have an exceptionally strong record of safe use and have led to the release of approximately 3,300 varieties to date, encompassing more than 100 major cereal, grain legume, oil, fruit, nut and herb species (FAO/IAEA mutant variety database). This includes several well-known cultivars, such as Rio Red and Star Ruby grapefruit, Shamrock apple, Illini Super Sweet corn and various high-yielding semi-dwarf rice varieties.
IV. Targeted genetic manipulation via the insertion of exogenous DNA
Conventional breeding approaches are complicated by linkage drag, a lack of natural genetic diversity and the polyploid nature of many crop species, which buffers spontaneous and induced mutations. In an attempt to overcome these issues, recombinant DNA cassettes were successfully transferred into plant cells using A. tumefaciens in the early 1980s (e.g., Hernalsteens et al., Citation1980; Hooykaas-Van Slogteren et al., Citation1984). This paved the way for new breeding platforms that allowed for the introduction of a specific desirable gene(s) that may not be available within breeding populations, without the co-transfer of the undesirable alleles that typically occurs during traditional breeding (reviewed by Saini et al., Citation2020). By far the most common form of this technology involves the transfer of a genetic cassette (including a promoter, coding sequence and transcriptional terminator) from an unrelated organism(s) in a process termed transgenesis, and this has formed the basis of what we now consider “GM” crops. While the thought of introducing a gene from one organism to another is typically considered to be specific to “GM” crops, such a phenomenon also occurs through the wide crosses between distinct species and even genera that often form part of traditional breeding programs in many crops (e.g., Alvarez and Guzmán, Citation2018), as well as through spontaneous horizontal gene transfer, which has, for example, led to the presence of Agrobacterium-derived genes in cultivated sweet potato (e.g., Kyndt et al., Citation2015; Fang et al., Citation2017). Therefore, the term “GM” only encompasses a subset of what are technically transgenic plants - specifically those where recombinant DNA was used to elicit the transfer of genetic material (Duensing et al., Citation2018).
Since the development of the first transgenic plants, where a chosen coding sequence was expressed to produce a protein leading to the trait of interest, or an antisense version of the coding sequence was expressed to down-regulate expression of the corresponding gene, several other iterations of this technology have been adapted for use in crop improvement. RNA interference (RNAi)-mediated gene silencing for example, is achieved through the introduction of a genetic cassette expressing a small double stranded RNA (dsRNA) hairpin that is homologous to a particular target gene. In cases where the homology occurs within a coding region of the target gene, the dsRNA triggers the post-transcriptional cleavage of the cognate mRNA, whereas targeting of a promoter region can suppress transcription, which both lead to gene silencing (Sinha, Citation2010). Unlike transgenic plants involving the heterologous expression of a chosen gene, marker-free plants derived from RNAi do not result in the production of a novel protein. To date, several crop cultivars possessing RNAi-mediated traits resulting from the down-regulation of endogenous genes, including high oleic acid and low linoleic acid soybean (Vistive® Gold), low lignin alfalfa (HarvXtra®), and nonbrowning apple (ArcticTM apple), have been commercialized (Barros et al., Citation2019; Schiemann et al., Citation2019). A related technology termed host-induced gene silencing (HIGS), whereby the dsRNA specifically targets essential genes in pathogens or pests, has also been used to stimulate disease/insect resistance in host plants (Nowara et al., Citation2010; Mamta et al., Citation2016; Tiwari et al., Citation2017).
Cisgenics, on the other hand, involves the transfer of a gene (including its native promoter and transcriptional terminator) or gene fragment from the species itself or a closely related plant into the recipient crop genotype. Similarly, intragenics also requires that only genetic material from related species capable of sexual hybridization is transferred into the recipient plant, but new combinations of promoters, genes and terminators are utilized (Holme et al., Citation2013). These two platforms allow the exploitation of the same gene pool that is available for traditional breeding purposes. However, unlike the traditional breeding practice of crossing through sexual hybridization, cisgenics/intragenics allows the introduction of the desirable gene without any associated linkage drag, and can easily be used to stack multiple traits into a single cultivar, thus enhancing the pace and efficiency of breeding substantially. This approach can be especially useful in plants that propagate vegetatively, such as apple, potato and banana. While cisgenics/intragenics has been used to generate potato resistant to late blight (Haverkort et al., Citation2009), apple resistant to fire blight (Kost et al., Citation2015), and barley with improved phytase activity (Holme et al., Citation2012), for example, these have yet to reach market. Progress is being made in this field, however, and intragenic potatoes exhibiting multiple traits including low acrylamide and resistance to bruising have now been commercialized (Innate® and Innate® Generation 2; Waltz, Citation2015).
The generation of transgenic, cisgenic and RNAi plants often (but not always) requires plant tissue culture for the introduction of the genetic cassette, which can induce heritable genetic changes through somaclonal variation. The introduction of an exogenous DNA construct will also interrupt an endogenous sequence of DNA, which typically occurs in a random manner and can be genic or inter-genic in nature. In the case of Agrobacterium-mediated introduction of the exogenous DNA cassette, integration takes place at existing DSBs through MMEJ repair (van Kregten et al., Citation2016). During this process, small deletions ranging from 11- to 100-bp are often elicited at the site of integration (Forsbach et al., Citation2003), along with the insertion of short filler DNA sequences at T-DNA junctions (Wei et al., Citation2016; Schouten et al., Citation2017). Novel SNVs are rare in transgenic plants, and it has been suggested that most identified substitutions were likely due to spontaneous or somaclonal mutations rather than transformation (Kawakatsu et al., Citation2013; Anderson et al., Citation2016; Park et al., Citation2019). However, it is possible that this could differ based on the plant species, SNV calling method and threshold, tissue culture conditions, transformation method, adjustments for intra-cultivar heterogeneity, and the sample number (Anderson et al., Citation2016). Similarly, structural variations in transgenic plants derived from Agrobacterium-mediated transformation have also been found to be rare, occurring at rates that are two orders of magnitude lower than those observed between different cultivars of the same species (Anderson et al., Citation2016). When they do occur, such events have been suggested to result due to microhomology across DNA break points (Anderson et al., Citation2016). Biolistic transformation of plants through particle bombardment, on the other hand, can provoke widespread DSBs within the genome and fragmentation of the introduced DNA, although this is not always the case. This can induce relatively large numbers of small regions of the introduced DNA being integrated throughout the genome, along with structural variations including chromosomal truncations, duplications, rearrangements or large deletions that may be comparable in frequency to those induced via irradiation with fast neutrons (Liu et al., Citation2019). While insertion at multiple loci can also occur with both Agrobacterium-mediated and biolistic transformation (Olhoft et al., Citation2004; Liu et al., Citation2019), such events, along with more complex rearrangements, are typically identified through molecular screening and/or poor agronomic performance (Clark and Krysan, Citation2010), and these genotypes are discarded.
V. Precision editing of plant genomes
Meganucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and CRISPR/Cas all comprise distinct genome editing tools for site-directed mutagenesis that can be used to elicit DSBs at precise, predefined genomic sites in many organisms, including plants (e.g., Bortesi and Fischer, Citation2015; Mohanta et al., Citation2017; Songstad et al., Citation2017; Graham et al., Citation2020). As is the case with other sources of DSBs, those induced by genome editing machinery are also repaired by the plant’s own DNA repair mechanisms, which often provoke the creation of a mutation at the targeted locus. Genome editing-induced DSBs are typically repaired using either error-prone NHEJ, which is the predominant route in the context of genome editing and typically leads to the production of small insertions or deletions (indels) that disrupt the targeted gene, or HDR processes (Vats et al., Citation2019). Due to the fact that these mutations are incurred through inherent DSB-repair mechanisms, many mutations induced via genome editing are indecipherable from those occurring spontaneously or through conventional breeding methods; however, the pace and precision with which they can be achieved is incomparable.
While the general mechanisms driving all of these site-specific nuclease-mediated genome editing platforms are similar, as are their genetic outcomes, CRISPR/Cas has become the most popular as of late due to its user-friendliness, time-effectiveness, low cost, remarkable versatility, and facility for targeting multiple genes/gene copies simultaneously (Braatz et al., Citation2017; van de Wiel et al., Citation2017; ). This affords remarkable value for the breeding of many polyploid crop species, which are difficult to improve using conventional methods, and allows the generation of a range of heterozygous, monoallelic and biallelic mutants, and a consequent allelic series of phenotypes, in the first generation – something that is typically not possible using conventional breeding methods.
Figure 4. CRISPR/Cas applications in plants that do not require the extended presence of a transgene. Red base pairs denote random indel mutations at the targeted site, whereas green base pairs represent specific alterations made at targeted loci. Cas, CRISPR-associated protein; dCas, catalytically dead Cas; DNA, deoxyribonucleic acid; HDR, homology-dependent repair; nCas, Cas nickase; NHEJ, nonhomologous end-joining; PAM, protospacer adjacent motif; pegRNA, prime editing guide RNA; RNA, ribonucleic acid; sgRNA, single guide RNA; uORF, upstream open reading frame.
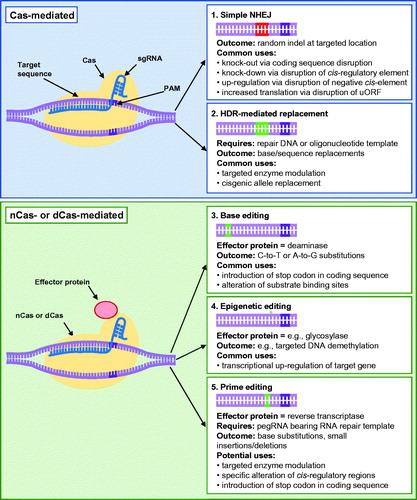
The potential of CRISPR/Cas as a tool for genome editing was first demonstrated in 2012 (Jinek et al., Citation2012), with successful targeted mutagenesis achieved in a variety of other organisms, including plants, shortly thereafter (e.g., Feng et al., Citation2013; Li, Norville, et al., Citation2013; Nekrasov et al., Citation2013; Shan et al., Citation2013; Xie and Yang, Citation2013). This technology was derived from an RNA-mediated adaptive immune system in bacteria and archaea, which protects mainly against invading phages by integrating small DNA fragments from the virus into their own genome, and then transcribing them into short RNAs that act as recognition signals to prevent subsequent attacks through the cleavage of homologous viral DNA by Cas proteins (Bortesi and Fischer, Citation2015). In its simplest form as a genome editing tool, the CRISPR/Cas platform comprises a Cas nuclease, which is responsible for inducing the DSB, along with a small, customizable, noncoding guide RNA (gRNA) that specifically directs Cas to the selected genomic locus. Most often, the gRNA takes the form of a chimeric single gRNA (sgRNA) that consists of an approximately 20-nt CRISPR RNA (crRNA), which is complementary to a target region (protospacer) and acts as a guide for the Cas protein, as well as a fixed trans-activating CRISPR-RNA (tracrRNA) that is required for the recruitment of Cas. In order for Cas enzymes to successfully cleave DNA, the crRNA component must be designed to anneal immediately upstream of a short protospacer adjacent motif (PAM). In the case of systems based on Cas9 from Streptomyces pyogenes, which is currently the most widely used Cas protein for genome editing in plants, the PAM sequence consists of 5′-NGG-3′ (Jinek et al., Citation2012) and less frequently ‘NAG’ (Meng et al., Citation2018), with cleavage typically occurring approximately 3-nt upstream of this site (Jinek et al., Citation2012). However, Cas9 enzymes have been engineered to recognize different PAMs (e.g., Gao et al., Citation2017; Hu et al., Citation2018), and other types of Cas proteins with distinct PAM sequences and cleavage characteristics have also been successfully used to elicit mutations at targeted sites in plants. For example, the Cas12a enzyme (previously known as Cpf1), which requires a T-rich PAM sequence and elicits cleavage distally and downstream of the PAM, has been gaining popularity recently (Zetsche et al., Citation2015).
Most commonly, Cas and sgRNA editing components are introduced into plants through the use of a plant binary vector and A. tumefaciens-mediated transformation or particle bombardment, which like transgenic/cisgenic/RNAi technology, results in the stable insertion of a transgenic cassette into the plant’s genome and often requires the use of tissue culture steps (Sandhya et al., Citation2020). Although this approach initially generates plants possessing “foreign” DNA, the transgene is unlinked to the edit and in instances where its presence is not required long-term, it can therefore simply be segregated out while maintaining the targeted edit. As a means of avoiding the production of stable transgenic lines altogether, transgene-free edited genotypes have also been produced through the transient introduction of DNA or RNA encoding Cas/sgRNA into plant cells, followed by tissue-culture mediated plant regeneration (e.g., Zhang et al., Citation2016; Andersson et al., Citation2017). However, editing frequencies using this approach tend to be lower than with the stable integration of a transgenic cassette, and in the case of the transient introduction of DNA, the potential for fragments to be incorporated into the genome exists (Andersson et al., Citation2017). The direct introduction of ribonucleoprotein (RNP) complexes comprising in vitro assembled sgRNA and Cas protein into plant cells, followed by the regeneration of edited genotypes, has also been gaining momentum as an alternative delivery method in recent years (Woo et al., Citation2015; Liang et al., Citation2017; Andersson et al., Citation2018). This approach circumvents the use of exogenous DNA altogether, and the RNPs simply degrade within a short period of time (Kim et al., Citation2014). While this strategy certainly holds promise, its feasibility is currently constrained by our ability (or lack thereof) to regenerate whole plants from protoplasts or other tissue types, which remains very challenging in many agronomically important crop cultivars. Indeed, this limitation can hinder the application of CRISPR/Cas in general (Atkins and Voytas, Citation2020), and the development/optimization of genotype-independent transformation protocols for these plants are underway in many instances and will be of paramount importance for the successful implementation of such breeding platforms in mainstream elite cultivars, as well as underutilized crop species, going forward.
A. CRISPR platforms not requiring the prolonged presence of a transgene
In terms of CRISPR/Cas-based tools that do not require the extended presence of a transgene, NHEJ-driven site-specific mutagenesis within coding regions is the most commonly used approach in the context of plant breeding to date. The aim of this strategy is for the introduced indel to disrupt gene function by causing the production of a null allele, which knocks out gene function. As such, potential target genes for breeding using this approach are limited to those that have a negative role with respect to the particular trait chosen for improvement. This strategy has been used to successfully improve a large number of desirable traits, such as those related to seed quality, nutritional status and disease resistance, for example, in a wide range of crop species (; e.g., Ma et al., Citation2015; Morineau et al., Citation2017; Nekrasov et al., Citation2017; Li, Wang, et al., Citation2018; Okuzaki et al., Citation2018; Do et al., Citation2019; Karunarathna et al., Citation2020; Subedi et al., Citation2020; Tian et al., Citation2020). Similarly, the knock-down of target gene expression can be achieved by targeting cis-regulatory regions rather than coding sequences using this same technology (e.g., Rodríguez-Leal et al., Citation2017; Zeng, Liu, et al., Citation2020), which allows for the fine-tuning of transcriptional/functional activity, while larger targeted deletions of genomic loci can also be incurred through the use of two sgRNAs simultaneously (e.g., Zhou et al., Citation2014). In addition to its potential use for the improvement of crop species that are already grown extensively, indels generated through CRISPR/Cas NHEJ-mediated targeting also holds promise to advance the de novo domestication of wild or underutilized species by targeting multiple genes implicated in the domestication process, including those involved in plant architecture, flower production, fruit size, seed shattering and yield (Khan et al., Citation2019). Such a feat has been undertaken in stress-tolerant tomato wild relatives (Li et al., Citation2018c; Zsögön et al., Citation2018), rice landraces (Lacchini et al., Citation2020), pennycress (Thlaspi arvense; McGinn et al., Citation2019), and groundcherry (Physalis pruinosa; Lemmon et al., Citation2018).
Table 2. Examples of CRISPR/Cas-mediated improvement of various traits in crop species through targeted gene disruption via the generation of NHEJ-derived indels.
While the knock-out/knock-down of target genes can elicit desirable traits in certain instances, in some cases very little progress has been made as of yet with respect to identifying negative regulators within pathways, and the up-regulation of target gene expression or enhancement of enzyme activity would allow for the targeting of substantially more genes of interest. While achieving such a feat without the lasting presence of a transgene can be technically challenging, several options have shown promise in this area. For example, the disruption of repressor elements within target gene promoters, as has been demonstrated previously in tomato (Rodríguez-Leal et al., Citation2017), or the NHEJ-based mutation of upstream open reading frames within 5′ untranslated regions of a target gene, which has been demonstrated to increase translation of the associated mRNA (Zhang et al., Citation2018), are both valuable options in this area provided the necessary regulatory elements exist in a given gene. NHEJ-mediated mutation of coding sequences is also beginning to be used in plants as a means of inducing directed evolution through the introduction of a Cas protein and a gene-specific sgRNA library (reviewed by Butt et al., Citation2019; Kuang et al., Citation2020), which can allow the identification of mutations that positively modulate the activity of the protein encoded by the target gene, thus increasing genetic diversity and accelerating trait improvement substantially.
Furthermore, various systems have also been developed to take advantage of Cas enzymes in which one or both catalytic domains have been ablated to yield nickase (nCas) or complete deactivation of cleavage function (dCas), respectively. This type of Cas enzyme serves as a scaffold to recruit other effector proteins, which are guided to specific genomic loci by a sgRNA without eliciting a DSB (reviewed by Kumlehn et al., Citation2018). For example, the fusion of dCas to either the catalytic domain of the Arabidopsis REPRESSOR OF SILENCING 1 (ROS1) glycosylase, or a C-terminal tail that is recognized and bound by a separate module containing the catalytic domain of a demethylase, can be used to trigger cytosine demethylation at a targeted location, at least in instances where transcriptional silencing of a gene is directed by DNA methylation. This has been shown to result in transcriptional up-regulation of the associated gene (Gallego-Bartolomé et al., Citation2018; Papikian et al., Citation2019; Devesa-Guerra et al., Citation2020), and since such epigenetic alterations appear to be heritable, it would allow for the transgene to be segregated out (Gallego-Bartolomé et al., Citation2018; Papikian et al., Citation2019) and no alteration in the genome sequence would be incurred. While these tools are not as simple to implement as NHEJ-based CRISPR/Cas knock-outs elicited through mutations within coding sequences, and have therefore not been used to the same extent as of yet, they certainly have the potential to broaden CRISPR/Cas-editing capacity substantially in the future.
Above and beyond knocking-out, down-regulating or up-regulating target genes, it is also possible to obtain more precise edits in some cases. For example, specific edits can be incurred through directed base substitutions, which can allow for the generation of stop codons and the rational improvement of protein function, or allele replacements. One way in which this can be achieved is by exploiting the error-free HDR mechanism (involving Cas, sgRNA(s) and a donor repair template DNA or oligonucleotide template), which has been used to achieve nucleotide or allele replacements (e.g., Sauer et al., Citation2016; de Pater et al., Citation2018; Li, Zhang, et al., Citation2018) in plants. However, the efficiency of this system is far lower than NHEJ-based editing, which has restricted its application thus far. The size of the replacement and organism from which the new allele was derived would also be important considerations as these factors would influence downstream regulatory decisions if the replacement was deemed “transgenic”.
To further facilitate specific nucleotide-level changes, several NHEJ-mediated alternatives have been developed in recent years to yield base substitutions. One such platform includes CRISPR/Cas-mediated cytosine and adenine base-editor systems (CBEs and ABEs) comprising either nCas or dCas fused to a cytidine or adenosine deaminase (e.g., Zong et al. Citation2017; Li, Zong, et al., 2018), which allow C-to-T or A-to-G substitutions in plants, respectively. Such a base editing approach has been used to successfully attain a specific C to T transition in genes encoding acetolactate synthase or acetyl-coenzyme A carboxylase, which confers resistance to particular herbicides in rice (Shimatani et al., Citation2017; Li, Zong, et al., 2018), watermelon (Tian et al., Citation2018), wheat (Zhang, Liu, et al., Citation2019), and maize (Li et al., Citation2020), for example. However, the widespread use of base editing has been limited due to the fact that the targeted bases must be present within a relatively small window relative to the PAM sequence, substitutions are limited to C-to-T and A-to-G transitions, and all C or A bases, respectively, within the window will be converted (Shimatani et al., Citation2017). Various efforts are currently underway to develop novel base editor systems using Cas9 variants that recognize alternative PAMs (e.g., Hua et al., Citation2019; Nishimasu et al., Citation2018; Wang et al., Citation2019), as well as those with altered catalytic windows (Jiang et al., Citation2018), which could expand the potential utility of this platform in crops. Prime editing, which is one of the newest additions to the CRISPR/Cas toolbox, could also provide a more widely applicable alternative. This technology allows for the generation of small insertions (up to approximately 15 nucleotides) and deletions (up to approximately 40 nucleotides), as well as all possible single nucleotides substitutions, and even the replacement of three consecutive nucleotides (Lin et al., Citation2020) at a targeted locus. This is achieved through the use of nCas fused to an engineered reverse transcriptase, along with a prime editing sgRNA that not only specifies the target site, but also acts as a template for the chosen edit (Anzalone et al., Citation2019). This approach has recently been shown to be successful in both wheat and rice (Lin et al., Citation2020), and while editing efficiencies were relatively low in this case, technical improvements are almost certainly on the horizon.
B. CRISPR platforms requiring the prolonged presence of a transgene
In addition to all of these CRISPR/Cas-derived edits that can be achieved in a manner that allows the ultimate development of transgene-free genotypes, there are also a number of other functionalities that would require the long-term persistence of the CRISPR/Cas transgene. For example, genes can be repressed or activated through the use of dCas9 paired with an activator (e.g., VP64) or repressor (e.g., SRDX) domain, along with a sgRNA specific to the target sequence (e.g., Piatek et al., Citation2015; Li, Wang, et al., Citation2019; Papikian et al., Citation2019), in order to elicit the adjustment of target gene expression. In addition, Cas13 systems are guided by a single crRNA and cleave single-stranded RNA targets rather than double-stranded DNA, which allows for the knock-down of gene expression and is particularly useful for eliciting resistance to viruses (Aman et al., Citation2018; Zhan et al., Citation2019). While these platforms have their uses, the fact that they necessitate the presence of a transgene currently complicates their commercial implementation in many countries.
C. Potential for off-target effects in plants
In terms of the genetic changes that are incurred through genome editing platforms, it is clear that there will not be a one-size fits all outcome given the vast array of applications. In the case of simple NHEJ-mediated editing of coding or cis-regulatory regions, random, short indels are typically seen at the precise target site in plants. Beyond targeted genetic alterations, there was initially some alarm with respect to the potential for unintended mutations at off-target sites in mice (Schaefer et al., Citation2017); however, it has since been proposed that such variation was more likely the result of preexisting genetic variation rather than the process of genome editing (e.g., Lareau et al., Citation2018), and this paper was later retracted. Furthermore, similar findings have not been corroborated in animal systems subsequently (Iyer et al., Citation2018; Dong et al., Citation2019; Thomas et al., Citation2019). A prevalence for off-target effects stemming from CRISPR/Cas technology also does not appear to be the case in plants, and data are now accumulating from experiments assessing high probability off-target sites (biased approach), as well as using whole genome sequencing (unbiased approach), which indicates that if present, CRISPR/Cas-derived off-target mutations are very rare, and in many cases cannot be detected whatsoever (; e.g., Zhang, Zhang, et al., Citation2014; Peterson et al., Citation2016; Nekrasov et al., Citation2017; Feng et al., Citation2018; Tang et al., Citation2018; Lee et al., Citation2019; Li, Manghwar, et al., Citation2019; Young et al., Citation2019).
In keeping with this, potential off-target mutations resulting from NHEJ-based, indel-producing, CRISPR/Cas systems are very unlikely to occur at genomic sites without sequence homology to the target site in plants. This is because the specificity of sgRNAs are highly dependent on the so-called “seed region” of crRNAs, which in the case of Cas9, encompasses approximately 8–12 nucleotides upstream of the PAM. Mismatches of 2 or more nucleotides in this sequence of the protospacer generally prevent Cas9 cleavage (Tang et al., Citation2018; Young et al., Citation2019; Gerashchenkov et al., Citation2020). This means that possible off-target mutations are highly predictable and can very easily be minimized through the careful selection of target sites. Several freely accessible web-based tools are now available to assist with crRNA design, which allows the in silico assessment of sgRNA specificity to be carried out prior to in planta analyses (e.g., Bae et al., Citation2014; Lei et al., Citation2014; Xie et al., Citation2014; Michno et al., Citation2015; Minkenberg et al., Citation2019). In addition, the use of truncated gRNAs (Fu et al., Citation2014), RNP delivery (Hahn and Nekrasov, Citation2019), paired Cas9 nickases with paired sgRNAS (Mikami et al., Citation2016), and the fusion of dCas to the FokI nuclease (Guilinger et al., Citation2014) can also increase specificity of CRISPR/Cas systems. The use of alternative Cas enzymes has also been shown to reduce off-target effects, and various high-fidelity Cas9 variants have been engineered to this effect in recent years (Zhong et al., Citation2019; Zeng, Li, et al., Citation2020). However, given their tendency to exhibit relatively low activities, along with the high level of specificity provided by traditional Cas9 enzymes in plants, such an approach will likely be unnecessary in crop applications.
In the case of other CRISPR/Cas platforms, the prevalence of off-target effects is slightly more variable, and depends upon the particular technology used. For example, the use of HDR to achieve allele replacement would result in what is essentially a transgenic or cisgenic plant with insertion of the “foreign” DNA occurring at a known, predetermined locus. Furthermore, in what appears to be an unusual case with respect to CRISPR/Cas-based platforms, CBEs have been found to result in the production of unexpected mutations in rice, typically in the form of C-to-T substitutions (Jin et al., Citation2019). In this instance, unlike other CRISPR/Cas platforms where rare off-target mutations are predictable based on sequence homology, mutations occurred in a more random manner. This phenomenon appears to be specific to CBEs, because off-target effects were not noted with ABEs (Jin et al., Citation2019).
Therefore, the vast majority of CRISPR/Cas platforms assessed thus far in plants appear to be highly precise, eliciting very few, if any, off-target effects from the editing technology itself. However, most genome editing endeavors in plants currently involve plant regeneration via tissue culture, and as such, mutations that are equivalent to those observed using somaclonal variation as a conventional breeding technique will be apparent in the resulting genotypes. In agreement with this, the vast majority of mutations in CRISPR/Cas-derived plants that have been found to occur at loci other than those targeted were the result of background mutations that were incurred during seed amplification (spontaneous mutations) or tissue culture (somaclonal mutations), rather than from the editing components themselves (Tang et al., Citation2018; Li, Manghwar, et al., Citation2019).
VI. Comparison of the possible unanticipated “risks” among breeding techniques
There has been much discussion concerning the possibility of unintended risks associated with crop improvement, and for the most part this consideration has been rather specific to crops derived using biotechnological approaches, such as the introduction of an exogenous DNA cassette or genome editing. This focus on biotechnology-derived genotypes fuels concerns regarding the safety of the novel trait itself, as well as apprehension that any new genetic variant might potentially alter genes or pathways in a manner that could lead to the production of a harmful by-product. What should instead be at the forefront of such debates is that unexpected/unknown genetic alterations are unavoidable and observable in every crop, regardless of the breeding technology (Council for Agricultural Science and Technology [CAST], 2018). These even occur spontaneously from one generation to the next, and are an absolute requisite for evolution itself, which means that they have laid the foundation for plant survival and adaptation throughout history (Weng et al., Citation2019). Furthermore, since genetic variation in the form of mutations that have either been incurred spontaneously over time or through biotechnological means are the basis for achieving alterations in traits, the development of improved crop cultivars would not be possible without it.
As mentioned previously, it is typical to see millions of SNVs and indels between individuals of two different conventionally bred cultivars of the same crop species, as well as thousands of differences in terms of the presence/absence of particular genes (e.g., Anderson et al., Citation2016). Even within cultivars, it is unlikely that two plants in a field will be genetically identical (e.g., Haun et al., Citation2011). This is in comparison to the very small number of off-target mutations that might be elicited through more modern breeding technologies, such as genome editing. In any case, despite the considerable genetic differences between conventionally bred plants, virtually none of which are anticipated or identified during the breeding process, these cultivars are highly similar phenotypically, typically only differ in a handful of agronomic traits, and have a very long history of safe use.
Plants have an extraordinary capacity to withstand major genetic changes without exhibiting detrimental effects, and even in instances where high loads of SNVs or deletions of large chromosomal regions have been incurred, drastic changes in phenotypes are often not observed (e.g., Bolon et al., Citation2011; Sevanthi et al., 2018). This is because in order for a phenotype to arise due to a mutation, it typically must occur within either a coding sequence or cis-regulatory region to have an effect. The occurrence of genic regions varies from species to species, but in barley, rice and maize, for examples, they only account for approximately 10–25% of the genome (Barakat et al., Citation1997; Messing et al., Citation2004), which means that the vast majority of mutations (spontaneous and induced) occur outside of the gene space (e.g., Tock and Henderson, Citation2018). Even when mutations do occur within a gene, many are silent or missense mutations (Cooper et al., Citation2008) that do not change the level, activity or function of the encoded protein. Furthermore, since high levels of gene redundancy exist in plant genomes, and particularly in many crop species due to their polyploid nature (Adams and Wendel, Citation2005; Comai, Citation2005), mutations that inactivate gene function often still do not translate into an unanticipated trait as a result of this buffering effect (Parry et al., Citation2009). This means that the vast majority of potential off-target mutations that may occur through any breeding method would have no effect on plant phenotype. Correspondingly, more than 90% of the induced mutations in gamma irradiated and EMS-mutagenized tomato plants were found to be located in intergenic regions, and only 0.2% of mutations were detrimental (Shirasawa et al., Citation2016).
Even in instances where genetic variation introduced during the breeding process does yield an unanticipated trait, the likelihood of this impacting the safety of the crop in the context of food, feed or the environment is very low. In terms of plant breeding-related changes that may affect food and feed safety, most concerns typically encompass the potential production of toxins, anti-nutrients or allergens (reviewed by Schnell et al., Citation2015). While conventional breeding methods have only very rarely resulted in the development of cultivars with hazardous attributes, in these cases they have almost exclusively led to increases in the levels of well-characterized, endogenous compounds that the plants were already known to produce rather than an entirely novel compound (Steiner et al., Citation2013). For example, certain varieties of potato achieved using conventional breeding methods (e.g., Akeley et al., Citation1968) were withdrawn from both U.S. and Swedish markets as a result of increased glycoalkaloid content (Zitnak and Johnston, Citation1970; Hellenás et al., Citation1995). Despite this, one of these high-glycoalkaloid potatoes (Lenape) continues to be used as a parent in conventional breeding programs due to its other positive attributes (e.g., Bethke, Citation2018). As a result of these issues relating to glycoalkaloid levels in white potatoes, all new varieties (regardless of the breeding method used) are now screened in this context. Similarly, canola breeders monitor levels of glucosinolates prior to commercialization (e.g., Spasibionek et al., Citation2020) – but these types of assessments are specific to the particular crop and relevant compounds (reviewed by Herman and Price, Citation2013). In spite of these few incidences where crop safety has been compromised as a result of conventional breeding, cultivars derived using these methods have a long and proven track record of safety overall.
Given that the incidence of “off-target”/unknown genetic effects resulting from the use of transgenic, cisgenic and RNAi technologies has been found to be very low compared to conventional breeding methods and/or spontaneous mutations that have occurred over time, one would assume that the risk associated with such changes would be negligible. The random insertion of an exogenous DNA cassette, which remains within the plant cultivar and is a cornerstone of these biotechnological breeding approaches, could feasibly lead to the disruption of a gene depending on the genomic site of integration. While this could knock-out the function of an endogenous gene, resulting in an apparent negative agronomic trait, it is unlikely that this would compromise the safety of the plant due to the plasticity of plant genomes (Sevanthi et al., Citation2018). Indeed, instead of the compositional differences that were feared in cultivars derived from the integration of an exogenous DNA cassette (i.e. transgenic, cisgenic and RNAi), almost 25 years worth of research has shown that compositional variation stemming from conventional breeding (which has an overwhelming history of safe use) and environmental factors dwarf any potential changes induced in biotechnology-derived crops (e.g., Esposito et al., Citation2002; Herman et al., Citation2004; McCann et al., Citation2006; Harrigan et al., Citation2009; Herman et al., Citation2010; Ridley et al., Citation2011; Kim et al., Citation2012; Muccilli et al., Citation2020). Transcriptomic, proteomic and metabolomic comparisons of transgenic crops and their nontransgenic counterparts have corroborated these results, with substantial differences being environmentally-induced or noted between conventionally bred cultivars rather than between transgenic vs. conventional lines (e.g., Corpillo et al., Citation2004; Catchpole et al., Citation2005; Lehesranta et al., Citation2005; Baker et al., Citation2006; Baudo et al., Citation2006; Abdeen et al., Citation2010; Coll et al., Citation2010, Kogel et al., Citation2010; Coll et al., Citation2011; Ricroch et al., Citation2011; Gong et al., Citation2012; Wang, Zhang, et al., Citation2018). Since DNA and RNA themselves do not constitute a food safety risk, the greatest theoretical risk associated with these plants then comes from the novel protein(s) derived from the introduced genetic cassettes themselves, and in this case, the level of risk would be specific to the particular protein(s) being produced. These proteins consist of those that elicit the desired trait, and may also include protein(s) associated with a selectable marker if present (typically providing antibiotic or herbicide resistance). Since these proteins are purposefully produced in these plants, assessing their specific risk should be a rather straightforward and simple endeavor.
In the case of NHEJ-mediated disruption or substitution of a few base pairs within a coding/cis-regulatory region using CRISPR/Cas, no exogenous protein would be produced once the transgene is segregated out. Furthermore, with appropriately designed sgRNAs, the genetic change(s) incurred in the target gene and possible off-target mutations are negligible compared to those derived from traditional breeding and naturally occurring diversity in plants (reviewed by Young et al., Citation2019). In addition, unlike conventional breeding methods, any possible off-target mutations are very easy to predict as a result of their homology to the crRNA target site, which means that the identification of off-target alterations can be anticipated, screened for and identified. Furthermore, if such an off-target mutation were recognized, its unlinked nature with the targeted mutation means that its removal via segregation prior to commercialization would be straightforward, which again differs substantially from conventionally bred crops, where the location of “off-target” mutations is typically never known. As such, unanticipated risks from this breeding technology are extremely low compared to other methods due to its highly precise nature (reviewed by Troadec and Pagès, Citation2019). However, as discussed previously, CRISPR/Cas-related technology is expanding at a rapid pace, and the level of risk will likely be case-specific. For example, HDR-mediated insertion of an exogenous DNA cassette from an unrelated organism would be a very different case than simple NHEJ-mediated disruption of a coding sequence, and would instead be more similar to transgenics due to the presence of a new protein product. However, any potential issues related to insertional effects would be minimized with HDR-mediated insertion compared to traditional transgenics as a result of the nonrandom nature of CRISPR/Cas technology.
VII. Breeding platforms and crop regulation
The majority of countries currently require premarket safety assessment of all transgenic and RNAi plants, which are typically shrouded under the umbrella term “GM”, while those derived from conventional breeding techniques such as wide crosses and induced mutagenesis do not require such evaluations (Mejia et al., Citation2017). Unfortunately, such discrepancies in regulatory requirements between new crop varieties that have been developed using conventional and biotechnological breeding approaches have stifled the commercialization of many transgenic crops due to the exorbitant cost and duration of de-regulation (Macnaghten and Habets, Citation2020). While a plethora of traits have been improved in crop species to date using a transgenic approach (e.g., Xue et al., Citation2004; Li, Liu, et al., Citation2013; Usher et al., Citation2015; Paul et al., Citation2017; Singer et al., Citation2019), the market is dominated by two main transgenic traits – herbicide tolerance (e.g., RoundUp Ready) and/or insect resistance (e.g., through the expression of Cry proteins from Bacillus thuringiensis [Bt]). Neither of these traits were designed explicitly to benefit the consumer, but instead were directed at increasing profits for large producer outfits and seed companies (Macnaghten and Habets, Citation2020). In spite of this, the adoption of “GM” crops by growers has been overwhelming, and as of 2018, such crops were grown by 17 million farmers in 26 countries on 192 million hectares of land, with dozens of further countries importing “GM” crops for various purposes (Schiemann et al., Citation2019; Qaim, Citation2020). Indeed, it has been estimated that on average, such crops have increased crop yields by 22% and increased profit gains for farmers by 68% overall (Klümper and Qaim, Citation2014), leading to cumulative increases in gross farm incomes of 225 billion USD between 1996 and 2018 in both developing and industrial countries (Brookes and Barfoot, Citation2020a). To put this in perspective, in order to maintain current agricultural production outputs without “GM” crops, nearly 25 million hectares of additional arable land would need to be cultivated worldwide (Qaim, Citation2016). The adoption of “GM” crops has also led to positive health (Smyth, Citation2020) and environmental impacts, including reductions in the amount of chemicals applied, as well as a decrease in fuel use and facilitation of reduced-tillage practices, which minimizes erosion and greenhouse gas emissions (Brookes and Barfoot, Citation2020b; Qaim, Citation2020).
Although the commercialization of transgenic crops has largely been limited to larger corporations that can afford the financial investment required for de-regulation thus far, this is not the result of any inherent characteristic of transgenic crops, and it is highly unfortunate that the vast majority of traits developed by scientists in noncorporate settings (e.g., Xue et al., Citation2004; Li, Liu, et al., Citation2013; Usher et al., Citation2015; Paul et al., Citation2017) do not make it into farmers’ fields under current regulatory frameworks. While there is no doubt that the safety of new crop cultivars should be at the forefront of deployment, the incredibly broad compositional study costs that are required of transgenic, but typically not conventionally-bred, crops do not appear to provide any benefits in terms of safety since differences in compositional equivalence have never been noted (reviewed by Herman and Price, Citation2013). Instead, it seems evident that focusing on the levels of known, endogenous toxins or allergens that are present in some crops, as is the case with conventionally-bred crops, along with the particular protein product(s) derived from the transgene itself, would be a sensible and prudent alternative.
Until recently, it was fairly clear whether a crop was to be subject to premarket safety assessment in most jurisdictions. However, with the advent of new technologies such as genome editing, which can yield cultivars with genetic alterations that could theoretically also have been generated using traditional breeding techniques, many countries are aiming to modernize their frameworks and/or clarify the regulatory requirements for these crops (for reviews see Metje-Sprink et al., Citation2020; Parrott et al., Citation2020; Schmidt et al., Citation2020). Since the genetic outcomes of genome editing can vary widely depending on the specific platform, one system being used as a template for adapting biosafety legislation is to distinguish between three different types of alterations derived from site-directed nucleases (SDNs). SDN-1 applications involve the production of a DSB and the NHEJ-mediated generation of a random indel or point mutation at a predetermined locus without the use of a repair template or the lasting presence of any foreign DNA. Conversely, SDN-2 involves the use of a short nucleotide template to alter up to 20 nucleotides via HDR, while SDN-3 includes the insertion of larger foreign DNA elements using HDR or NHEJ (reviewed by CAST, Citation2018; Schmidt et al., Citation2020). Unfortunately, as genome editing technology evolves, gaps in this system are becoming apparent. For instance, while base and prime editing could feasibly fall within SDN-1 and SDN-2 classifications, respectively, it is currently unclear where epigenetic editing would be positioned, and the system does not make any distinction between transgenic and cisgenic sequences.
In countries where decisions have been made regarding the regulation of crops derived from genome editing, asynchrony still exists. For example, the U.S. Department of Agriculture (USDA) Animal and Plant Health Inspection Service (APHIS) has stated that plants bearing a single NHEJ-mediated mutation achieved in the absence of an exogenous repair template, base pair substitution, gene introduced from a sexually compatible species or alteration of a gene to correspond with an allele known to be present in the gene pool (i.e. SDN-1, some forms of SDN-2, and cisgenic versions of SDN-3) will not require de-regulation prior to commercial implementation as long as they do not demonstrate pest-related characteristics, because such alterations could have been achieved using traditional breeding techniques. Furthermore, plants bearing a trait(s) with a mechanism of action that is the same as in a previously de-regulated plant in the United States are also exempt (USDA-APHIS Department of Agriculture, Citation2020). Some regulatory authorities in other countries (e.g., Japan, Israel, Australia and several South American countries) have also come to resolutions whereby genome edited plants that do not bear foreign DNA and/or use substantial template to guide genome repair (i.e. SDN-1, or SDN-1 and SDN-2) are not regulated as “GM”. Discussions are currently in progress to introduce comparable regulatory transformations in many other countries as well (reviewed by Schmidt et al., Citation2020; Schulman et al., Citation2020). However, differences exist in terms of what information is required by relevant authorities, and in the case of Japan, for example, it is necessary to indicate the method used, gene modified, trait altered, and possible effects on biological diversity, as well as to provide confirmation that the plant does not possess foreign DNA; all of which would be made available to the public in a manner that respected confidentiality considerations (Ministry of the Environment, Citation2018).
Conversely, Canada’s regulatory system currently differs from those of many other countries, with the trigger for premarket assessment being focused on the trait introduced (for example, as with plants with novel traits; PNTs) rather than the breeding method used to achieve the trait (reviewed by Schmidt et al., Citation2020). Although there are clear benefits to having a product-based system, at present there is a lack of clearly delineated standards that specify what traits would be considered novel. This deficit in clarity surrounding the regulatory process itself, as well as its divergence from regulatory laws in other countries, can be problematic. In addition, while the term “PNT” can be applied to even conventionally-bred cultivars, there is a tendency to assume that such plants are in all cases “GM”, which can complicate trade (reviewed by Eriksson et al., Citation2019).
The European Union and New Zealand, on the other hand, have opted to subject genome edited crops to strict “GM” regulatory laws, regardless of the genetic alteration or trait (Callaway, Citation2018; Schmidt et al., Citation2020; Schulman et al., Citation2020). Such decisions are in no way evidence-based, because crop varieties developed using induced random mutagenesis are not subjected to the same regulatory barriers despite the presence of a large number of unknown genetic alterations (Schulman et al., Citation2020). To further complicate matters, such a law is technically unenforceable since cultivars bearing genome editing-based SDN-1 alterations are indistinguishable from spontaneously occurring variants, or those resulting from conventional breeding (Schmidt et al., Citation2020). Such a regulatory approach will not only lead to a substantial impediment to both innovation and progress in crop breeding in these countries (reviewed by Wolter et al., Citation2019), but could also prove extremely detrimental for farmers, the general public, member states, international trade and the environment (reviewed by Eriksson et al., Citation2019; Lemay and Moineau, Citation2020; Schmidt et al., Citation2020; Schulman et al., Citation2020).
Unfortunately, decisions such as these tend to be based in part on unsubstantiated opinions, politics and misinformation (Macnaghten and Habets, Citation2020), and there is thus a critical need to increase understanding related to biotechnological breeding platforms, as well as public awareness, in order to elicit rational, evidence-based, flexible and trade-facilitative amendments to our current crop regulatory laws as a means of keeping pace with evolving breeding technologies. While several countries appear to be gearing toward regulatory approaches whereby insertions/replacements derived from a compatible source plant, indels, and the modulation of one or a small number of base pairs would not require premarket safety assessment, the question will arise as to precisely how many nucleotide substitutions, sequential or not, can be introduced before it will be considered necessary? What, exactly, constitutes “foreign DNA”? This is especially relevant as it becomes more apparent how much genetic variation has arisen spontaneously and exists among conventionally bred cultivars. While there is no doubt that this threshold will be completely arbitrary, it will be a necessary component of deriving a clear and straightforward regulatory system (reviewed by Custers et al., Citation2018).
Furthermore, although a focus on the scientific bases of genetic alterations and consequent phenotypic outcomes will undoubtedly be a cornerstone of premarket assessments and decision-making by policymakers, it may also be wise to consider wider ethical and socio-economic impacts of subjecting new crop varieties to burdensome and costly premarket safety assessments as a result of the breeding technology used for their development. Unlike potential economic impacts, these considerations have often been left largely unacknowledged in past “GM” safety and risk discourse, which has likely contributed to current controversies surrounding “GM” crops (Macnaghten and Habets, Citation2020).
VIII. Conclusions
For over 10,000 years, humans have been using the genetic diversity engendered by spontaneous mutations and recombination as the basis for “improving” crops to better suit our needs. Traditional breeding methods have been very successful in terms of generating elite crop cultivars that are widely grown today, and such approaches remain a foundation of crop improvement. Although the development of marker-assisted (Collard and Mackill, Citation2008) and genomic (Desta and Ortiz, Citation2014) selection have increased the efficiency of such breeding strategies, germplasm improvement remains a lengthy and laborious endeavor, where linkage drag poses a serious challenge (reviewed by Wolter et al., Citation2019). In addition, since functional diversity is often limited in elite varieties that have undergone various genetic bottlenecks both before and during domestication (Shi and Lai, Citation2015), the outcome of breeding using such methods can be unpredictable at best, and in some cases impossible. Given the rapid changes that are currently occurring in the context of crop demand and climate change, there is a vital need to increase both the pace and precision of breeding, and introduce new genetic variation, through the complementary use of modern techniques. While many such techniques now exist, they are evolving rapidly, and further innovation in this area will provide one piece of the puzzle in terms of achieving food security in the future.
Variation in DNA sequences provides the basis for all breeding approaches, both conventional and biotechnological. However, such changes are not in any way specific to crop breeding since plant genomes are constantly incurring novel mutations, structural variations, transposon insertions and new alleles/genes through processes that are not human-directed (McClintock, Citation1984; Weber et al., Citation2012). Due to the exceptional plasticity of plant genomes, the vast majority of these mutations, as well as those genomic changes that may be elicited through breeding, have an incredibly low potential to lead to a phenotypic alteration that will adversely affect crop safety (reviewed by Schnell et al., Citation2015). While there have been enduring concerns regarding the safety of biotechnologically-derived crops compared to those bred conventionally, these worries have not been substantiated. Similarly, genome edited plants have demonstrated few, if any, off-target mutations stemming from the technology itself.
Although an enormous number of scientific studies have overwhelmingly supported the safety of biotechnologically-derived crops, there remains an undercurrent of anxiety regarding “GM” crops in a substantial proportion of the general public (reviewed by CitationTabei, 2019). Such concerns are largely driven by a growing educational gap in the context of basic biology, genetics and breeding practices, since this has in no way kept pace with the rapid growth in science that has occurred over the last few decades (Kausch et al., Citation2019). These discrepancies translate into a fear of the unknown (CAST, Citation2013), and have contributed to the poor public perception surrounding “GM” crops that are prevalent today despite the fact that very little apprehension is exhibited when the same technologies are used in pharmacological and medical applications (Malyska et al., Citation2016; Cui and Shoemaker, Citation2018). The notion that breeding techniques that make use of “natural” processes to elicit targeted genetic alterations are “unnatural” is nonsensical, as is the perception that a single mutation will pose a large risk when the same mutation mixed with thousands of other unknown mutations will not (reviewed by Schulman et al., Citation2020).
We are at a point where it is imperative that we provide a means of filling in these knowledge gaps and at the same time restructure existing crop regulatory practices to take into consideration the recent accumulation of knowledge regarding the genetic changes that occur spontaneously, as well as those introduced through biotechnology and that have accumulated via conventional breeding. In order for these changes to provide maximal capacity to achieve substantial improvements in crop productivity and quality in a relatively short timeframe, updated legislation should be evidence-based, internationally coherent, and allow flexibility in terms of adapting to continuously emerging technologies (reviewed by Duensing et al., Citation2018). Reducing the regulatory burden of genome edited crops, and potentially also those derived using other technologies such as cisgenesis, will mean an expedited and more cost-efficient path to commercialization. This would allow both academic and public sector research institutes to diversify the array of improved traits to include those with consumer and/or environmental benefits, rather than those solely aimed at generating profits. Indeed, in Argentina, there is evidence that such an approach toward genome edited crops has already led to an increase in the proportion of product applications from the public sector, as well as a wider range of products submitted for review (Schmidt et al., Citation2020). If carried out synchronously across nations, this would lead to a considerable expansion in the innovation and advancement of crop breeding endeavors in a manner that would benefit growers, the general public and the environment, while at the same time easing international trade barriers, which would not only provide economic advantages, but would also help to equalize accessibility in a global context.
Acknowledgments
The authors are grateful for the support provided by Agriculture and Agri-Food Canada (S.D.S., J.D.L., A.B., S.K.), as well as financial support provided by the National Sciences and Engineering Research Council of Canada Collaborative Research and Training Experience (CREATE) program (J.S.). We also acknowledge Isabelle Dépault (AAFC, Senior Trade Policy Analyst, Technical Trade Policy Division) and Étienne Lepage (AAFC, Deputy Director, Science Policy and Partnerships Division) for their indispensable guidance and support.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Abdeen, A., Schnell, J., and Miki, B. 2010. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genom. 11: 1–21.
- Abe, A., Takagi, H., Yaegashi, H., Natsume, S., Utsushi, H., Tamiru, M., and Terauchi, R. 2018. Next-generation breeding of rice by whole-genome approaches. In Rice Genomics, Genetics and Breeding; Sasaki, T. and Ashikari, M., Eds. Singapore: Springer, pp 511–522.
- Abe, F., Haque, E., Hisano, H., Tanaka, T., Kamiya, Y., Mikami, M., Kawaura, K., Endo, M., Onishi, K., Hayashi, T., and Sato, T. 2019. Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 28: 1362–1369. doi:10.1016/j.celrep.2019.06.090
- Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. 2018. Production of high oleic/low linoleic rice by genome editing. Plant Physiol. Biochem. 131: 58–62. doi:10.1016/j.plaphy.2018.04.033
- Adams, K. L., and Wendel, J. F. 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. doi:10.1016/j.pbi.2005.01.001
- Agnieska, A., Bayer, P. E., Barker, G. C., Edger, P. P., Kim, H. R., Martinez, P. A., Chan, C. K. K., Severn-Ellis, A., McCombie, W. R., Parkin, I. A. P., Paterson, A. H., Pires, J. C., Sharpe, A. G., Tang, H., Teakle, G. R., Town, C. D., Batley, J., and Edwards, D. 2016. The pangenome of an agronomically important crop plant, Brassica oleracea. Nat. Commun. 7: 13390.
- Akeley, R. V., Mills, W. R., Cunningham, C. E., and Watts, J. 1968. Lenape: a new potato variety high in solids and chipping quality. Amer. Pot. J. 45: 142–145. doi:10.1007/BF02863068
- Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., Bo, X., Wang, N., Sharif, R., and Wang, P. 2019. CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max. L). BMC Biotechnol. 19: 9. doi:10.1186/s12896-019-0501-2
- Ali, N., Heslop-Harrison, J. S., Ahmad, H., Graybosch, R. A., Hein, G. L., and Schwarzacher, T. 2016. Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance. Heredity. (Edinb) 117: 114–123. doi:10.1038/hdy.2016.36
- Ali, S., Liu, Y., Ishaq, M., Shah, T., Abdullah , Ilyas, A., and Din, I. U. 2017. Climate change and its impact on the yield of major food crops: evidence from Pakistan. Foods 6: 39. doi:10.3390/foods6060039
- Alvarez, J. B., and Guzmán, C. 2018. Interspecific and intergeneric hybridization as a source of variation for wheat grain quality improvement. Theor. Appl. Genet. 131: 225–251. doi:10.1007/s00122-017-3042-x
- Aman, R., Mahas, A., Butt, H., Ali, Z., Aljedaani, F., and Mahfouz, M. 2018. Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses 10: 732. doi:10.3390/v10120732
- Anderson, J. E., Michno, J.-M., Kono, T. J. Y., Stec, A. O., Campbell, B. W., Curtin, S. J., and Stupar, R. M. 2016. Genomic variation and DNA repair associated with soybean transgenesis: a comparison to cultivars and mutagenized plants. BMC Biotechnol. 16: 41. doi:10.1186/s12896-016-0271-z
- Andersson, M., Turesson, H., Nicolia, A., Fält, A. S., Samuelsson, M., and Hofvander, P. 2017. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36: 117–128. doi:10.1007/s00299-016-2062-3
- Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., Samuelsson, M., and Hofvander, P. 2018. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164: 378–384. doi:10.1111/ppl.12731
- Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., Chen, P. J., Wilson, C., Newby, G. A., Raguram, A., and Liu, D. R. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576: 149–157. doi:10.1038/s41586-019-1711-4
- Arai-Kichise, Y., Shiwa, Y., Ebana, K., Shibata-Hatta, M., Yoshikawa, H., Yano, M., and Wakasa, K. 2014. Genome-wide DNA polymorphisms in seven rice cultivars of temperate and tropical japonica groups. PLoS One. 9: e86312. doi:10.1371/journal.pone.0086312
- Arai-Kichise, Y., Shiwa, Y., Nagasaki, H., Ebana, K., Yoshikawa, H., Yano, M., and Wakasa, K. 2011. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol. 52: 274–282. doi:10.1093/pcp/pcr003
- Ashikari, M., Sasaki, A., Kitano, H., Matsuoka, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Datta, S., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G. S., Kitano, H., and Matsuoka, M. 2002. Loss-of-function of a rice gibberellin biosynthetic gene GA20 oxidase (GA20ox-2) led to the rice ‘Green Revolution. Breed. Sci. 52: 143–150. doi:10.1270/jsbbs.52.143
- Asseng, S., Martre, P., Maiorano, A., Rötter, R. P., O'Leary, G. J., Fitzgerald, G. J., Girousse, C., Motzo, R., Giunta, F., Babar, M. A., Reynolds, M. P., Kheir, A. M. S., Thorburn, P. J., Waha, K., Ruane, A. C., Aggarwal, P. K., Ahmed, M., Balkovič, J., Basso, B., Biernath, C., Bindi, M., Cammarano, D., Challinor, A. J., De Sanctis, G., Dumont, B., Eyshi Rezaei, E., Fereres, E., Ferrise, R., Garcia-Vila, M., Gayler, S., Gao, Y., Horan, H., Hoogenboom, G., Izaurralde, R. C., Jabloun, M., Jones, C. D., Kassie, B. T., Kersebaum, K.-C., Klein, C., Koehler, A.-K., Liu, B., Minoli, S., Montesino San Martin, M., Müller, C., Naresh Kumar, S., Nendel, C., Olesen, J. E., Palosuo, T., Porter, J. R., Priesack, E., Ripoche, D., Semenov, M. A., Stöckle, C., Stratonovitch, P., Streck, T., Supit, I., Tao, F., Van der Velde, M., Wallach, D., Wang, E., Webber, H., Wolf, J., Xiao, L., Zhang, Z., Zhao, Z., Zhu, Y., and Ewert, F. 2019. Climate change impact and adaptation for wheat protein. Glob. Chang. Biol. 25: 155–173. doi:10.1111/gcb.14481
- Atkins, P. A. P., and Voytas, D. F. 2020. Overcoming bottlenecks in plant gene editing. Curr. Opin. Plant Biol. 54: 79–84. doi:10.1016/j.pbi.2020.01.002
- Bae, S., Park, J., and Kim, J. S. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 30: 1473–1475. doi:10.1093/bioinformatics/btu048
- Baker, J. M., Hawkins, N. D., Ward, J. L., Lovegrove, A., Napier, J. A., Shewry, P. R., and Beale, M. H. A. 2006. A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol. J. 4: 381–392. doi:10.1111/j.1467-7652.2006.00197.x
- Ballini, E., Berruyer, R., Morel, J.-B., Lebrun, M.-H., Nottéghem, J.-L., and Tharreau, D. 2007. Modern elite rice varieties of the 'Green Revolution' have retained a large introgression from wild rice around the Pi33 rice blast resistance locus. New Phytol. 175: 340–350. doi:10.1111/j.1469-8137.2007.02105.x
- Barakat, A., Carels, N., and Bernardi, G. 1997. The distribution of genes in the genomes of Gramineae. Proc. Natl. Acad. Sci. USA. 94: 6857–6861. doi:10.1073/pnas.94.13.6857
- Bari, V. K., Nassar, J. A., Kheredin, S. M., Gal-On, A., Ron, M., Britt, A., Steele, D., Yoder, J., and Aly, R. 2019. CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 9: 11438. doi:10.1038/s41598-019-47893-z
- Barros, J., Temple, S., and Dixon, R. A. 2019. Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotechnol. 56: 48–54. doi:10.1016/j.copbio.2018.09.003
- Baudo, M. M., Lyons, R., Powers, S., Pastori, G. M., Edwards, K. J., Holdsworth, M. J., and Shewry, P. R. 2006. Transgenesis has less impact on the transcriptome of wheat grain than conventional breeding. Plant Biotechnol. J. 4: 369–380. doi:10.1111/j.1467-7652.2006.00193.x
- Bedő, Z., and Láng, L. 2015. Wheat breeding: current status and bottlenecks. In Alien Introgression in Wheat; Molnár-Láng, M., Ceoloni, C., and Doležel, J., Eds. Cham: Springer, pp 77–101.
- Belfield, E. J., Gan, X., Mithani, A., Brown, C., Jiang, C., Franklin, K., Alvey, E., Wibowo, A., Jung, M., Bailey, K., Kalwani, S., Ragoussis, J., Mott, R., and Harberd, N. P. 2012. Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 22: 136–1315.
- Berger, J. D., Shrestha, D., and Ludwig, C. 2017. Reproductive strategies in Mediterranean legumes: trade-offs between phenology, seed size and vigor within and between wild and domesticated Lupinus species collected along aridity gradients. Front Plant Sci. 8: 548. doi:10.3389/fpls.2017.00548
- Bethke, P. C. 2018. Progress and successes of the specialty crop research initiative on acrylamide reduction in processed potato products. Am. J. Potato Res. 95: 328–337. doi:10.1007/s12230-018-9660-2
- Blakeslee, A. F., and Avery, A. G. 1937. Methods of inducing doubling of chromosome in plants by treatment with colchicine. J. Heredity. 28: 393–411. doi:10.1093/oxfordjournals.jhered.a104294
- Bolon, Y.-T., Haun, W. J., Xu, W. W., Grant, D., Stacey, M. G., Nelson, R. T., Gerhardt, D. J., Jeddeloh, J. A., Stacey, G., Muehlbauer, G. J., Orf, J. H., Naeve, S. L., Stupar, R. M., and Vance, C. P. 2011. Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 156: 240–253. doi:10.1104/pp.110.170811
- Bortesi, L., and Fischer, R. 2015. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33: 41–52. doi:10.1016/j.biotechadv.2014.12.006
- Braatz, J., Harloff, H.-J., Mascher, M., Stein, N., Himmelbach, A., and Jung, C. 2017. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in Polyploid oilseed rape (Brassica napus). Plant Physiol. 174: 935–942. doi:10.1104/pp.17.00426
- Brisson, N., Gate, P., Gouache, D., Charmet, G., Oury, F.-X., and Huard, F. 2010. Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crops Res. 119: 201–212. doi:10.1016/j.fcr.2010.07.012
- Brookes, G., and Barfoot, P. 2020a. GM crop technology use 1996-2018: farm income and production impacts. GM Crops Food. 11: 242–261. doi:10.1080/21645698.2020.1779574
- Brookes, G., and Barfoot, P. 2020b. Environmental impacts of genetically modified (GM) crop use 1996-2018: impacts on pesticide use and carbon emissions. GM Crops Food. 11: 215–241. doi:10.1080/21645698.2020.1773198
- Bruinsma, J. 2009. The resource outlook to 2050. By how much do land, water use and crop yields need to increase by 2050? Presented at the Proceedings of the FAO Expert Meeting on How to Feed the World in 2050, Rome, 24–26 June 2009. FAO.
- Burr, B., Evola, S. V., Burr, F. A., and Beckmann, J. S. 1983. The application of restriction fragment length polymorphism to plant breeding. In Genetic Engineering (Principles and Methods), vol 8; Setlow, J. K. and Hollaender, A., Eds. Boston: Springer, pp 45–59.
- Butt, H., Eid, A., Momin, A. A., Bazin, J., Crespi, M., Arold, S. T., and Mahfouz, M. M. 2019. CRISPR-directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 20: 236–240. doi:10.1186/s13059-019-1680-9
- Calafiore, R., Ruggieri, V., Raiola, A., Rigano, M. M., Sacco, A., Hassan, M. I., Frusciante, L., and Barone, A. 2016. Exploiting genomics resources to identify candidate genes underlying antioxidants content in tomato fruit. Front. Plant Sci. 7: 397. doi:10.3389/fpls.2016.00397
- Callaway, E. 2018. CRISPR plants now subject to tough GM laws in European Union. Nature 560: 16. doi:10.1038/d41586-018-05814-6
- Caplan, A., Herrera-Estrella, L., Inzé, D., Van Haute, E., Van Montagu, M., Schell, J., and Zambryski, P. 1983. Introduction of genetic material into plant cells. Science 222: 815–821. doi:10.1126/science.222.4625.815
- Carlson, P. S., Smith, H. H., and Dearing, R. D. 1972. Parasexual interspecific plant hybridization. Proc. Natl. Acad. Sci. USA. 69: 2292–2294. doi:10.1073/pnas.69.8.2292
- Catchpole, G. S., Beckmann, M., Enot, D. P., Mondhe, M., Zywicki, B., Taylor, J., Hardy, N., Smith, A., King, R. D., Kell, D. B., Fiehn, O., and Draper, J. 2005. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. USA. 102: 14458–14462. doi:10.1073/pnas.0503955102
- Causse, M., Desplat, N., Pascual, L., Le Paslier, M.-C., Sauvage, C., Bauchet, G., Bérard, A., Bounon, R., Tchoumakov, M., Brunel, D., and Bouchet, J.-P. 2013. Whole genome resequencing in tomato reveals variation associated with introgression and breeding events. BMC Genomics. 14: 791. doi:10.1186/1471-2164-14-791
- Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., Sherman, A., Arazi, T., and Gal-On, A. 2016. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17: 1140–1153. doi:10.1111/mpp.12375
- Chen, L., Huang, L., Min, D., Phillips, A., Wang, S., Madgwick, P. J., Parry, M. A. J., and Hu, Y.-G. 2012. Development and characterization of a new TILLING population of common bread wheat (Triticum aestivum L.). PLoS One. 7: e41570. doi:10.1371/journal.pone.0041570
- Cheng, Z., Lin, J., Lin, T., Xu, M., Huang, Z., Yang, Z., Huang, X., and Zheng, J. 2014. Genome-wide analysis of radiation-induced mutations in rice (Oryza sativa L. ssp. indica). Mol. Biosyst. 10: 795–805. doi:10.1039/c3mb70349e
- Ching, A., Caldwell, K. S., Jung, M., Dolan, M., Smith, O. S., Tingey, S., Morgante, M., and Rafalski, A. J. 2002. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. 3: 19. doi:10.1186/1471-2156-3-19
- Clark, K. A., and Krysan, P. J. 2010. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 64: 990–1001. doi:10.1111/j.1365-313X.2010.04386.x
- Coll, A., Nadal, A., Collado, R., Capellades, G., Kubista, M., Messeguer, J., and Pla, M. 2010. Natural variation explains most transcriptomic changes among maize plants of MON810 and comparable non-GM varieties subjected to two N-fertilization farming practices. Plant Mol. Biol. 73: 349–362. doi:10.1007/s11103-010-9624-5
- Coll, A., Nadal, A., Rossignol, M., Puigdomènech, P., and Pla, M. 2011. Proteomic analysis of MON810 and comparable non-GM maize varieties grown in agricultural fields. Transgenic Res. 20: 939–949. doi:10.1007/s11248-010-9453-y
- Collard, B. C. Y., and Mackill, D. J. 2008. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R Soc. Lond. B Biol. Sci. 363: 557–572. doi:10.1098/rstb.2007.2170
- Comai, L. 2005. The advantages and disadvantages of being polyploidy. Nat. Rev. Genet. 6: 836–846. doi:10.1038/nrg1711
- Cooper, J. L., Till, B. J., Laport, R. G., Darlow, M. C., Kleffner, J. M., Jamai, A., El-Mellouki, T., Liu, S., Ritchie, R., Nielsen, N., Bilyeu, K. D., Meksem, K., Comai, L., and Henikoff, S. 2008. TILLING to detect induced mutations in soybean. BMC Plant Biol. 8: 9. doi:10.1186/1471-2229-8-9
- Corpillo, D., Gardini, G., Vaira, A., Basso, M., Aime, S., Accotto, G., and Fasano, M. 2004. Proteomics as a tool to improve investigation of substantial equivalence in genetically modified organisms: the case of a virus-resistant tomato. Proteomics 4: 193–200. doi:10.1002/pmic.200300540
- Council for Agricultural Science and Technology (CAST). 2013. Impact of the Precautionary Principle on Feeding Current and Future Generations. Issue Paper 52. CAST, Ames, IA.
- Council for Agricultural Science and Technology (CAST). 2018. Gene Editing in Agriculture: Methods, Applications, and Governance – A Paper in the Series on the Need for Agricultural Innovation to Sustainably Feed the World by 2050. Issue Paper 60. CAST, Ames, IA.
- Cowling, W. A., Buirchell, B. J., and Falk, D. E. 2009. A model for incorporating novel alleles from the primary gene pool into elite crop breeding programs while reselecting major genes for domestication or adaptation. Crop Pasture Sci. 60: 1009–1015. doi:10.1071/CP08223
- Cui, K., and Shoemaker, S. P. 2018. Public perception of genetically-modified (GM) food: a nationwide Chinese consumer study. NPJ Sci. Food. 2: 10. doi:10.1038/s41538-018-0018-4
- Custers, R., Casacuberta, J. M., Eriksson, D., Sági, L., and Schiemann, J. 2018. Genetic alterations that do or do not occur naturally; consequences for genome edited organisms in the context of regulatory oversight. Front. Bioeng. Biotechnol. 6: 213. doi:10.3389/fbioe.2018.00213
- Darwin, C. 1859. On the Origin of species by Means of Natural Selection, 1st ed., vol 9; J. Murray, Ed. London, UK: W. Clowes and Sons.
- De Pater, S., Klemann, B. J. P. M., and Hooykaas, P. J. J. 2018. True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci. Rep. 8: 3338. doi:10.1038/s41598-018-21697-z
- DeBolt, S. 2010. Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome. Biol. Evol. 2: 441–453. doi:10.1093/gbe/evq033
- Deryng, D., Conway, D., Ramankutty, N., Price, J., and Warren, R. 2014. Global crop yield response to extreme heat stress under multiple climate change futures. Environ. Res. Lett. 9: 034011. doi:10.1088/1748-9326/9/3/034011
- Desta, Z. A., and Ortiz, R. 2014. Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 19: 592–601. doi:10.1016/j.tplants.2014.05.006
- Devesa-Guerra, I., Morales-Ruiz, T., Pérez-Roldán, J., Parrilla-Doblas, J. T., Dorado-León, M., García-Ortiz, M. V., Ariza, R. R., and Roldán-Arjona, T. 2020. DNA methylation editing by CRISPR-guided excision of 5-methylcytosine. J. Mol. Biol. 432: 2204–2216. doi:10.1016/j.jmb.2020.02.007
- Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T. N., Stacey, G., Gillman, J. D., Zhang, Z. J., and Stacey, M. G. 2019. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19. doi:10.1186/s12870-019-1906-8
- Dong, C., Dalton-Morgan, J., Vincent, K., and Sharp, P. 2009. A modified TILLING method for wheat breeding. Plant Genome 2: 39–47. doi:10.3835/plantgenome2008.10.0012
- Dong, Y., Li, H., Zhao, L., Koopman, P., Zhang, F., and Huang, J. X. 2019. Genome-wide off-target analysis in CRISPR-Cas9 modified mice and their offspring. G3 (Bethesda) 9: 3645–3651. doi:10.1534/g3.119.400503
- Driever, S. M., Simkin, A. J., Alotaibi, S., Fisk, S. J., Madgwick, P. J., Sparks, C. A., Jones, H. D., Lawson, T., Parry, M. A. J., and Raines, C. A. 2017. Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Phil. Trans. Royal Soc. B Biol. Sci. 372: 20160374.
- Du, H., Yu, Y., Ma, Y., Gao, Q., Cao, Y., Chen, Z., Ma, B., Qi, M., Li, Y., Zhao, X., Wang, J., Liu, K., Qin, P., Yang, X., Zhu, L., Li, S., and Liang, C. 2017. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 8: 15324. doi:10.1038/ncomms15324
- Du, X.-Z., Ge, X.-H., Zhao, Z.-G., and Li, Z.-Y. 2008. Chromosome elimination and fragment introgression and recombination producing intertribal partial hybrids from Brassica napus x Lesquerella fendleri crosses. Plant Cell Rep. 27: 261–271. doi:10.1007/s00299-007-0452-2
- Duensing, N., Sprink, T., Parrott, W. A., Fedorova, M., Lema, M. A., Wolt, J. D., and Bartsch, D. 2018. Novel features and considerations for ERA and regulation of crops produced by genome editing. Front. Bioengineer. Biotechnol. 6: 79.
- Eriksson, D., Kershen, D., Nepomuceno, A., Pogson, B. J., Prieto, H., Purnhagen, K., Smyth, S., Wesseler, J., and Whelan, A. 2019. A comparison of the EU regulatory approach to directed mutagenesis with that of other jurisdictions, consequences for international trade and potential steps forward. New Phytol. 222: 1673–1684. doi:10.1111/nph.15627
- Esposito, F., Fogliano, V., Cardi, T., Carputo, D., and Filippone, E. 2002. Glycoalkaloid content and chemical composition of potatoes improved with nonconventional breeding approaches. J. Agric. Food Chem. 50: 1553–1561. doi:10.1021/jf010520t
- Evans, D. A. 1989. Somaclonal variation – genetic basis and breeding applications. Trends Genet. 5: 46–50. doi:10.1016/0168-9525(89)90021-8
- Fang, H., Huangfu, L., Chen, R., Li, P., Xu, S., Zhang, E., Cao, W., Liu, L., Yao, Y., Liang, G., Xu, C., Zhou, Y., and Yang, Z. 2017. Ancestor of land plants acquired the DNA-3-methyladenine glycosylase (MAG) gene from bacteria through horizontal gene transfer. Sci. Rep. 7: 9324. doi:10.1038/s41598-017-05066-w
- FAO, IFAD, UNICEF, WFP, and WHO. 2017. The state of food security and nutrition in the world 2017. Building resilience for peace and food security. Rome. http://www.fao.org/3/i7695en/i7695en.pdf.
- Fedak, G., Cao, W., Wolfe, D., Chi, D., and Xue, A. 2017. Molecular characterization of Fusarium resistance from Elymus repens introgressed into bread wheat. Cytol. Genet. 51: 130–133. doi:10.3103/S0095452717020025
- Feng, C., Su, H., Bai, H., Wang, R., Liu, Y., Guo, X., Liu, C., Zhang, J., Yuan, J., Birchler, J. A., and Han, F. 2018. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol. J. 16: 1848–1857. doi:10.1111/pbi.12920
- Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.-L., Wang, Z., Zhang, Z., Zheng, R., Yang, L., Zeng, L., Liu, X., and Zhu, J.-K. 2014. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA. 111: 4632–4637. doi:10.1073/pnas.1400822111
- Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., Cao, F., Zhu, S., Zhang, F., Mao, Y., and Zhu, J.-K. 2013. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. doi:10.1038/cr.2013.114
- Food and Agriculture Organization (FAO). 2017. The future of food and agriculture – trends and challenges. Rome. http://www.fao.org/3/a-i6881e.pdf.
- Forsbach, A., Schubert, D., Lechtenberg, B., Gils, M., and Schmidt, R. 2003. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant. Mol. Biol. 52: 161–176. doi:10.1023/a:1023929630687
- Fu, C.-Y., Liu, W.-G., Liu, D.-L., Li, J.-H., Zhu, M.-S., Liao, Y.-L., Liu, Z.-R., Zeng, X.-Q., and Wang, F. 2016. Genome-wide DNA polymorphism in the indica rice varieties RGD-7S and Taifeng B as revealed by whole genome re-sequencing. Genome. 59: 197–207. doi:10.1139/gen-2015-0101
- Fu, Y., Sander, J. D., Reyon, D., Cascio, M., and Joung, J. K. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. doi:10.1038/nbt.2808
- Gabur, I., Chawla, H. S., Snowdon, R. J., and Parkin, I. A. P. 2019. Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 132: 733–750. doi:10.1007/s00122-018-3233-0
- Gallego-Bartolomé, J., Gardiner, J., Liu, W., Papikian, A., Ghoshal, B., Kuo, H. Y., Zhao, J. M.-C., Segal, D. J., and Jacobsen, S. E. 2018. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA. 115: E2125–E2134. doi:10.1073/pnas.1716945115
- Gan, X., Stegle, O., Behr, J., Steffen, J. G., Drewe, P., Hildebrand, K. L., Lyngsoe, R., Schultheiss, S. J., Osborne, E. J., Sreedharan, V. T., Kahles, A., Bohnert, R., Jean, G., Derwent, P., Kersey, P., Belfield, E. J., Harberd, N. P., Kemen, E., Toomajian, C., Kover, P. X., Clark, R. M., Rätsch, G., and Mott, R. 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 477: 419–423. doi:10.1038/nature10414
- Gao, H., Gadlage, M. J., Lafitte, H. R., Lenderts, B., Yang, M., Schroder, M., Farrell, J., Snopek, K., Peterson, D., Feigenbutz, L., Jones, S., St Clair, G., Rahe, M., Sanyour-Doyel, N., Peng, C., Wang, L., Young, J. K., Beatty, M., Dahlke, B., Hazebroek, J., Greene, T. W., Cigan, A. M., Chilcoat, N. D., and Meeley, R. B. 2020. Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat. Biotechnol. 38: 579–581. doi:10.1038/s41587-020-0444-0
- Gao, L., Cox, D. B. T., Yan, W. X., Manteiga, J. C., Schneider, M. W., Yamano, T., Nishimasu, H., Nureki, O., Crosetto, N., and Zhang, F. 2017. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 35: 789–792. doi:10.1038/nbt.3900
- Gerashchenkov, G. A., Rozhnova, N. A., Kuluev, B. R., Kiryanova, O. Y., Gumerova, G. R., Knyazev, A. V., Vershinina, Z. R., Mikhailova, E. V., Chemeris, D. A., Matniyazov, R. T., Baimiev, A. K., Gubaidullin, I. M., Baimiev, A. K., and Chemeris, A. V. 2020. Design of guide RNA for CRISPR/Cas plant genome editing. Mol. Biol. 54: 24–42. doi:10.1134/S0026893320010069
- Gill, S. S., and Tuteja, N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48: 909–930. doi:10.1016/j.plaphy.2010.08.016
- Golicz, A., Bayer, P. E., Barker, G. C., Edger, P. P., Kim, H., and Martinez, P. A. 2016. The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Comm. 7: 13390.
- Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., Beyene, G., Taylor, N. J., Carrington, J. C., Staskawicz, B. J., and Bart, R. S. 2019. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17: 421–434. doi:10.1111/pbi.12987
- Gong, C. Y., Li, Q., Yu, H. T., Wang, Z., and Wang, T. 2012. Proteomics insight into the biological safety of transgenic modification of rice as compared with conventional genetic breeding and spontaneous genotypic variation. J. Proteome Res. 11: 3019–3029. doi:10.1021/pr300148w
- González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A.-S., Storani, L., Oneto, C. A. D., Hofvander, P., and Feingold, S. E. 2019. Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front Plant Sci. 10: 1649. doi:10.3389/fpls.2019.01649
- Gorbunova, V., and Levy, A. A. 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4: 263–269. doi:10.1016/S1360-1385(99)01430-2
- Gornicki, P., and Faris, J. D. 2014. Rewiring the wheat reproductive system to harness heterosis for the next wave of yield improvement. Proc. Natl. Acad. Sci. USA. 111: 9024–9025. doi:10.1073/pnas.1407956111
- Gouel, C., and Guimbard, H. 2019. Nutrition transition and the structure of global food demand. Amer. J. Agric. Econ. 101: 383–403. doi:10.1093/ajae/aay030
- Graham, N., Patil, G. B., Bubeck, D. M., Dobert, R. C., Glenn, K. C., Gutsche, A. T., Kumar, S., Lindbo, J. A., Maas, L., May, G. D., Vega-Sanchez, M. E., Stupar, R. M., and Morrell, P. L. 2020. Plant genome editing and the relevance of off-target changes. Plant Physiol. 183: 1453–1471. doi:10.1104/pp.19.01194
- Greene, E. A., Codomo, C. A., Taylor, N. E., Henikoff, J. G., Till, B. J., Reynolds, S. H., Enns, L. C., Burtner, C., Johnson, J. E., Odden, A. R., Comai, L., and Henikoff, S. 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740.
- Gross, B. L., and Olsen, K. M. 2010. Genetic perspectives on crop domestication. Trends Plant Sci. 15: 529–537. doi:10.1016/j.tplants.2010.05.008
- Guilinger, J. P., Thompson, D. B., and Liu, D. R. 2014. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32: 577–582. doi:10.1038/nbt.2909
- Hada, M., and Georgakilas, A. G. 2008. Formation of clustered DNA damage after high-LET irradiation: a review. J. Radiat. Res. 49: 203–210. doi:10.1269/jrr.07123
- Hahn, F., and Nekrasov, V. 2019. CRISPR/Cas precision: do we need to worry about off-targeting in plants? Plant Cell Rep. 38: 437–441. doi:10.1007/s00299-018-2355-9
- Hajjar, R., and Hodgkin, T. 2007. The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156: 1–13. doi:10.1007/s10681-007-9363-0
- Hall, A. J., and Richards, R. A. 2013. Prognosis for genetic improvement of yield potential and water-limited yield of major grain crops. Field Crops Res. 143: 18–33. doi:10.1016/j.fcr.2012.05.014
- Harrigan, G. G., Ridley, W. P., Miller, K. D., Sorbet, R., Riordan, S. G., Nemeth, M. A., Reeves, W., and Pester, T. A. 2009. The forage and grain of MON 87460, a drought-tolerant corn hybrid, are compositionally equivalent to that of conventional corn. J. Agric. Food Chem. 57: 9754–9763. doi:10.1021/jf9021515
- Haun, W. J., Hyten, D. L., Xu, W. W., Gerhardt, D. J., Albert, T. J., Richmond, T., Jeddeloh, J. A., Jia, G., Springer, N. M., Vance, C. P., and Stupar, R. M. 2011. The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol. 155: 645–655. doi:10.1104/pp.110.166736
- Haverkort, A. J., Struik, P. C., Visser, R. G. F., and Jacobsen, E. 2009. Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Res. 52: 249–264. doi:10.1007/s11540-009-9136-3
- Hayta, S., Smedley, M. A., Demir, S. U., Blundell, R., Hinchliffe, A., Atkinson, N., and Harwood, W. A. 2019. An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods. 15: 121. doi:10.1186/s13007-019-0540-7
- Hellenás, K.-E., Branzell, C., Johnsson, H., and Slanina, P. 1995. High levels of glycoalkaloids in the established Swedish potato variety magnum bonum. J. Sci. Food Agric. 68: 249–255. doi:10.1002/jsfa.2740680217
- Herman, R. A., and Price, W. D. 2013. Unintended compositional changes in genetically modified (GM) crops: 20 years of research. J. Agric. Food Chem. 61: 11695–11701. doi:10.1021/jf400135r
- Herman, R. A., Phillips, A. M., Collins, R. A., Tagliani, L. A., Claussen, F. A., Graham, C. D., Bickers, B. L., Harris, T. A., and Prochaska, L. M. 2004. Compositional equivalency of Cry1F corn event TC6275 and conventional corn (Zea mays L.). J. Agric. Food Chem. 52: 2726–2734. doi:10.1021/jf049969n
- Herman, R. A., Phillips, A. M., Lepping, M. D., Fast, B. J., and Sabbatini, J. 2010. Compositional safety of event DAS-40278-9 (AAD-1) herbicide-tolerant maize. GM Crops. 1: 294–311. doi:10.4161/gmcr.1.5.14285
- Hernalsteens, J.-P., Van Vliet, F., De Beuckeleer, M., Depicker, A., Engler, G., Lemmers, M., Holsters, M., Van Montagu, M., and Schell, J. 1980. The Agrobacterium tumefaciens Ti plasmid as a host vector system for introducing foreign DNA in plant cells. Nature 287: 654–656. doi:10.1038/287654a0
- Hillman, G. C., and Davies, M. S. 1990. Domestication rates in wild-type wheats and barley under primitive cultivation. Biol. J. Linnean Soc. 39: 39–78. doi:10.1111/j.1095-8312.1990.tb01611.x
- Hirano, T., Kazama, Y., Ishii, K., Ohbu, S., Shirakawa, Y., and Abe, T. 2015. Comprehensive identification of mutations induced by heavy-ion beam irradiation in Arabidopsis thaliana. Plant J. 82: 93–104. doi:10.1111/tpj.12793
- Hirsch, C. D., and Springer, N. M. 2017. Transposable element influences on gene expression in plants. Biochim. Biophys. Acta. Gene Regul. Mech. 1860: 157–165. doi:10.1016/j.bbagrm.2016.05.010
- Holme, I. B., Dionisio, G., Brinch-Pedersen, H., Wendt, T., Madsen, C. K., Vincze, E., and Holm, P. B. 2012. Cisgenic barley with improved phytase activity. Plant Biotechnol. J. 10: 237–247. doi:10.1111/j.1467-7652.2011.00660.x
- Holme, I. B., Wendt, T., and Holm, P. B. 2013. Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 11: 395–407. doi:10.1111/pbi.12055
- Hooykaas-Van Slogteren, G. M. S., Hooykaas, P. J. J., and Schilperoort, R. A. 1984. Expression of Ti plasmid genes in monocotyledonous plants infected with Agrobacterium tumefaciens. Nature 311: 763–764. doi:10.1038/311763a0
- Hoßfeld, U., Jacobsen, H.-J., Plass, C., Brors, B., and Wackernagel, W. 2017. 150 years of Johann Gregor Mendel's "Versuche über Pflanzen-Hybriden"“. Mol. Genet. Genomics. 292: 1–3. doi:10.1007/s00438-016-1254-4
- Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., Zeina, C. M., Gao, X., Rees, H. A., Lin, Z., and Liu, D. R. 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556: 57–63. doi:10.1038/nature26155
- Hua, K., Tao, X., and Zhu, J. K. 2019. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 17: 499–504. doi:10.1111/pbi.12993
- Hussain, M., Iqbal, M. A., Till, B. J., and Rahman, M. U. 2018. Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLoS One. 13: e0201918. doi:10.1371/journal.pone.0201918
- Hwang, S.-G., Lee, S. C., Lee, J., Lee, J. W., Kim, J.-H., Choi, S. Y., Kim, J.-B., Choi, H.-I., and Jang, C. S. 2020. Resequencing of a core rice mutant population induced by gamma-ray irradiation and its application in a genome-wide association study. J. Plant Biol. 63: 463–472. doi:10.1007/s12374-020-09266-2
- International HapMap Consortium. 2005. A haplotype map of the human genome. Nature. 437: 1299–1320.
- International HapMap 3 Consortium. 2010. Integrating common and rare genetic variation in diverse human populations. Nature. 467: 52–58.
- Iyer, V., Boroviak, K., Thomas, M., Doe, B., Riva, L., Ryder, E., and Adams, D. J. 2018. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 14: e1007503. doi:10.1371/journal.pgen.1007503
- Jankowicz-Cieslak, J., and Till, B. J. 2015. Forward and reverse genetics in crop breeding. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J. M., Jain, S. M., and Johnson D. V., Eds. Cham: Springer, pp 215–240.
- Jansson, C., Vogel, J., Hazen, S., Brutnell, T., and Mockler, T. 2018. Climate-smart crops with enhanced photosynthesis. J. Exp. Bot. 69: 3801–3809. doi:10.1093/jxb/ery213
- Jeong, I.-S., Yoon, U.-H., Lee, G.-S., Ji, H.-S., Lee, H.-J., Han, C.-D., Hahn, J.-H., An, G., and Kim, T.-H. 2013. SNP-based analysis of genetic diversity in anther-derived rice by whole genome sequencing. Rice (NY) 6: 6. doi:10.1186/1939-8433-6-6
- Jiang, W., Feng, S., Huang, S., Yu, W., Li, G., Yang, G., Liu, Y., Zhang, Y., Zhang, L., Hou, Y., Chen, J., Chen, J., and Huang, X. 2018. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res. 28: 855–861. doi:10.1038/s41422-018-0052-4
- Jiang, C., Mithani, A., Belfield, E. J., Mott, R., Hurst, L. D., and Harberd, N. P. 2014. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 24: 1821–1829. doi:10.1101/gr.177659.114
- Jiang, C., Mithani, A., Gan, X., Belfield, E. J., Klingler, J. P., Zhu, J. K., Ragoussis, J., Mott, R., and Harberd, N. P. 2011. Regenerant Arabidopsis lineages display a distinct genome-wide spectrum of mutations conferring variant phenotypes. Curr. Biol. 21: 1385–1390. doi:10.1016/j.cub.2011.07.002
- Jiang, G.-L. 2013. Molecular markers and marker-assisted breeding in plants. In Plant Breeding from Laboratories to Fields; S. B. Andersen, Ed. Croatia: InTech, pp 45–83.
- Jiang, G.-L. 2015. Molecular marker-assisted breeding: A plant breeder’s review. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J., Jain, S., and Johnson, D., Eds. Cham: Springer, pp 431–472.
- Jiang, N., and Panaud, O. 2013. Transposable element dynamics in rice and its wild relatives. In Genetics and Genomics of Rice, Plant Genetics and Genomics: Crops and Models, vol. 5; Zhang, Q. and Wing, R. A., Eds. New York: Springer, pp 55–70.
- Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., and Weeks, D. P. 2017. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15: 648–657. doi:10.1111/pbi.12663
- Jin, S., Zong, Y., Gao, Q., Zhu, Z., Wang, Y., Qin, P., Liang, C., Wang, D., Qiu, J.-L., Zhang, F., and Gao, C. 2019. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364: 292–295.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. doi:10.1126/science.1225829
- Jo, Y. D., and Kim, J.-B. 2019. Frequency and spectrum of radiation-induced mutations revealed by whole-genome sequencing analyses of plants. Qubs. 3: 7. doi:10.3390/qubs3020007
- Jones, S. S., Murray, T. D., and Allan, R. E. 1995. The development of disease resistance in wheat. Annu. Rev. Phytopathol. 33: 429–443. doi:10.1146/annurev.py.33.090195.002241
- Joos, H., Timmerman, B., Montagu, M. V., and Schell, J. 1983. Genetic analysis of transfer and stabilization of Agrobacterium DNA in plant cells. Embo J. 2: 2151–2160. doi:10.1002/j.1460-2075.1983.tb01716.x
- Kamthan, A., Chaudhuri, A., Kamthan, M., and Datta, A. 2016. Genetically modified (GM) crops: milestones and new advances in crop improvement. Theor. Appl. Genet. 129: 1639–1655. doi:10.1007/s00122-016-2747-6
- Karp, A., and Maddock, S. E. 1984. Chromosome variation in wheat plants regenerated from cultured immature embryos. Theor. Appl. Genet. 67: 249–255. doi:10.1007/BF00317047
- Karunarathna, N. L., Wang, H., Harloff, H.-J., Jiang, L., and Christian J. 2020. Elevating seed oil content in a polyploidy crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 18: 2251–2266.
- Kashima, K., Mejima, M., Kurokawa, S., Kuroda, M., Kiyono, H., and Yuki, Y. 2015. Comparative whole-genome analyses of selection marker-free rice-based cholera toxin B-subunit vaccine lines and wild-type lines . BMC Genomics. 16: 48. doi:10.1186/s12864-015-1285-y
- Kasulin, L., Rowan, B. A., Leon, R. J. C., Schuenemann, V. J., Weigel, D., and Botto, J. F. 2017. A single haplotype hyposensitive to light and requiring strong vernalization dominates Arabidopsis thaliana populations in Patagonia, Argentina. Mol. Ecol. 26: 3389–3404. doi:10.1111/mec.14107
- Katche, E., Quezada-Martinez, D., Katche, E. I., Vasquez-Teuber, P., and Mason, A. S. 2019. Interspecific hybridization for Brassica crop improvement. Crop Breed. Genet. Genom. 1: e190007.
- Kaur, N., Alok, A., Shivani , Kumar, P., Kaur, N., Awasthi, P., Chaturvedi, S., Pandey, P., Pandey, A., Pandey, A. K., and Tiwari, S. 2020. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Met. Engineer. 59: 76–86. doi:10.1016/j.ymben.2020.01.008
- Kausch, A. P., Nelson-Vasilchik, K., Hague, J., Mookkan, M., Quemada, H., Dellaporta, S., Fragoso, C., and Zhang, Z. J. 2019. Edit at will: genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci. 281: 186–205. doi:10.1016/j.plantsci.2019.01.006
- Kawakatsu, T., Kawahara, Y., Itoh, T., and Takaiwa, F. 2013. A whole-genome analysis of a transgenic rice seed-based edible vaccine against cedar pollen allergy. DNA Res. 20: 623–631. doi:10.1093/dnares/dst036
- Kazama, Y., Ishii, K., Hirano, T., Wakana, T., Yamada, M., Ohbu, S., and Abe, T. 2017. Different mutational function of low- and high-linear energy transfer heavy-ion irradiation demonstrated by whole-genome resequencing of Arabidopsis mutants. Plant J. 92: 1020–1030. doi:10.1111/tpj.13738
- Khan, M. Z., Zaidi, S. S., Amin, I., and Mansoor, S. 2019. A CRISPR way for fast-forward crop domestication. Trends Plant Sci. 24: 293–296. doi:10.1016/j.tplants.2019.01.011
- Kim, J. K., Park, S.-Y., Ha, S.-H., Lee, S. M., Lim, S.-H., Kim, H. J., Ko, H.-S., Oh, S.-D., Park, J.-S., and Suh, S.-C. 2012. Compositional assessment of carotenoid-biofortified rice using substantial equivalence. Afr. J. Biotechnol. 11: 9330–9335.
- Kim, S., Kim, D., Cho, S. W., Kim, J., and Kim, J. S. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24: 1012–1019. doi:10.1101/gr.171322.113
- Kirik, A., Salomon, S., and Puchta, H. 2000. Species-specific double-strand break repair and genome evolution in plants. Embo J. 19: 5562–5566. doi:10.1093/emboj/19.20.5562
- Klümper, W., and Qaim, M. 2014. A meta-analysis of the impacts of genetically modified crops. PLoS One. 9: e111629. doi:10.1371/journal.pone.0111629
- Kogel, K.-H., Voll, L. M., Schäfer, P., Jansen, C., Wu, Y., Langen, G., Imani, J., Hofmann, J., Schmiedl, A., Sonnewald, S., von Wettstein, D., Cook, R. J., and Sonnewald, U. 2010. Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances. Proc. Natl. Acad. Sci. U S A. 107: 6198–6203. doi:10.1073/pnas.1001945107
- Kost, T. D., Gessler, C., Jänsch, M., Flachowsky, H., Patocchi, A., and Broggini, G. A. L. 2015. Development of the first cisgenic apple with increased resistance to fire blight. PLoS One. 10: e0143980. doi:10.1371/journal.pone.0143980
- Kuang, Y., Li, S., Ren, B., Yan, F., Spetz, C., Li, X., Zhou, X., and Zhou, H. 2020. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant. 13: 565–572. doi:10.1016/j.molp.2020.01.010
- Kumar, A. P. K., McKeown, P. C., Boualem, A., Ryder, P., Brychkova, G., Bendahmane, A., Sarkar, A., Chatterjee, M., and Spillane, C. 2017. TILLING by Sequencing (TbyS) for targeted genome mutagenesis in crops. Mol. Breeding. 37: 14.
- Kumlehn, J., Pietralla, J., Hensel, G., Pacher, M., and Puchta, H. 2018. The CRISPR/Cas revolution continues: from efficient gene editing for crop breeding to plant synthetic biology. J. Integr. Plant Biol. 60: 1127–1153. doi:10.1111/jipb.12734
- Kyndt, T., Quispe, D., Zhai, H., Jarret, R., Ghislain, M., Liu, Q., Gheysen, G., and Kreuze, J. F. 2015. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc. Natl. Acad. Sci. USA. 112: 5844–5849. doi:10.1073/pnas.1419685112
- Lacchini, E., Kiegle, E., Castellani, M., Adam, H., Jouannic, S., Gregis, V., and Kater, M. M. 2020. CRISPR-mediated accelerated domestication of African rice landraces. PLoS One. 15: e0229782. doi:10.1371/journal.pone.0229782
- Laibach, F. 1925. Das Taubwerden von Bastardsamen und die Künstliche Aufzucht früh absterbender Bastardembryonen. Z. Bot. 17: 417–459.
- Lam, H.-M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F.-L., Li, M.-W., He, W., Qin, N., Wang, B., Li, J., Jian, M., Wang, J., Shao, G., Wang, J., Sun, S. S.-M., and Zhang, G. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42: 1053–1059. doi:10.1038/ng.715
- Lareau, C. A., Clement, K., Hsu, J. Y., Pattanayak, V., Joung, J. K., Aryee, M. J., and Pinello, L. 2018. Response to “unexpected mutations after CRISPR-Cas9 editing in vivo”. Nat. Methods. 15: 238–239. doi:10.1038/nmeth.4541
- Lee, K., Zhang, Y., Kleinstiver, B. P., Guo, J. A., Aryee, M. J., Miller, J., Malzahn, A., Zarecor, S., Lawrence-Dill, C. J., Joung, J. K., Qi, Y., and Wang, K. 2019. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 17: 362–372. doi:10.1111/pbi.12982
- Lee, M., and Phillips, R. L. 1987. Genomic rearrangements in maize induced by tissue culture. Genome 29: 122–128. doi:10.1139/g87-021
- Lehesranta, S. J., Davies, H. V., Shepherd, L. V. T., Nunan, N., McNicol, J. W., Auriola, S., Koistinen, K. M., Suomalainen, S., Kokko, H. I., and Karenlampi, S. O. 2005. Comparison of tuber proteomes of potato varieties, landraces, and genetically modified lines. Plant Physiol. 138: 1690–1699. doi:10.1104/pp.105.060152
- Lei, Y., Lu, L., Liu, H.-Y., Li, S., Xing, F., and Chen, L.-L. 2014. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant. 7: 1494–1496. doi:10.1093/mp/ssu044
- Lemay, M.-L., and Moineau, S. 2020. How are genes modified? Crossbreeding, mutagenesis, and CRISPR-Cas9. In Genetically Modified and Irradiated Food. Controversial Issues: Facts versus Perceptions; Andersen, V., Ed. Vienna, Austria: Academic Press, pp 39–54.
- Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., Van Eck, J., and Lippman, Z. B. 2018. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants. 4: 766–770. doi:10.1038/s41477-018-0259-x
- Lenaerts, B., Collard, B. C. Y., and Demont, M. 2019. Review: improving global food security through accelerated plant breeding. Plant Sci. 287: 110207. doi:10.1016/j.plantsci.2019.110207
- Leng, P.-F., Lübberstedt, T., and Xu, M.-L. 2017. Genomics-assisted breeding – A revolutionary strategy for crop improvement. J. Integrat. Agric. 16: 2674–2685. doi:10.1016/S2095-3119(17)61813-6
- Li, B., Liu, H., Zhang, Y., Kang, T., Zhang, L., Tong, J., Xiao, L., and Zhang, H. 2013. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnol. J. 11: 1080–1091. doi:10.1111/pbi.12102
- Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., Zhang, R., and Gao, C. 2018e. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19: 59. doi:10.1186/s13059-018-1443-z
- Li, F., Numa, H., Hara, N., Sentoku, N., Ishii, T., Fukuta, Y., Nishimura, N., and Kato, H. 2019. Identification of a locus for seed shattering in rice (Oryza sativa L.) by combining bulked segregant analysis with whole-genome sequencing. Mol. Breeding. 39: 36.
- Li, F., Shimizu, A., Nishio, T., Tsutsumi, N., and Kato, H. 2019. Comparison and characterization of mutations induced by gamma-ray and carbon-ion irradiation in rice (Oryza sativa L.) using whole-genome resequencing. G3 (Bethesda) 9: 3743–3751. doi:10.1534/g3.119.400555
- Li, J., Chitwood, J., Menda, N., Mueller, L., and Hutton, S. F. 2018. Linkage between the I-3 gene for resistance to Fusarium wilt race 3 and increased sensitivity to bacterial spot in tomato. Theor. Appl. Genet. 131: 145–155. doi:10.1007/s00122-017-2991-4
- Li, J., Manghwar, H., Sun, L., Wang, P., Wang, G., Sheng, H., Zhang, J., Liu, H., Qin, L., Rui, H., Li, B., Lindsey, K., Daniell, H., Jin, S., and Zhang, X. 2019. Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR/Cas9-edited cotton plants. Plant Biotechnol. J. 17: 858–868. doi:10.1111/pbi.13020
- Li, J., Zhang, X., Sun, Y., Zhang, J., Du, W., Guo, X., Li, S., Zhao, Y., and Xia, L. 2018. Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. J Integr Plant Biol. 60: 536–540. doi:10.1111/jipb.12650
- Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., Church, G. M., and Sheen, J. 2013. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31: 688–691. doi:10.1038/nbt.2654
- Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., Lin, Q., Luo, W., Wu, G., and Li, H. 2016. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci. 7: 377. doi:10.3389/fpls.2016.00377
- Li, S., Zheng, Y. C., Cui, H. R., Fu, H. W., Shu, Q. Y., and Huang, J.-Z. 2016a. Frequency and type of inheritable mutations induced by γ rays in rice as revealed by whole genome sequencing. J. Zhejiang Univ. Sci. B. 17: 905–915. doi:10.1631/jzus.B1600125
- Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., Dong, W., Gao, C., and Xu, C. 2018c. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36: 1160–1163. doi:10.1038/nbt.4273
- Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., Luo, Y., and Zhu, H. 2018. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9: 559. doi:10.3389/fpls.2018.00559
- Li, Y., Zhu, J., Wu, H., Liu, C., Huang, C., Lan, J., Zhao, Y., and Xie, C. 2020. Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J. 8: 449–456. doi:10.1016/j.cj.2019.10.001
- Li, Z., McKibben, M. T. W., Finch, G. S., Blischak, P. D., Sutherland, B. L., and Barker, M. S. in press. Patterns and processes of diploidization in land plants. doi: 10.5281/zenodo.3964504.
- Li, Z., Wang, F., and Li, J.-F. 2019. Targeted transcriptional activation in plants using a potent dead Cas9-derived synthetic gene activator. Curr. Protocols. 127: e89.
- Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., Liu, J., Zhang, H., Liu, C., Ran, Y., and Gao, C. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Comm. 8: 14261.
- Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., Wang, Y., Anzalone, A. V., Raguram, A., Doman, J. L., Liu, D. R., and Gao, C. 2020. Prime genome editing in rice and wheat. Nat. Biotechnol. 38: 582–585. doi:10.1038/s41587-020-0455-x
- Lin, T., Zhu, G., Zhang, J., Xu, X., Yu, Q., Zheng, Z., Zhang, Z., Lun, Y., Li, S., Wang, X., Huang, Z., Li, J., Zhang, C., Wang, T., Zhang, Y., Wang, A., Zhang, Y., Lin, K., Li, C., Xiong, G., Xue, Y., Mazzucato, A., Causse, M., Fei, Z., Giovannoni, J. J., Chetelat, R. T., Zamir, D., Städler, T., Li, J., Ye, Z., Du, Y., and Huang, S. 2014. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46: 1220–1226. doi:10.1038/ng.3117
- Lisch, D. 2013. How important are transposons for plant evolution? Nat. Rev. Genet. 14: 49–61. doi:10.1038/nrg3374
- Liu, J., Nannas, N. J., Fu, F.-F., Shi, J., Aspinwall, B., Parrott, W. A., and Dawe, R. K. 2019. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell. 31: 368–383. doi:10.1105/tpc.18.00613
- Liu, L., Zhang, J., Xu, J., Li, Y., Guo, L., Wang, Z., Zhang, X., Zhao, B., Guo, Y.-D., and Zhang, N. 2020. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 301: 110683. doi:10.1016/j.plantsci.2020.110683
- Liu, M., and Li, Z.-Y. 2007. Genome doubling and chromosome elimination with fragment recombination leading to the formation of Brassica rapa-type plants with genomic alterations in crosses with Orychophragmus violaceus. Genome. 50: 985–993. doi:10.1139/g07-071
- Llewellyn, D. 2018. Does global agriculture need another green revolution. Engineering. 4: 449–451. doi:10.1016/j.eng.2018.07.017
- Lobaton, J. D., Miller, T., Gil, J., Ariza, D., Hoz, J. F., Soler, A., Beebe, S., Duitama, J., Gepts, P., and Raatz, B. 2018. Resequencing of common bean identifies regions of inter-gene pool introgression and provides comprehensive resources for molecular breeding. Plant Genome. 11: 170068–170021. doi:10.3835/plantgenome2017.08.0068
- Lu, H.-P., Luo, T., Fu, H.-W., Wang, L., Tan, Y.-Y., Huang, J.-Z., Wang, Q., Ye, G.-Y., Gatehouse, A. M. R., Lou, Y.-G., and Shu, Q.-Y. 2018. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants. 4: 338–344. doi:10.1038/s41477-018-0152-7
- Lu, P., Han, X., Qi, J., Yang, J., Wijeratne, A. J., Li, T., and Ma, H. 2012. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22: 508–518. doi:10.1101/gr.127522.111
- Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., Wang, B., Yang, Z., Li, H., Lin, Y., Xie, Y., Shen, R., Chen, S., Wang, Z., Chen, Y., Guo, J., Chen, L., Zhao, X., Dong, Z., and Liu, Y.-G. 2015. A robust CRISPR/cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 8: 1274–1284. doi:10.1016/j.molp.2015.04.007
- Macnaghten, P., and Habets, M. G. J. L. 2020. Breaking the impasse: towards a forward-looking governance framework for gene editing with plants. Plants. People. Planet. 2: 353–365. doi:10.1002/ppp3.10107
- Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., Voytas, D. F., Choi, I.-R., and Chadha-Mohanty, P. 2018. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16: 1918–1927. doi:10.1111/pbi.12927
- Malyska, A., Bolla, R., and Twardowski, T. 2016. The role of public opinion in shaping trajectories of agricultural biotechnology. Trends Biotechnol. 34: 530–534. doi:10.1016/j.tibtech.2016.03.005
- Mamta , Reddy, K. R. K., and Rajam, M. V. 2016. Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol. Biol. 90: 281–292. doi:10.1007/s11103-015-0414-y
- Matthews, E. (ed.). 2019. Creating a sustainable food future: A menu of solutions to feed nearly 10 billion people by 2050. World Resources Institute Final Report. http://agritrop.cirad.fr/593176/1/WRR_Food_Full_Report_0.pdf.
- McCann, M. C., Rogan, G. J., Fitzpatrick, S., Trujillo, W. A., Sorbet, R., Hartnell, G. F., Riodan, S. G., and Nemeth, M. A. 2006. Glyphosate-tolerant alfalfa is compositionally equivalent to conventional alfalfa (Medicago sativa L.). J. Agric. Food Chem. 54: 7187–7192. doi:10.1021/jf061482m
- McClintock, B. 1984. The significance of responses of the genome to challenge. Science 226: 792–801. doi:10.1126/science.15739260
- McGinn, M., Phippen, W. B., Chopra, R., Bansal, S., Jarvis, B. A., Phippen, M. E., Dorn, K. M., Esfahanian, M., Nazarenus, T. J., Cahoon, E. B., Durrett, T. P., Marks, M. D., and Sedbrook, J. C. 2019. Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnol. J. 17: 776–788. doi:10.1111/pbi.13014
- McHale, L. K., Haun, W. J., Xu, W. W., Bhaskar, P. B., Anderson, J. E., Hyten, D. L., Gerhardt, D. J., Jeddeloh, J. A., and Stupar, R. A. 2012. Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 159: 1295–1308. doi:10.1104/pp.112.194605
- Mejia, L. A., Dary, O., and Boukerdenna, H. 2017. Global regulatory framework for production and marketing of crops biofortified with vitamins and minerals. Ann. N Y Acad. Sci. 1390: 47–58. doi:10.1111/nyas.13275
- Meng, X., Hu, X., Liu, Q., Song, X., Gao, C., Li, J., and Wang, K. 2018. Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice. Sci. China Life Sci. 61: 122–125. doi:10.1007/s11427-017-9247-9
- Mercati, F., De Lorenzis, G., Brancadoro, L., Lupini, A., Abenavoli, M. R., Barbagallo, M. G., Di Lorenzo, R., Scienza, A., and Sunseri, F. 2016. High-throughput 18K SNP array to assess genetic variability of the main grapevine cultivars from Sicily. Tree Genet. Genomes. 12: 59.
- Messing, J., Bharti, A. K., Karlowski, W. M., Gundlach, H., Kim, H. R., Yu, Y., Wei, F., Fuks, G., Soderlund, C. A., Mayer, K. F. X., and Wing, R. A. 2004. Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA. 101: 14349–14354. doi:10.1073/pnas.0406163101
- Metje-Sprink, J., Sprink, T., and Hartung, F. 2020. Genome-edited plants in the field. Curr. Opin. Biotechnol. 61: 1–6. doi:10.1016/j.copbio.2019.08.007
- Meyer, R. S., and Purugganan, M. D. 2013. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14: 840–852. doi:10.1038/nrg3605
- Michno, J. M., Wang, X., Liu, J., Curtin, S. J., Kono, T. J., and Stupar, R. M. 2015. CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food. 6: 243–252. doi:10.1080/21645698.2015.1106063
- Miguel, C., and Marum, L. 2011. An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J. Exp. Bot. 62: 3713–3725. doi:10.1093/jxb/err155
- Mikami, M., Toki, S., and Endo, M. 2016. Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 57: 1058–1068. doi:10.1093/pcp/pcw049
- Ministry of the Environment. 2018. To genome editing technologies users. Government of Japan. https://www.env.go.jp/press/2_2_%20genome%20editing_En.pdf.
- Minkenberg, B., Zhang, J., Xie, K., and Yang, Y. 2019. CRISPR-PLANT v2: an online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol. J. 17: 5–8. doi:10.1111/pbi.13025
- Miyao, A., Nakagome, M., Ohnuma, T., Yamagata, H., Kanamori, H., Katayose, Y., Takahashi, A., Matsumoto, T., and Hirochika, H. 2012. Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol. 53: 256–264. doi:10.1093/pcp/pcr172
- Mohanta, T., Bashir, T., Hashem, A., Abd_Allah, E., and Bae, H. 2017. Genome editing tools in plants. Genes. 8: 399. doi:10.3390/genes8120399
- Mohd-Yusoff, N. F., Ruperao, P., Tomoyoshi, N. E., Edwards, D., Gresshoff, P. M., Biswas, B., and Batley, J. 2015. Scanning the effects of ethyl methanesulfonate on the whole genome of Lotus japonicus using second-generation sequencing analysis. G3 (Bethesda) 5: 559–567. doi:10.1534/g3.114.014571
- Molin, W. T., Yaguchi, A., Blenner, M., and Saski, C. A. 2020. The EccDNA replicon: a heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri. Plant Cell. 32: 2132–2140. doi:10.1105/tpc.20.00099
- Montenegro, J. D., Golicz, A. A., Bayer, P. E., Hurgobin, B., Lee, H., Chan, C.-K K., Visendi, P., Lai, K., Doležel, J., Batley, J., and Edwards, D. 2017. The pangenome of hexaploid bread wheat. Plant J. 90: 1007–1013. doi:10.1111/tpj.13515
- Morineau, C., Bellec, Y., Tellier, F., Gissot, L., Kelemen, Z., Nogué, F., and Faure, J.-D. 2017. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15: 729–739. doi:10.1111/pbi.12671
- Morita, R., Kusaba, M., Iida, S., Yamaguchi, H., Nishio, T., and Nishimura, M. 2009. Molecular characterization of mutations induced by gamma irradiation in rice. Genes Genet. Syst. 84: 361–370. doi:10.1266/ggs.84.361
- Muccilli, V., Vitale, A., Sheng, L., Gentile, A., Cardullo, N., Tringali, C., Oliveri, C., La Rosa, R., Di Guardo, M., La Malfa, S., Deng, Z., and Distefano, G. 2020. Substantial equivalence of a transgenic lemon fruit showing postharvest fungal pathogens resistance. J. Agric. Food Chem. 68: 3806–3816. doi:10.1021/acs.jafc.9b07925
- Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 691–693. doi:10.1038/nbt.2655
- Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. 2017. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7: 482. doi:10.1038/s41598-017-00578-x
- Neuffer, M. G., and Ficsor, G. 1963. Mutagenic action of ethyl methanesulfonate in maize. Science. 139: 1296–1297. doi:10.1126/science.139.3561.1296
- Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., Noda, T., Abudayyeh, O. O., Gootenberg, J. S., Mori, H., Oura, S., Holmes, B., Tanaka, M., Seki, M., Hirano, H., Aburatani, H., Ishitani, R., Ikawa, M., Yachie, N., Zhang, F., and Nureki, O. 2018. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 361: 1259–1262. doi:10.1126/science.aas9129
- Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. 2017. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7: 7057. doi:10.1038/s41598-017-06400-y
- Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., Hensel, G., Kumlehn, J., and Schweizer, P. 2010. HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 22: 3130–3141. doi:10.1105/tpc.110.077040
- Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., and Koizuka, N. 2018. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131: 63–69. doi:10.1016/j.plaphy.2018.04.025
- Olhoft, P. M., Flagel, L. E., and Somers, D. A. 2004. T-DNA locus structure in a large population of soybean plants transformed using the Agrobacterium-mediated cotyledonary-node method. Plant Biotechnol. J. 2: 289–300. doi:10.1111/j.1467-7652.2004.00070.x
- Olsen, K. M., and Wendel, J. F. 2013. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant. Biol. 64: 47–70. doi:10.1146/annurev-arplant-050312-120048
- Olsen, O., Wang, X., and von Wettstein, D. 1993. Sodium azide mutagenesis: preferential generation of A.T->G.C transitions in the barley Ant18 gene. Proc. Natl. Acad. Sci. USA. 90: 8043–8047. doi:10.1073/pnas.90.17.8043
- Ossowski, S., Schneeberger, K., Clark, R. M., Lanz, C., Warthmann, N., and Weigel, D. 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033. doi:10.1101/gr.080200.108
- Ossowski, S., Schneeberger, K., Lucas-Lledó, J. I., Warthmann, N., Clark, N. W., Shaw, R. G., Weigel, D., and Lynch, M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. doi:10.1126/science.1180677
- Papikian, A., Liu, W., Gallego-Bartolomé, J., and Jacobsen, S. E. 2019. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 10: 729. doi:10.1038/s41467-019-08736-7
- Park, D., Park, S.-H., Kim, H. S., Choi, B.-S., Kim, J.-K., Kim, N.-S., and Choi, I.-Y. 2019. NGS sequencing reveals that many of the genetic variations in transgenic rice plants match the variations found in natural rice population. Genes Genomics. 41: 213–222. doi:10.1007/s13258-018-0754-5
- Park, J.-S., Park, J.-H., Kim, S.-J., and Park, Y.-D. 2020. Genome analysis of tissue culture-derived variations in regenerated Brassica rapa ssp. pekinensis plants using next-generation sequencing. Hortic. Environ. Biotechnol. 61: 549–558. doi:10.1007/s13580-020-00237-7
- Parrott, W. A., Harbell, J., Kaeppler, H., Jones, T., Tomes, D., Van Eck, J., Wang, K., and Wenck, A. 2020. The proposed APHIS regulation modernization could enhance agriculture biotechnology research and development in the USA. In Vitro Celldevbiol-Plant. 56: 1–7. doi:10.1007/s11627-019-10039-x
- Parry, M. A., and Hawkesford, M. J. 2012. An integrated approach to crop genetic improvement. J. Integr. Plant Biol. 54: 250–259. doi:10.1111/j.1744-7909.2012.01109.x
- Parry, M. A. J., Madgwick, P. J., Bayon, C., Tearall, K., Hernandez-Lopez, A., Baudo, M., Rakszegi, M., Hamada, W., Al-Yassin, A., Ouabbou, H., Labhilili, M., and Phillips, A. L. 2009. Mutation discovery for crop improvement. J. Exp. Bot. 60: 2817–2825. doi:10.1093/jxb/erp189
- Paul, J.-Y., Khanna, H., Kleidon, J., Hoang, P., Geijskes, J., Daniells, J., Zaplin, E., Rosenberg, Y., James, A., Mlalazi, B., Deo, P., Arinaitwe, G., Namanya, P., Becker, D., Tindamanyire, J., Tushemereirwe, W., Harding, R., and Dale, J. 2017. Golden bananas in the field: elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 15: 520–532. doi:10.1111/pbi.12650
- Pellicer, J., Hidalgo, O., Dodsworth, S., and Leitch, I. J. 2018. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes (Basel) 9: 88.
- Pennisi, E. 2018. Human mutation rate a legacy from our past. Science 360: 143. doi:10.1126/science.360.6385.143
- Peterson, B. A., Haak, D. C., Nishimura, M. T., Teixeira, P. J. P. L., James, S. R., Dangl, J. L., and Nimchuk, Z. L. 2016. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One. 11: e0162169. doi:10.1371/journal.pone.0162169
- Piatek, A., Ali, Z., Baazim, H., Li, L., Abulfaraj, A., Al-Shareef, S., Aouida, M., and Mahfouz, M. M. 2015. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 13: 578–589. doi:10.1111/pbi.12284
- Pikaard, C. S., and Scheid, O. M. 2014. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6: a019315. doi:10.1101/cshperspect.a019315
- Pompili, V., Costa, L. D., Piazza, S., Pindo, M., and Malnoy, M. 2020. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 18: 845–858. doi:10.1111/pbi.13253
- Pratap, A., Chaturvedi, S. K., Tomar, R., Rajan, N., Malviya, N., Thudi, M., Saabale, P. R., Prajapati, U., Varshney, R. K., and Singh, N. P. 2017. Marker-assisted introgression of resistance to fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol. Genet. Genomics. 292: 1237–1245. doi:10.1007/s00438-017-1343-z
- Pucker, B., Holtgräwe, D., Rosleff Sörensen, T., Stracke, R., Viehöver, P., and Weisshaar, B. 2016. A de novo genome sequence assembly of the Arabidopsis thaliana accession Niederzenz-1 displays presence/absence variation and strong synteny. PLoS One. 11: e0164321. doi:10.1371/journal.pone.0164321
- Purugganan, M. D., and Fuller, D. Q. 2009. The nature of selection during plant domestication. Nature. 457: 843–848. doi:10.1038/nature07895
- Puttick, M. N., Clark, J., and Donoghue, P. C. J. 2015. Size is not everything: rates of genome size evolution, not C-value, correlate with speciation in angiosperms. Proc. Biol. Sci. 282: 20152289.
- Qaim, M. 2016. Genetically Modified Crops and Agricultural Development. Palgrave Macmillan, New York.
- Qaim, M. 2020. Role of new plant breeding technologies for feed security and sustainable agricultural development. Appl. Econ. Persp. Pol. 42: 129–150. doi:10.1002/aepp.13044
- Qin, Y., Shin, K.-S., Woo, H.-J., and Lim, M.-H. 2018. Genomic variations of rice regenerants from tissue culture revealed by whole genome re-sequencing. Plant Breed. Biotech. 6: 426–433. doi:10.9787/PBB.2018.6.4.426
- Rahmatov, M., Rouse, M. N., Steffenson, B. J., Andersson, S. C., Wanyera, R., Pretorius, Z. A., Houben, A., Kumarse, N., Bhavani, S., and Johansson, E. 2016. Sources of stem rust resistance in wheat-alien introgression lines. Plant Dis. 100: 1101–1109. doi:10.1094/PDIS-12-15-1448-RE
- Ray, D. K., Ramankutty, N., Mueiller, N. D., West, P. C., and Foley, J. A. 2012. Recent patterns of crop yield growth and stagnation. Nat. Comm. 3: 1293.
- Ray, D. K., West, P. C., Clark, M., Gerber, J. S., Prishchepov, A. V., and Chatterjee, S. 2019. Climate change has likely already affected global food production. PLoS One. 14: e0217148. doi:10.1371/journal.pone.0217148
- Renny-Byfield, S., and Wendel, J. F. 2014. Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 101: 1711–1725. doi:10.3732/ajb.1400119
- Renny-Byfield, S., Rodgers-Melnick, E., and Ross-Ibarra, J. 2017. Gene fractionation and function in the ancient subgenomes of maize. Mol. Biol. Evol. 34: 1825–1832. doi:10.1093/molbev/msx121
- Ricroch, A. E., Bergé, J. B., and Kuntz, M. 2011. Evaluation of genetically engineered crops using transcriptomic, proteomic, and metabolomic profiling techniques. Plant Physiol. 155: 1752–1761. doi:10.1104/pp.111.173609
- Ridley, W. P., Harrigan, G. G., Breeze, M. L., Nemeth, M. A., Sidhu, R. S., and Glenn, K. C. 2011. Evaluation of compositional equivalence for multitrait biotechnology crops. J. Agric. Food Chem. 59: 5865–5876. doi:10.1021/jf103874t
- Roach, J. C., Glusman, G., Smit, A. F. A., Huff, C. D., Hubley, R., Shannon, P. T., Rowen, L., Pant, K. P., Goodman, N., Bamshad, M., Shendure, J., Drmanac, R., Jorde, L. B., Hood, L., and Galas, D. J. 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 328: 636–639. doi:10.1126/science.1186802
- Robbins, W. J. 1922. Cultivation of excised root tips and stem tips under sterile conditions. Int. J. Plant Sci. 73: 376–390.
- Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. 2017. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 171: 470–480. doi:10.1016/j.cell.2017.08.030
- Saini, P., Saini, P., Kaur, J. J., Francies, R. M., Gani, M., Rajendra, A. A., Negi, N., Jagtap, A., Kadam, A., Singh, C., and Chauhan, S. S. 2020. Molecular approaches for harvesting natural diversity for crop improvement. In Rediscovery of Genetic and Genomic Resources for Future Food Security; Salgotra, R. K. and Zargar S. M., Eds. Singapore: Springer Nature, pp 67–169.
- Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., and Barro, F. 2018. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16: 902–910. doi:10.1111/pbi.12837
- Sanchez-Moran, E., Santos, J. L., Jones, G. H., and Franklin, F. C. H. 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis . Genes Dev. 21: 2220–2233. doi:10.1101/gad.439007
- Sandhya, D., Jogam, P., Allini, V. R., Abbagani, S., and Alok, A. 2020. The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Engin. Biotechnol. 18: 25.
- Sashidhar, N., Harloff, H. J., Potgieter, L., and Jung, C. 2020. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 18: 2241–2250. doi:10.1111/pbi.13380
- Satoh, H., Matsusaka, H., and Kumamaru, T. 2010. Use of N-methyl-N-nitrosourea treatment of fertilized egg cells for saturation mutagenesis in rice. Breed. Sci. 60: 475–485. doi:10.1270/jsbbs.60.475
- Sauer, N. J., Mozoruk, J., Miller, R. B., Warburg, Z. J., Walker, K. A., Beetham, P. R., Schöpke, C. R., and Gocal, G. F. W. 2016. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol. J. 14: 496–502. doi:10.1111/pbi.12496
- Schaefer, K. A., Wu, W. H., Colgan, D. F., Tsang, S. H., Bassuk, A. G., and Mahajan, V. B. 2017. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods. 14: 547–548. doi:10.1038/nmeth.4293
- Scheelbeek, P. F. D., Bird, F. A., Tuomisto, H. L., Green, R., Harris, F. B., Joy, E. J. M., Chalabi, Z., Allen, E., Haines, A., and Dangour, A. D. 2018. Effect of environmental changes on vegetable and legume yields and nutritional quality. Proc. Natl. Acad. Sci. USA. 115: 6804–6809. doi:10.1073/pnas.1800442115
- Schiemann, J., Dietz-Pfeilstetter, A., Hartung, F., Kohl, C., Romeis, J., and Sprink, T. 2019. Risk assessment and regulation of plants modified by modern biotechniques: current status and future challenges. Annu Rev Plant Biol. 70: 699–726. doi:10.1146/annurev-arplant-050718-100025
- Schleussner, C.-F., Deryng, D., Müller, C., Elliott, J., Saeed, F., Folberth, C., Liu, W., Wang, X., Pugh, T. A. M., Thiery, W., Seneviratne, S. I., and Rogelj, J. 2018. Crop productivity changes in 1.5 °C and 2 °C worlds under climate sensitivity uncertainty. Environ. Res. Lett. 13: 064007. doi:10.1088/1748-9326/aab63b
- Schmidt, C., Pacher, M., and Puchta, H. 2019. DNA break repair in plants and its application for genome engineering. In Transgenic Plants. Methods in Molecular Biology, vol 1864; Kumar, S., Barone, P., Smith, M., Eds. NY: Humana Press, pp 237–266.
- Schmidt, S. M., Belisle, M., and Frommer, W. F. 2020. The evolving landscape around genome editing in agriculture: many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO Rep. 21: e50680. doi:10.15252/embr.202050680
- Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., Hyten, D. L., Song, Q., Thelen, J. J., Cheng, J., Xu, D., Hellsten, U., May, G. D., Yu, Y., Sakurai, T., Umezawa, T., Bhattacharyya, M. K., Sandhu, D., Valliyodan, B., Lindquist, E., Peto, M., Grant, D., Shu, S., Goodstein, D., Barry, K., Futrell-Griggs, M., Abernathy, B., Du, J., Tian, Z., Zhu, L., Gill, N., Joshi, T., Libault, M., Sethuraman, A., Zhang, X.-C., Shinozaki, K., Nguyen, H. T., Wing, R. A., Cregan, P., Specht, J., Grimwood, J., Rokhsar, D., Stacey, G., Shoemaker, R. C., and Jackson, S. A. 2010. Genome sequence of the palaeopolyploid soybean. Nature. 463: 178–183. doi:10.1038/nature08670
- Schnable, J. C., Springer, N. M., and Freeling, M. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA. 108: 4069–4074. doi:10.1073/pnas.1101368108
- Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., Liang, C., Zhang, J., Fulton, L., Graves, T. A., Minx, P., Reily, A. D., Courtney, L., Kruchowski, S. S., Tomlinson, C., Strong, C., Delehaunty, K., Fronick, C., Courtney, B., Rock, S. M., Belter, E., Du, F., Kim, K., Abbott, R. M., Cotton, M., Levy, A., Marchetto, P., Ochoa, K., Jackson, S. M., Gillam, B., Chen, W., Yan, L., Higginbotham, J., Cardenas, M., Waligorski, J., Applebaum, E., Phelps, L., Falcone, J., Kanchi, K., Thane, T., Scimone, A., Thane, N., Henke, J., Wang, T., Ruppert, J., Shah, N., Rotter, K., Hodges, J., Ingenthron, E., Cordes, M., Kohlberg, S., Sgro, J., Delgado, B., Mead, K., Chinwalla, A., Leonard, S., Crouse, K., Collura, K., Kudrna, D., Currie, J., He, R., Angelova, A., Rajasekar, S., Mueller, T., Lomeli, R., Scara, G., Ko, A., Delaney, K., Wissotski, M., Lopez, G., Campos, D., Braidotti, M., Ashley, E., Golser, W., Kim, HRan., Lee, S., Lin, J., Dujmic, Z., Kim, W., Talag, J., Zuccolo, A., Fan, C., Sebastian, A., Kramer, M., Spiegel, L., Nascimento, L., Zutavern, T., Miller, B., Ambroise, C., Muller, S., Spooner, W., Narechania, A., Ren, L., Wei, S., Kumari, S., Faga, B., Levy, M. J., McMahan, L., Van Buren, P., Vaughn, M. W., Ying, K., Yeh, C.-T., Emrich, S. J., Jia, Y., Kalyanaraman, A., Hsia, A.-P., Barbazuk, W. B., Baucom, R. S., Brutnell, T. P., Carpita, N. C., Chaparro, C., Chia, J.-M., Deragon, J.-M., Estill, J. C., Fu, Y., Jeddeloh, J. A., Han, Y., Lee, H., Li, P., Lisch, D. R., Liu, S., Liu, Z., Nagel, D. H., McCann, M. C., SanMiguel, P., Myers, A. M., Nettleton, D., Nguyen, J., Penning, B. W., Ponnala, L., Schneider, K. L., Schwartz, D. C., Sharma, A., Soderlund, C., Springer, N. M., Sun, Q., Wang, H., Waterman, M., Westerman, R., Wolfgruber, T. K., Yang, L., Yu, Y., Zhang, L., Zhou, S., Zhu, Q., Bennetzen, J. L., Dawe, R. K., Jiang, J., Jiang, N., Presting, G. G., Wessler, S. R., Aluru, S., Martienssen, R. A., Clifton, S. W., McCombie, W. R., Wing, R. A., and Wilson, R. K. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science. 326: 1112–1115. doi:10.1126/science.1178534
- Schnell, J., Steele, M., Bean, J., Neuspiel, M., Girard, C., Dormann, N., Pearson, C., Savoie, A., Bourbonnière, L., and Macdonald, P. 2015. A comparative analysis of insertional effects in genetically engineered plants: considerations for pre-market assessments. Transgenic Res. 24: 1–17. doi:10.1007/s11248-014-9843-7
- Schouten, H. J., Geest, H. V., Papadimitriou, S., Bemer, M., Schaart, J. G., Smulders, M. J. M., Sanchez Perez, G., and Schijlen, E. 2017. Re-sequencing transgenic plants revealed rearrangements at T-DNA inserts, and integration of a short T-DNA fragment, but no increase of small mutations elsewhere. Plant Cell Rep. 36: 493–504. doi:10.1007/s00299-017-2098-z
- Schulman, A. H., Oksman-Caldentey, K.-M., and Teeri, T. H. 2020. European Court of Justice delivers no justice to Europe on genome-edited crops. Plant Biotechnol. J. 18: 8–10. doi:10.1111/pbi.13200
- Sevanthi, A. M. V., Kandwal, P., Kale, P. B., Prakash, C., Ramkumar, M. K., Yadav, N., Mahato, A. K., Sureshkumar, V., Behera, M., Deshmukh, R. K., Jeyaparakash, P., Kar, M. K., Manonmani, S., Muthurajan, R., Gopala, K. S., Neelamraju, S., Sheshshayee, M. S., Swain, P., Singh, A. K., Singh, N. K., Mohapatra, T., and Sharma, R. P. 2018. Whole genome characterization of a few EMS-induced mutants of upland rice variety nagina 22 reveals a staggeringly high frequency of SNPs which show high phenotypic plasticity towards the wild-type. Front. Plant Sci. 9: 1179. doi:10.3389/fpls.2018.01179
- Sfeir, A., and Symington, L. S. 2015. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40: 701–714. doi:10.1016/j.tibs.2015.08.006
- Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., Zhang, K., Liu, J., Xi, J. J., Qiu, J.-L., and Gao, C. 2013. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686–688. doi:10.1038/nbt.2650
- Sharma, D. R., Kaur, R., and Kumar, K. 1996. Embryo rescue in plants – a review. Euphytica 89: 325–337.
- Shi, J., and Lai, J. 2015. Patterns of genomic changes with crop domestication and breeding. Curr. Opin. Plant. Biol. 24: 47–53. doi:10.1016/j.pbi.2015.01.008
- Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., Teramura, H., Yamamoto, T., Komatsu, H., Miura, K., Ezura, H., Nishida, K., Ariizumi, T., and Kondo, A. 2017. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35: 441–443. doi:10.1038/nbt.3833
- Shirasawa, K., Hirakawa, H., Nunome, T., Tabata, S., and Isobe, S. 2016. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 14: 51–60. doi:10.1111/pbi.12348
- Shirley, B. W., Hanley, S., and Goodman, H. M. 1992. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 4: 333–347. doi:10.2307/3869544
- Shull, G. H. 1908. The composition of a field of maize. Am. Breeders Assoc. Rep. os-4: 296–301. doi:10.1093/jhered/os-4.1.296
- Singer, S. D., Soolanayakanahally, R. Y., Foroud, N. A., and Kroebel, R. 2019. Biotechnological strategies for improved photosynthesis in a future of elevated atmospheric CO2. Planta. 251: 24. doi:10.1007/s00425-019-03301-4
- Singh, J., Zhang, S., Chen, C., Cooper, L., Bregitzer, P., Sturbaum, A. K., Hayes, P. M., and Lemaux, P. G. 2006. High-frequency Ds remobilization over multiple generations in barley facilitates gene tagging in large genome cereals. Plant. Mol. Biol. 62: 937–950. doi:10.1007/s11103-006-9067-1
- Singh, M., and Singh, J. 2012. Seed development-related expression of ARGONAUTE4_9 class of genes in barley: possible role in seed dormancy. Euphytica. 188: 123–129. doi:10.1007/s10681-012-0624-1
- Singh, M., Singh, S., Randhawa, H., and Singh, J. 2013. Polymorphic homoeolog of key gene of RdDM pathway, ARGONAUTE4_9 class is associated with pre-harvest sprouting in wheat (Triticum aestivum L.). Plos One. 8: e77009. doi:10.1371/journal.pone.0077009
- Singh, S., Tan, H.-Q., and Singh, J. 2012. Mutagenesis of barley malting quality QTLs with Ds transposons. Funct. Integr. Genomics. 12: 131–141. doi:10.1007/s10142-011-0258-8
- Sinha, S. K. 2010. RNAi induced gene silencing in crop improvement. Physiol. Mol. Biol. Plants 16: 321–332. doi:10.1007/s12298-010-0036-4
- Slade, A. J., Fuerstenberg, S. I., Loeffler, D., Steine, M. N., and Facciotti, D. 2005. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23: 75–81. doi:10.1038/nbt1043
- Smýkal, P., Nelson, M. N., Berger, J. D., and von Wettberg, E. J. B. 2018. The impact of genetic changes during crop domestication. Agronomy. 8: 119. doi:10.3390/agronomy8070119
- Smyth, S. J. 2020. The human health benefits from GM crops. Plant Biotechnol. J. 18: 887–888. doi:10.1111/pbi.13261
- Somers, D. J., Kirkpatrick, R., Moniwa, M., and Walsh, A. 2003. Mining single-nucleotide polymorphisms from hexaploid wheat ESTs. Genome. 46: 431–437. doi:10.1139/g03-027
- Songstad, D. D., Petolino, J. F., Voytas, D. F., and Reichert, N. A. 2017. Genome editing of plants. Crit. Rev. Plant Sci. 36: 1–23. doi:10.1080/07352689.2017.1281663
- Soto-Cerda, B. J., Peñaloza, E. H., Montenegro, A. B., Rupayan, A. R., Gallardo, M. H., and Salvo-Garrido, H. 2013. An efficient marker-assisted backcrossing strategy for enhancing barley (Hordeum vulgare L.) production under acidity and aluminium toxicity. Mol. Breeding 31: 855–866. doi:10.1007/s11032-013-9839-7
- Spampinato, C. P. 2017. Protecting DNA from errors and damage: an overview of DNA repair mechanisms in plants compared to mammals. Cell Mol. Life Sci. 74: 1693–1709. doi:10.1007/s00018-016-2436-2
- Spasibionek, S., Mikołajczyk, K., Ćwiek-Kupczyńska, H., Piętka, T., Krótka, K., Matuszczak, M., Nowakowska, J., Michalski, K., and Bartkowiak-Broda, I. 2020. Marker assisted selection of new high oleic and low linolenic winter oilseed rape (Brassica napus L.) inbred lines revealing good agricultural value. PLoS One. 15: e0233959. doi:10.1371/journal.pone.0233959
- Springer, N. M., Ying, K., Fu, Y., Ji, T., Yeh, C.-T., Jia, Y., Wu, W., Richmond, T., Kitzman, J., Rosenbaum, H., Iniguez, A. L., Barbazuk, W. B., Jeddeloh, J. A., Nettleton, D., and Schnable, P. S. 2009. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. doi:10.1371/journal.pgen.1000734
- Stadler, L. J. 1928a. Genetic effects of x-rays in maize. Proc. Natl. Acad. Sci. USA. 14: 69–75. doi:10.1073/pnas.14.1.69
- Stadler, L. J. 1928b. Mutations in barley induced by X-rays and radium. Science. 68: 186–187. doi:10.1126/science.68.1756.186
- Steiner, H.-Y., Halpin, C., Jez, J. M., Kough, J., Parrott, W., Underhill, L., Weber, N., and Hannah, C. 2013. Editor's choice: evaluating the potential for adverse interactions within genetically engineered breeding stacks. Plant Physiol. 161: 1587–1594. doi:10.1104/pp.112.209817
- Subbaiyan, G. K., Waters, D. L. E., Katiyar, S. K., Sadananda, A. R., Vaddadi, S., and Henry, R. J. 2012. Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotechnol. J. 10: 623–634. doi:10.1111/j.1467-7652.2011.00676.x
- Subedi, U., Ozga, J. A., Chen, G., Foroud, N. A., and Singer, S. D. 2020. CRISPR/Cas-mediated genome editing for the improvement of oilseed crop productivity. Crit. Rev. Plant Sci. 39: 195–221. doi:10.1080/07352689.2020.1782568
- Sun, S., Zhou, Y., Chen, J., Shi, J., Zhao, H., Zhao, H., Song, W., Zhang, M., Cui, Y., Dong, X., Liu, H., Ma, X., Jiao, Y., Wang, B., Wei, X., Stein, J. C., Glaubitz, J. C., Lu, F., Yu, G., Liang, C., Fengler, K., Li, B., Rafalski, A., Schnable, P. S., Ware, D. H., Buckler, E. S., and Lai, J. 2018. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 50: 1289–1295. doi:10.1038/s41588-018-0182-0
- Tabata, R., Kamiya, T., Shigenobu, S., Yamaguchi, K., Yamada, M., Hasebe, M., Fujiwara, T., and Sawa, S. 2013. Identification of an EMS-induced causal mutation in a gene required for boron-mediated root development by low-coverage genome re-sequencing in Arabidopsis. Plant Signal Behav. 8: e22534. doi:10.4161/psb.22534
- Tabei, Y. 2019. Risk and safety considerations 2: genetic variations and potential risks-traditional breeding and genome editing. Transgenic Res. 28: 119–124. doi:10.1007/s11248-019-00144-3
- Takagi, H., Uemura, A., Yaegashi, H., Tamiru, M., Abe, A., Mitsuoka, C., Utsushi, H., Natsume, S., Kanzaki, H., Matsumura, H., Saitoh, H., Yoshida, K., Cano, L. M., Kamoun, S., and Terauchi, R. 2013. MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 200: 276–283. doi:10.1111/nph.12369
- Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., He, H., Wang, W., Zeng, X., Shao, Y., Pan, Y., Hu, Y., Peng, Y., Fu, X., Li, H., Xia, S., and Zhao, B. 2017. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7: 14438. doi:10.1038/s41598-017-14832-9
- Tang, X., Liu, G., Zhou, J., Ren, Q., You, Q., Tian, L., Xin, X., Zhong, Z., Liu, B., Zheng, X., Zhang, D., Malzahn, A., Gong, Z., Qi, Y., Zhang, T., and Zhang, Y. 2018. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19: 84. doi:10.1186/s13059-018-1458-5
- Thomas, M., Burgio, G., Adams, D. J., and Iyer, V. 2019. Collateral damage and CRISPR genome editing. PLoS Genet. 15: e1007994. doi:10.1371/journal.pgen.1007994
- Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., Zhang, H., Ren, Y., Gong, G., Zong, M., Liu, F., Chen, Q., and Xu, Y. 2018. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37: 1353–1356. doi:10.1007/s00299-018-2299-0
- Tian, Y., Chen, K., Li, X., Zheng, Y., and Chen, F. 2020. Design of high-oleic tobacco (Nicotiana tabacum L.) seed oil by CRISPR-Cas9-mediated knockout of NtFAD2-2. BMC Plant Biol. 20: 233. doi:10.1186/s12870-020-02441-0
- Tilman, D., and Clark, M. 2015. Food, agriculture & the environment: can we feed the world and save the Earth? Daedalus. 144: 8–23. doi:10.1162/DAED_a_00350
- Tiwari, I. M., Jesuraj, A., Kamboj, R., Devanna, B. N., Botella, J. R., and Sharma, T. R. 2017. Host delivered RNAi, an efficient approach to increase rice resistance to sheath blight pathogen (Rhizoctonia solani). Sci. Rep. 7: 7521. doi:10.1038/s41598-017-07749-w
- Tiwari, J. K., Devi, S., Ali, N., Luthra, S. K., Kumar, V., Bhardwaj, V., Singh, R. K., Rawat, S., and Chakrabarti, S. K. 2018. Progress in somatic hybridization research in potato during the past 40 years. Plant Cell. Tiss. Organ Cult. 132: 225–238. doi:10.1007/s11240-017-1327-z
- Tock, A. J., and Henderson, I. R. 2018. Hotspots for Initiation of Meiotic Recombination. Front. Genet. 9: 521. doi:10.3389/fgene.2018.00521
- Troadec, M.-B., and Pagès, J.-C. 2019. Where are we with unintended effects in genome editing applications from DNA to phenotype: focus on plant applications. Transgenic Res. 28: 125–133. doi:10.1007/s11248-019-00146-1
- Uauy, C., Paraiso, F., Colasuonno, P., Tran, R. K., Tsai, H., Berardi, S., Comai, L., and Dubcovsky, L. 2009. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9: 115. doi:10.1186/1471-2229-9-115
- Uchida, N., Sakamoto, T., Kurata, T., and Tasaka, M. 2011. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 52: 716–722. doi:10.1093/pcp/pcr029
- United Nations Department of Economic and Social Affairs. 2017. World population prospects 2019: highlights. http://population.un.org/wpp/Publications/Files/WPP2019_10Key. Findings.pdf.
- USDA-APHIS Department of Agriculture. 2020. Federal register, final rule, movement of certain genetically engineered organisms. service 7 CFR parts 330, 340 and 372 (Docket no. APHIS-2018-0034) RIN 0579-AE47. 85: 29790-29838. https://www.aphis.usda.gov/brs/fedregister/BRS_2020518.pdf.
- Usher, S., Haslam, R. P., Ruiz-Lopez, N., Sayanova, O., and Napier, J. A. 2015. Field trial evaluation of the accumulation of omega-3 long chain polyunsaturated fatty acids in transgenic Camelina sativa: making fish oil substitutes in plants. Metab. Eng. Commun. 2: 93–98. doi:10.1016/j.meteno.2015.04.002
- van de Wiel, C. C. M., Schaart, J. G., Lotz, L. A. P., and Smulders, M. J. M. 2017. New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol. Rep. 11: 1–8. doi:10.1007/s11816-017-0425-z
- Van Kregten, M., de Pater, S., Romejin, R., van Schendel, R., Hooykaas, P. J. J., and Tijsterman, M. 2016. T-DNA integration in plants results from polymerase-ɵ-mediated DNA repair. Nat. Plant. 2:
- Van Tuyl, J. M., and De Jeu, M. J. 1997. Methods for overcoming interspecific crossing barriers. In Pollen Biotechnology for Crop Production and Improvement; Sawhney, V. K. and Shivanna, K. R., Eds. Cambridge: Cambridge University Press, pp 273–293.
- Van, K., Hwang, E.-Y., Kim, M. Y., Park, H. J., Lee, S.-H., and Cregan, P. B. 2005. Discovery of SNPs in soybean genotypes frequently used as the parents of mapping populations in the United States and Korea. J. Hered. 96: 529–535. doi:10.1093/jhered/esi069
- Vats, S., Kumawat, S., Kumar, V., Patil, G. B., Joshi, T., Sonah, H., Sharma, T. R., and Deshmukh, R. 2019. Genome editing in plants: exploration of technological advancements and challenges. Cells. 8: 1386. doi:10.3390/cells8111386
- Vietmeyer, N. 2010. Borlaug: Bread Winner, 1960-1969, Vol. 3. Bracing Books, Lorton, VA.
- Vikram, P., Swamy, B. P. M., Dixit, S., Singh, R., Singh, B. P., Miro, B., Kohli, A., Henry, A., Singh, N. K., and Kumar, A. 2015. Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 5: 14799. doi:10.1038/srep14799
- Wallace, S. S. 1998. Enzymatic processing of radiation-induced free radical damage in DNA1. Radiat. Res. 150: S60–S79. doi:10.2307/3579809
- Waltz, E. 2015. USDA approves next-generation GM potato. Nat. Biotechnol. 33: 12–13. doi:10.1038/nbt0115-12
- Wan, D.-Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y., and Wen, Y.-Q. 2020. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hort. Res. 7:
- Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., Liu, Y. G., and Zhao, K. 2016. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 11: e0154027. doi:10.1371/journal.pone.0154027
- Wang, J., Meng, X., Hu, X., Sun, T., Li, J., Wang, K., and Yu, H. 2019. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol. J. 17: 70–711.
- Wang, N., Wang, Y., Tian, F., King, G. J., Zhang, C., Long, Y., Shi, L., and Meng, J. 2008. A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol. 180: 751–765. doi:10.1111/j.1469-8137.2008.02619.x
- Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., Li, M., Zheng, T., Fuentes, R. R., Zhang, F., Mansueto, L., Copetti, D., Sanciangco, M., Palis, K. C., Xu, J., Sun, C., Fu, B., Zhang, H., Gao, Y., Zhao, X., Shen, F., Cui, X., Yu, H., Li, Z., Chen, M., Detras, J., Zhou, Y., Zhang, X., Zhao, Y., Kudrna, D., Wang, C., Li, R., Jia, B., Lu, J., He, X., Dong, Z., Xu, J., Li, Y., Wang, M., Shi, J., Li, J., Zhang, D., Lee, S., Hu, W., Poliakov, A., Dubchak, I., Ulat, V. J., Borja, F. N., Mendoza, J. R., Ali, J., Li, J., Gao, Q., Niu, Y., Yue, Z., Naredo, M. E. B., Talag, J., Wang, X., Li, J., Fang, X., Yin, Y., Glaszmann, J.-C., Zhang, J., Li, J., Hamilton, R. S., Wing, R. A., Ruan, J., Zhang, G., Wei, C., Alexandrov, N., McNally, K. L., Li, Z., and Leung, H. 2018. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 557: 43–49. doi:10.1038/s41586-018-0063-9
- Wang, X.-J., Zhang, W., Yang, J.-T., and Wang, Z.-X. 2018. Effect on transcriptome and metabolome of stacked transgenic maize containing insecticidal cry and glyphosate tolerance epsps genes. Plant J. 3: 7106.
- Watson, J. D., and Crick, F. H. C. 1953. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 171: 737–738. doi:10.1038/171737a0
- Weber, N., Halpin, C., Hannah, L. C., Jez, J. M., Kough, J., and Parrott, W. 2012. Editor's choice: crop genome plasticity and its relevance to food and feed safety of genetically engineered breeding stacks. Plant Physiol. 160: 1842–1853. doi:10.1104/pp.112.204271
- Wei, F. J., Kuang, L. Y., Oung, H. M., Cheng, S. Y., Wu, H. P., Huang, L. T., Tseng, Y.-T., Chiou, W.-Y., Hsieh-Feng, V., Chung, C.-H., Yu, S.-M., Lee, L.-Y., Gelvin, S. B., and Hsing, Y.-L. 2016. Somaclonal variation does not preclude the use of rice transformants for genetic screening. Plant J. 85: 648–659. doi:10.1111/tpj.13132
- Weng, M.-L., Becker, C., Hildebrandt, J., Neumann, M., Rutter, M. T., Shaw, R. G., Weigel, D., and Fenster, C. B. 2019. Fine-grained analysis of spontaneous mutation spectrum and frequency in Arabidopsis thaliana. Genetics. 211: 703–714. doi:10.1534/genetics.118.301721
- Wicker, T., Gundlach, H., Spannagl, M., Uauy, C., Borrill, P., Ramírez-González, R. H., De Oliveira, R., Mayer, K. F. X., Paux, E., and Choulet, F. 2018. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 19: 103. doi:10.1186/s13059-018-1479-0
- Wicker, T., Yu, Y., Haberer, G., Mayer, K. F. X., Marri, P. R., Rounsley, S., Chen, M., Zuccolo, A., Panaud, O., Wing, R., and Roffler, S. 2016. DNA transposon activity is associated with increased mutation rates in genes of rice and other grasses. Nat. Commun. 7: 12790. doi:10.1038/ncomms12790
- Willing, E.-M., Piofczyk, T., Albert, A., Winkler, B., Scheeberger, K., and Pecinka, A. 2016. UVR2 ensures transgenerational genome stability under simulated natural UV-B in Arabidopsis thaliana. Nat. Comm. 7: 13522.
- Wolter, F., Schindele, P., and Puchta, H. 2019. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19: 176. doi:10.1186/s12870-019-1775-1
- Woo, J. W., Kim, J., Kwon, S., Corvalán, C., Cho, S. W., Kim, H., Kim, S.-G., Kim, S.-T., Choe, S., and Kim, J.-S. 2015. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33: 1162–1164. doi:10.1038/nbt.3389
- Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., and Rieseberg, L. H. 2009. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA. 106: 13875–13879. doi:10.1073/pnas.0811575106
- Xanthopoulou, A., Manioudaki, M., Bazakos, C., Kissoudis, C., Farsakoglou, A.-M., Karagiannis, E., Michailidis, M., Polychroniadou, C., Zambounis, A., Kazantzis, K., Tsaftaris, A., Madesis, P., Aravanopoulos, F., Molassiotis, A., and Ganopoulos, I. 2020. Whole genome re-sequencing of sweet cherry (Prunus avium L.) yields insights into genomic diversity of a fruit species. Horticulture Res. 7: 60.
- Xiao, B., Huang, Y., Tang, N., and Xiong, L. 2007. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115: 35–46. doi:10.1007/s00122-007-0538-9
- Xie, K., and Yang, Y. 2013. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant. 6: 1975–1983. doi:10.1093/mp/sst119
- Xie, K., Zhang, J., and Yang, Y. 2014. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant. 7: 923–926. doi:10.1093/mp/ssu009
- Xin, Z., Wang, M. L., Barkley, N. A., Burow, G., Franks, C., Pederson, G., and Burke, J. 2008. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 8: 103. doi:10.1186/1471-2229-8-103
- Xu, X., Liu, X., Ge, S., Jensen, J. D., Hu, F., Li, X., Dong, Y., Gutenkunst, R. N., Fang, L., Huang, L., Li, J., He, W., Zhang, G., Zheng, X., Zhang, F., Li, Y., Yu, C., Kristiansen, K., Zhang, X., Wang, J., Wright, M., McCouch, S., Nielsen, R., Wang, J., and Wang, W. 2011. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. doi:10.1038/nbt.2050
- Xue, Z.-Y., Zhi, D.-Y., Xue, G.-P., Zhang, H., Zhao, Y.-X., and Xia, G.-M. 2004. Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na. +. Plant Sci. 167: 849–859. doi:10.1016/j.plantsci.2004.05.034
- Yadav, C. B., Bhareti, P., Muthamilarasan, M., Mukherjee, J., Khan, Y., Rathi, P., and Prasad, M. 2015. Genome-wide SNP identification and characterization in two soybean cultivars with contrasting Mungbean Yellow Mosaic India Virus disease resistance traits. PLoS One. 10: e0123897. doi:10.1371/journal.pone.0123897
- Yamamoto, T., Nagasaki, H., Yonemaru, J., Ebana, K., Nakajima, M., Shibaya, T., and Yano, M. 2010. Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics. 11: 267. doi:10.1186/1471-2164-11-267
- Yang, G., Luo, W., Zhang, J., Yan, X., Du, Y., Zhou, L., Li, W., Wang, H., Chen, Z., and Guo, T. 2019. Genome-wide comparisons of mutations induced by carbon-ion beam and gamma-rays irradiation in rice via resequencing multiple mutants. Front. Plant Sci. 10: 1514. doi:10.3389/fpls.2019.01514
- Yates, J. L., Boerma, H. R., and Fasoula, V. A. 2012. SSR-marker analysis of the intracultivar phenotypic variation discovered within 3 soybean cultivars. J. Hered. 103: 570–578. doi:10.1093/jhered/ess015
- Yoon, Y.-J., Venkatesh, J., Lee, J.-H., Kim, J., Lee, H.-E., Kim, D.-S., and Kang, B.-C. 2020. Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus. Front. Plant Sci. 11: 1098. doi:10.3389/fpls.2020.01098
- Young, J., Zastrow-Hayes, G., Deschamps, S., Svitashev, S., Zaremba, M., Acharya, A., Paulraj, S., Peterson-Burch, B., Schwartz, C., Djukanovic, V., Lenderts, B., Feigenbutz, L., Wang, L., Alarcon, C., Siksnys, V., May, G., Chilcoat, N. D., and Kumar, S. 2019. CRISPR-Cas9 editing in maize: systematic evaluation of off-target activity and its relevance in crop improvement. Sci. Rep. 9: 6729. doi:10.1038/s41598-019-43141-6
- Yu, W., Wang, L., Zhao, R., Sheng, J., Zhang, S., Li, R., and Shen, L. 2019. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 19: 354. doi:10.1186/s12870-019-1939-z
- Zeng, D., Li, X., Huang, J., Li, Y., Cai, S., Yu, W., Li, Y., Huang, Y., Xie, X., Gong, Q., Tan, J., Zheng, Z., Guo, M., Liu, Y.-G., and Zhu, Q. 2020. Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol. J. 18: 1348–1350. doi:10.1111/pbi.13293
- Zeng, D., Liu, T., Ma, X., Wang, B., Zheng, Z., Zhang, Y., Xie, X., Yang, B., Zhao, Z., Zhu, Q., and Liu, Y.-G. 2020. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5-UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18: 2385–2387.
- Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E., Joung, J., van der Oost, J., Regev, A., Koonin, E. V., and Zhang, F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163: 759–771. doi:10.1016/j.cell.2015.09.038
- Zhai, Y., Cai, S., Hu, L., Yang, Y., Amoo, O., Fan, C., and Zhou, Y. 2019. CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132: 2111–2123. doi:10.1007/s00122-019-03341-0
- Zhan, X., Zhang, F., Zhong, Z., Chen, R., Wang, Y., Chang, L., Bock, R., Nie, B., and Zhang, J. 2019. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 17: 1814–1822. doi:10.1111/pbi.13102
- Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., Bi, J., Zhang, F., Luo, X., Wang, J., Tang, J., Yu, X., Liu, G., and Luo, L. 2019. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breeding. 39: 47.
- Zhang, D., Wang, Z., Wang, N., Gao, Y., Liu, Y., Wu, Y., Bai, Y., Zhang, Z., Lin, X., Dong, Y., Ou, X., Xu, C., and Liu, B. 2014. Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS One. 9: e96879. doi:10.1371/journal.pone.0096879
- Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., Wang, D., and Gao, C. 2018. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36: 894–898. doi:10.1038/nbt.4202
- Zhang, H., Zhang, J., Lang, Z., Botella, J. R., and Zhu, J.-K. 2017. Genome editing- principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 36: 291–309. doi:10.1080/07352689.2017.1402989
- Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., Mao, Y., Yang, L., Zhang, H., Xu, N., and Zhu, J.-K. 2014. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12: 797–807. doi:10.1111/pbi.12200
- Zhang, L., Sun, P. Y., Xie, H. K., Zhang, Y. H., Zhang, Y. Y., Peng, X. M., and Yang, Z. 2020. Characterization of γ-radiation-induced DNA polymorphisms in the M1 population of the japonica rice variety Gaogengnuo by whole-genome resequencing. Russ. J. Genet. 56: 693–705. doi:10.1134/S1022795420060149
- Zhang, M., Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., Ren, R., Xu, J., and Lou, L. 2020. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 39: 589–595. doi:10.1007/s00299-020-02516-0
- Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., Chen, K., Li, J., Jiang, L., and Gao, C. 2019. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants. 5: 480–485. doi:10.1038/s41477-019-0405-0
- Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., Qui, J. L., and Gao, C. 2016. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Comm 7: 12617.
- Zhao, T., and Schranz, M. E. 2019. Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc. Natl. Acad. Sci. USA. 116: 2165–2174. doi:10.1073/pnas.1801757116
- Zheng, M., Zhang, L., Tang, M., Liu, J., Liu, H., Yang, H., Fan, S., Terzaghi, W., Wang, H., and Hua, W. 2020. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18: 644–654. doi:10.1111/pbi.13228
- Zheng, Y., Li, S., Huang, J., Fu, H., Zhou, L., Furusawa, Y., and Shu, Q. 2020. Mutagenic effect of three ion beams on rice and identification of heritable mutations by whole genome sequencing. Plants 9: 551. doi:10.3390/plants9050551
- Zhenga, Z., Guoab, M., Wua, L., and Suna, H. 2017. Genome-wide analysis of a radiation-induced lesion mimic mutant in rice. Gmr. 16: gmr16039812. doi:10.4238/gmr16039812
- Zhong, Z., Sretenovic, S., Ren, Q., Yang, L., Bao, Y., Qi, C., Yuan, M., He, Y., Liu, S., Liu, X., Wang, J., Huang, L., Wang, Y., Baby, D., Wang, D., Zhang, T., Qi, Y., and Zhang, Y. 2019. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant. 12: 1027–1036. doi:10.1016/j.molp.2019.03.011
- Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H., and Yang, B. 2014. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42: 10903–10914. doi:10.1093/nar/gku806
- Zhou, W., Liang, G., Molloy, P. L., and Jones, P. A. 2020. DNA methylation enables transposable element-driven genome expansion. Proc. Natl. Acad. Sci. USA. 117: 19359–19366. doi:10.1073/pnas.1921719117
- Zitnak, A., and Johnston, G. R. 1970. Glycoalkaloid content of B5141-6 potatoes. Amer. Pot. J 47: 256–260. doi:10.1007/BF02864825
- Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., Qiu, J.-L., Wang, D., and Gao, C. 2017. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35: 438–440. doi:10.1038/nbt.3811
- Zsögön, A., Čermák, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., Freschi, L., Voytas, D. F., Kudla, J., and Peres, L. E. P. 2018. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36: 1211–1216. doi:10.1038/nbt.4272
