Abstract
The growing preference for producing cytochrome P450-mediated natural products in microbial systems stems from the challenging nature of the organic chemistry approaches. The P450 enzymes are redox-dependent proteins, through which they source electrons from reducing cofactors to drive their activities. Widely researched in biochemistry, most of the previous studies have extensively utilised expensive cell-free assays to reveal mechanistic insights into P450 functionalities in presence of commercial redox partners. However, in the context of microbial bioproduction, the synergic activity of P450- reductase proteins in microbial systems have not been largely investigated. This is mainly due to limited knowledge about their mutual interactions in the context of complex systems. Hence, manipulating the redox potential for natural product synthesis in microbial chassis has been limited. As the potential of redox state as crucial regulator of P450 biocatalysis has been greatly underestimated by the scientific community, in this review, we re-emphasize their pivotal role in modulating the in vivo P450 activity through affecting the product profile and yield. Particularly, we discuss the applications of widely used in vivo redox engineering methodologies for natural product synthesis to provide further suggestions for patterning on P450-based terpenoids production in microbial platforms.
Graphical Abstract
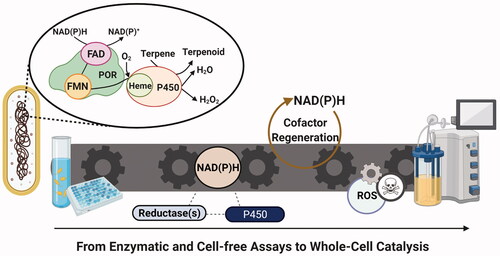
Introduction
Initially, cytochrome P450 oxidoreductase superfamily (P450s or CYPs) looks an outstanding catalyst for the biosynthesis of natural products like terpenoids. They alter the biological properties of the terpenoids through regio- and enantioselective hydroxylation, rendering them the largest structurally diverse group of plant natural products with distinct characteristics [Citation1–3]. However, while more than 300,000 P450 sequences have been reported from all trees of life up to 2018 [Citation4], these have not all been exploited for industrial production in versatile microbial systems. It is undeniable that our limited knowledge of the function of terpene synthesis networks in non-native environments has hindered this goal. Therefore, the majority of review papers have mainly focused on P450 enzyme and related metabolic engineering [Citation5–7]. However, a thorough discussion on the redox topic, particularly for terpenoid synthesis in microbial cells, is missing. Here, we highlight the importance of reductase partner engineering as a promising approach for feasible P450-based biocatalysis in microbial cells. Our main points include: assessing the P450-reductase fusion protein construction and reducing equivalents regeneration with regards to P450 activity and product profile. Conversely, P450 enzymes have also been recognised for generating redox signaling molecules, which exert a physiological impact on the cell [Citation8]. Consequently, as a precautionary approach, we highlight the drawbacks to excess cofactor and reductase expression, and we propose potential strategies to minimise the cause of oxidative stress for more efficient P450-based biocatalysis.
Classification of P450s by reductase partners
While widely known for their hydroxylation activity, the cytochrome P450 enzymes can catalyse many other natural reactions such as: N- and S-oxidation, dealkylation, hydroxylation, peroxidation, epoxidation, aromatic hydroxylation, oxidative and reductive dehalogenation, and deamination. Some of these have been extensively covered in other excellent reviews [Citation9–11]. These flexibilities make P450 enzymes superior to lengthy, low-yield and not eco-friendly organic chemical syntheses. There are several patents associated with P450-catalysed microbial biosynthesis of terpenoids such as: carotenes [Citation12], sesquiterpenes such as: isovalencene, spirovetiva-1(10), 7(11)-diene and valencene [Citation13], flavored nootkatone, vanillin, and valencene-containing alcoholic beverages [Citation14], and diterpenes [Citation15], with commercially well-known examples being: artemisinin and nootkatone by Amyris [Citation16] and Oxford Biotrans and Isobionics companies, respectively. Despite these success stories, the general complex activity of P450s and, potentially, the species-specific dependency on electron transfer proteins hamper their usage in wider areas. Based on these interacting proteins which shuttle the electron, the cytochrome P450 enzymes, including P450s, are categorised into ten different classes, among which the most widely studied are classes I, II, III, VII, and VIII as shown in . There are several past review papers about the classification of P450 enzymes [Citation22–27]. However, the important point and the common scenario is that, through redox partners such as: FAD-containing ferredoxin reductase (FdR), ferredoxin (Fdx), FAD- and flavin mononucleotide (FMN)-binding cytochrome P450 reductase (POR, formerly known as CPR), or FMN-dependent small redox protein (flavodoxin; Fld), the electron is shuttled from the reducing equivalent (NADPH) to the heme-iron of P450 enzyme, where the oxygen is activated to convert the terpenes to terpenoids.
Figure 1. Examples of several widely investigated P450 classes with examples. (A) Class I-P450cam [2GR6]; (B) Class II and Rat Liver POR [1AMO [Citation17]]; (C) Class III-P450cin [1T2B [Citation18]]; (D) Class VII-CYP116B46 [6LAA [Citation19]]; (E) Class VIII-P450BM3 [1BVY [Citation20]]. Fdx: ferredoxin; FMN: flavin mononucleotide; PFOR: phthalate family oxygenase reductase; POR: cytochrome P450 reductase; FAD: flavin adenine dinucleotide; CinA: P450cin heme active site; CinB: P450cin FAD containing protein cindoxin reductase; CinC: P450cin FMN containing flavodoxin [Citation21]. Figure created with BioRender.com.
![Figure 1. Examples of several widely investigated P450 classes with examples. (A) Class I-P450cam [2GR6]; (B) Class II and Rat Liver POR [1AMO [Citation17]]; (C) Class III-P450cin [1T2B [Citation18]]; (D) Class VII-CYP116B46 [6LAA [Citation19]]; (E) Class VIII-P450BM3 [1BVY [Citation20]]. Fdx: ferredoxin; FMN: flavin mononucleotide; PFOR: phthalate family oxygenase reductase; POR: cytochrome P450 reductase; FAD: flavin adenine dinucleotide; CinA: P450cin heme active site; CinB: P450cin FAD containing protein cindoxin reductase; CinC: P450cin FMN containing flavodoxin [Citation21]. Figure created with BioRender.com.](/cms/asset/f463960f-06cf-40e8-9ec0-688b42664814/ibty_a_1990210_f0001_c.jpg)
The regulatory effect of reductase partner on P450 function
In bacterial systems, the electrostatic interactions between the ferredoxin (Fdx), P450 heme and the reductase domains, as well as the negatively and positively charged amino acids on the Fdx iron-sulfur cluster and P450 proximal site, mediate the conformational changes of the Fdx for electron transfer to P450s [Citation28,Citation29]. In addition, the substrate binding to P450 induces P450 conformational change to increase its preference for Fdx through electrostatic and steric complementarity [Citation30].
However, the plants as one of the main abundant sources of terpenoids [Citation31,Citation32], employ a more complex, endoplasmic membrane-bound class II P450 enzymes. These systems are composed of a P450 and FAD- and flavin mononucleotide (FMN)-binding cytochrome P450 reductase (POR) [Citation22]. Similar to bacterial P450s, the interaction efficiency between POR and P450 is known to be modulated by the electrostatic potential of each protein and docking of acidic POR residues from the negatively charged FMN to the basic, positive charge region at P450 prosthetic heme site [Citation33–35], although it cannot be generalised to all P450s as reported with human microsomal P450s [Citation36]. Despite having a lower electron transfer rate to soluble bacterial P450s, POR binding can reorient the P450 catalytic domain, and vice versa, to decrease the interaction of P450 with the membrane, while the interaction of POR FMN and NADP domains with P450 can enable the electron transfer [Citation37]. Furthermore, the presence of several hydrophobic residues at the P450 might play a role in meditating P450-POR interaction and in POR recognition by P450 [Citation38,Citation39]. It has also been proposed that both POR upward orientation outward the membrane and FMN-binding domain rotation might co-occur to promote the POR and P450 binding and electron transfer to P450 heme iron [Citation40], and the latest studies have shown that the closed conformation of POR is also important for intramolecular electron transfer for heme oxygenases [Citation41]. Here, NADP+ binding and dissociation regulate the specific induced-fit-like changes in POR with highly interspecies-conserved FMN-domains to have an open, electron-donating conformation, while also influencing the interflavin and POR-P450 electron transfers [Citation36,Citation42–44]. This enhances P450 competing capability to interact with a specific POR among many other structurally diverse P450s [Citation45]. Various studies have highlighted the importance of POR in P450-mediated terpenoid biosynthesis. For example, the early attempt to build the paclitaxel biosynthetic pathway in Saccharomyces cerevisiae yielded a very low hydroxylated product titer of taxa-4(20),11-dien-5alpha-ol, being about 25 µg/L when only CYP725A4 was expressed [Citation46]. One claim was that the absence of a POR partner did not promote hydroxylation [Citation46]. This assumption was later confirmed in our recent study, where tandem, heterologous expression of both Taxus sp. CYP725A4 and POR in S. cerevisiae resulted in approximately 78 mg/L oxygenated taxanes produced at 1 L bioreactor scale [Citation47].
Some P450 enzymes require other interacting partners such as cytochrome b5 (Cb5), a small membrane-anchored haemoprotein, that forms a heterodimeric complex with P450 to provide additional redox support as an electron buffer for P450 and POR interaction [Citation16,Citation33,Citation48–51]. Cb5 reductases also exist in all human cell types, facilitating some P450-catalysed steroidogenesis [Citation52] and the functional deficiency of this enzyme has been associated with congenital methemoglobinemia [Citation53,Citation54]. Due to either being more thermodynamically favorable [Citation55] or because of its higher redox potential (by 320 mV), being 20 mV [Citation50], Cb5 can transfer the second electron to P450 ferric-superoxo heme species (). However, it does not need to necessarily act in electron transfer, but it can modify the P450 tertiary structure and its active site as an allosteric effector, which can increase the substrate affinity, coupling efficiency and breakdown from products [Citation55,Citation58–62]. Cb5 can also increase P450-peroxide anions stability and faster second electron transfer for more rapid product formation [Citation51,Citation58,Citation63]. For instance, Bart and Scott [Citation64] reported a 60% less NADPH consumption using wild type Cb5 and CYP2A6 for catalysing in vitro coumarin 7-hydroxylation, despite no increase in product yield.
Figure 2. Cytochrome b5 mechanism of action. Cytochrome b5 (Cb5) acts as an electron buffer for P450 enzyme and can transfer the second electron from cytochrome b5 reductase or cytochrome P450 reductase to P450 enzyme. Adapted with modifications from [Citation56,Citation57]. Figure created with BioRender.com.
![Figure 2. Cytochrome b5 mechanism of action. Cytochrome b5 (Cb5) acts as an electron buffer for P450 enzyme and can transfer the second electron from cytochrome b5 reductase or cytochrome P450 reductase to P450 enzyme. Adapted with modifications from [Citation56,Citation57]. Figure created with BioRender.com.](/cms/asset/3d45435e-c690-4625-addd-c291a3b39cff/ibty_a_1990210_f0002_c.jpg)
Estrada et al. [Citation65] reported that Cb5 promoted the androgen-forming, 17α-hydroxypregnenolone lyase activity of CYP17A1. Hence, they are likely to be useful for terpenoid biosynthesis applications. A recent study by Wang et al. [Citation66] showed that triterpenoid acid titer was increased by eight-fold upon integration of Glycyrrhiza uralensis Cb5 in S. cerevisiae strain with integrated CYP88D6, CYP72A154, B-amyrin synthase, and Arabidopsis thaliana POR1 genes and overexpressed key mevalonate pathway genes (ERG20-ERG9, ERG8, ERG10, ERG13, ERG1, and a truncated HMG1). Similarly, Paddon et al. [Citation16] reported up to 40% improvement in artemisinic aldehyde intermediate in artemisinic acid production by co-expressing A. annua Cb5 in S. cerevisiae expressing Artemisia annua CYP71AV1 and POR. However, in Sagwan-Barkdoll and Anterola [Citation67] study, co-expressing the Taxus x media Cb5 gene with CYP7254, POR, and other paclitaxel biosynthesis genes resulted in the complete absence of main product (taxa-4(20),11-dien-5alpha-ol), while decreasing the concentration of undesired oxygenated taxanes. These results denote that the Cb5s are P450-specific or the P450s have different numbers of positive Cb5 binding sites [Citation68]. Also, the competition of Cb5 and POR in binding to P450 proximal surface due to overlapping binding sites on P450 [Citation36,Citation64] should not be ignored. Despite this, it has been proposed that, at a highly coupled substrate metabolism rate, the reaction is not sensitive to Cb5 [Citation62] and the POR-P450 ionic strength can also modulate the Cb5 competing capability [Citation36].
Engineering the expression of the P450 and interacting reductase genes
The first rational step for increasing terpenoid productivity is to improve the substrate availability. However, this approach is at the risk of an irreversible mechanism-based inactivation of the enzyme [Citation69]. Instead, more studies have attempted to control the P450 expression and improve the product formation in terms of the reductase power. An example of such work is depicted in Emmerstorfer et al. [Citation70] study where overexpression of ICE2 led to POR (AtCPR1) stabilisation and enhanced (+)‐valencene conversion by 40% due to improved P450 enzyme and mRNA stability and increased S. cerevisiae endoplasmic reticulum membrane. In Chen et al. [Citation71] study, the hydrocortisone production was also improved by approximately 3.4-fold through: screening a variety of PORs, deleting the enzymes associated with C20-hydroxylation by-products formation NADPH-dependent aldo-keto reductase (YPR1) and glycerol dehydrogenase (GCY1) [Citation72], careful promoter selection, increasing the CYP5311B2 (R126D/Y398F) genomic copy numbers to two and genomic expression of Absidia orchidis Cb5. Although modulating the P450 and reductase gene dosages seems a common practice in P450 microbial research, here, increasing the gene copies of A. orchidis POR did not make any significant improvement to hydrocortisone production. This is beneficial considering the possibility of oxidative stress and reactive oxygen species (ROS) generation. This phenomenon can happen with overexpressing reductases or P450s, leading to toxicity and reduced growth rate as occurred with overexpressing S. cerevisiae endogenous POR, NCP1, for abscisic acid production [Citation73]. Similarly, according to Biggs et al. [Citation74], the optimum activity and highest oxygenated diterpene concentration by N-terminally truncated CYP725A4 was achieved using a low-copy number plasmid (five-copy) and a weak promoter (Trc). In contrast, in the Sarrade-Loucheur et al. [Citation75] study, the genomic overexpression of human and yeast POR genes did not impair the S. cerevisiae growth, and Zhao et al. [Citation76] obtained up to 96% efficiency in triterpene protopanaxadiol production in yeast using three copies of P450-POR fusion protein without affecting the cell growth. Modulation of P450 and POR copy numbers in the Gold et al. [Citation70] study also showed an increase in steviol production upon increasing the rate-limiting P450, KO, expression at the ratio of (KO[P450]: KAH[P450]: POR = 2:1:1). Therefore, the outcome is thought to be dependent on the final product and the P450 enzyme. As the risk of metabolic load through high expression of recombinant genes is undeniable [Citation77], a balance of heterologous productivity with cellular fitness needs to be achieved. We therefore suggest the readers refer to [Citation78] for an excellent review on advances in terpene production through microbial host engineering. Notwithstanding, we envisage the future of microbial biocatalysis will rely on microbial consortia to distribute the metabolic tasks, while using more sophisticated exoelectrogenic and riboflavin-secreting microorganisms like Shewanella sp. and Geobacter sp. to facilitate the P450 biocatalysis [Citation79,Citation80]. However, a few relevant studies have been conducted in P450 area. An example is the production of up to 33 and 30 mg/L of oxygenated taxanes or nootkatol, respectively, using microbial consortia of Escherichia coli and S. cerevisiae [Citation81].
Synthetic biology tools for P450 expression optimisation
Back to our main discussion, in a single microbial chassis, the P450 expression can be more effectively controlled via standard tools offered by synthetic biology such as pre-evaluated promoter libraries. Generally, the inducible promoter systems are preferred in P450 biotransformation, however strong constitutive promoters can favor higher terpenoid titers. For instance, a 3.1-fold increase in β-nootkatol and (+)-nootkatone titers in S. cerevisiae were achieved using HXT7 and CYC1 promoters instead of GAL1 for Hyoscyamus muticus premnaspirodiene oxygenase (HPO) and A. thaliana AtCPR, respectively [Citation82]. However, given a small set of characterised promoters and the lack of such promoter-terminator toolkits for P450 research, other regulatory elements can also be utilised to further modulate the expression of these genes in parallel. For instance, the carotenoids titers were increased by four-fold in E. coli using a small library of relatively short ribosome binding sites (RBS) [Citation83] or a 51% increased β-carotene production was achieved using degenerate RBS for tuning the genomic expression of highly-transcribing mevalonate pathway mvas and Hmg1 genes in E. coli [Citation84].
The transmembrane linkers of the membrane-bound eukaryotic P450s and reductases can regulate their possible interactions with adjacent macromolecules on endoplasmic reticulum, while the P450 linker assists in its folding to obtain its functional state at the cytoplasmic interphase [Citation85]. However, these insoluble P450s and PORs promote the inclusion body formation upon expression in bacterial cells like E. coli. Although P450 overexpression in E. coli have been shown to decrease the translation of upstream enzymes in methylerythritol 4-phosphate (MEP) pathway [Citation74], much research has been undertaken to circumvent their aggregation problem through N-terminus modification and truncation [Citation86], addition of small peptide tags like polyhistidine, 28-codon leader and small ubiquitin-related modifier and using bovine microsomal P450 17 alpha-hydroxylase membrane anchorage sequence (MALLLAVF), which however, do not guarantee the optimal enzymatic activity [Citation87–92]. In this regard, a list of N-terminal sequence modifications including bacterial membrane anchors and signal peptides for improving the P450 expression in E. coli cell factories have been suggested as P450-expression enhancing toolbox [Citation93] and have been thoroughly covered in other studies [Citation94,Citation95].
Immobilisation techniques for improved P450 activity
The replacing technologies such as immobilisation have been instrumental for supporting redox activity for P450s while rendering these endoplasmic reticulum-localised enzymes homogenous and soluble [Citation96]. On this point, nanodiscs, which are stable, self-assembled, nanoscale lipid bilayers of plants that are solubilised by amphipathic scaffold proteins, have been successfully used to immobilise P450s and reductases [Citation97,Citation98] (). For example, Rouck et al. [Citation100] used nanodiscs to incorporate (N-terminally modified) CYP725A4-(N-terminally truncated) POR chimera constructs to produce the hydroxylated product taxa-4(20),11-dien-5alpha-ol. In another interesting immobilisation study, Lee et al. [Citation99] harnessed E. coli specific inclusion bodies called “intracellular poly(3-hydroxybutyrate) (P(3HB)) granules” for P450 catalysis. Here, by attaching phasin (P(3HB)-associated protein)-P450 enzyme to the granules (), the stability and activity of the P450BM3 was improved regardless of changes in reaction conditions. The recovery and purification of the enzymes from bacteria was also easy and the enzyme preserved its activity upon repetitive use. Therefore, investigating this approach for long-term terpenoid biosynthesis would be promising. The optimisation of P450 enzymes can also be achieved through many other approaches such as: codon optimisation, expression of homologous P450s, protein engineering and bioprocess optimisation. However, since the focus of this review paper is on redox state and reductases rather than P450 enzymes, we invite the readers to refer to some very good P450-focused reviews [Citation48,Citation101,Citation102] to complement the knowledge.
Figure 3. Immobilisation techniques with potential for P450-mediated biocatalysis. (A) Nanodiscs resemble the plant lipid bilayer membranes for P450 and reductase immobilisation and can improve P450 solubility (B) Immobilisation of self-sufficient (reductase-free) P450s on E. coli poly(3-hydroxybutyrate) granules increases the P450 enzyme solubility and activity under harsh reaction conditions. Adapted with modifications from [Citation99]. Figure created with BioRender.com
![Figure 3. Immobilisation techniques with potential for P450-mediated biocatalysis. (A) Nanodiscs resemble the plant lipid bilayer membranes for P450 and reductase immobilisation and can improve P450 solubility (B) Immobilisation of self-sufficient (reductase-free) P450s on E. coli poly(3-hydroxybutyrate) granules increases the P450 enzyme solubility and activity under harsh reaction conditions. Adapted with modifications from [Citation99]. Figure created with BioRender.com](/cms/asset/a2a756fb-6b8c-4800-8c95-22765bc645b7/ibty_a_1990210_f0003_c.jpg)
The effect of surrogate reductases on P450 activity
There remain many unknowns regarding the P450s in natural product biosynthesis pathways. In fact, neither the specific biological activities nor the cognate electron transfer partners of all P450 enzymes have been elucidated [Citation103], given that the effect of evolution for selective interactions of P450s with particular reductases cannot be ignored [Citation25]. However, this cannot be generalised to more versatile P450s with detoxification activities in both bacteria and eukaryotes. Besides the traditional in vitro assays, there has been extensive research using omics tools to characterise more putative PORs in eukaryotic systems for improved natural products biosynthesis [Citation104]. Despite this, the research outcomes can sometimes show a better P450 activity using surrogate redox partners, particularly because the endogenous microbial redox state might not always adequately promote the P450 activity. Examples include E. coli flavodoxin (Fld) and flavodoxin reductase (Fpr) that weakly supported production of 15-hydroxyabietic by CYP105A1 [Citation105], or the overexpression or basal expression of S. cerevisiae NCP1 that did not favor sesquiterpenoid abscisic acid and 4-hydroxy-diclofenac production [Citation73,Citation75].
Utilising bacterial surrogate redox partners can also alter or control the site-selectivity and product profile of P450 enzymes. For example, Zhang et al. [Citation106] found that monooxygenase MycG activity was altered with standalone P450RhF reductase domain (RhFRED) which affected the substrate binding availability and rendered the enzyme capable of unnatural N-demethylation of mycinamicin substrates. Among the surrogate reductases, Pseudomonas putida putidaredoxin reductase (PdR; CamA) and putidaredoxin (Pdx; CamB) have been utilised for characterisation of the orphan P450s [Citation103]. This was exemplified by Lim et al. [Citation107] who characterised lauric acid regioselectivity using Sulfolobus acidocaldarius CYP119 with these redox partners. Likewise, using P. putida CamA/CamB for a mutant variant of myxobacterium Chondromyces apiculatus DSM436 versatile P450, CYP109Q5 (A280V/T229L), enabled the hydroxylation of β-ionone, valencene and fatty acids in E. coli. However, little or no activity was observed using E. coli JM109 (FAD-containing ferredoxin reductase) FdR/Fdx and Bos taurus FdR/AdX constructs [Citation108]. Apart from the more widely-used and smaller bacterial redox genes, several other studies have relied on eukaryotic PORs to control the P450 chemistry. For instance, Sun et al. [Citation109] used two PORs from G. uralensis and one POR from A. thaliana and Medicago truncatula along with a mutated CYP726A3 (T338S) and achieved higher aldehyde production in triterpenoid licorice biosynthesis pathway in S. cerevisiae.
Self-sufficient P450 systems: P450-reductase fusions
Fusing P450s to reductase genes can improve the biocatalysis
Although protein-protein interactions and gene expression levels are crucial for efficient P450-catalysed reactions, there is still a necessity to improve these enzymes in terms of the turnover rate, stability, electron supply source, substrate specificity and promiscuity in a non-native host [Citation110–112]. Fusing the genes in multigene biosynthetic pathways have been shown promising for increasing the terpene and oxidised terpenoid products [Citation113,Citation114] without necessarily impacting the protein production level. Following them, the P450s can be fused to reductase. However, it is argued that fusing P450 and redox enzymes might have little or no influence on: enzyme performance, electron coupling and product formation [Citation48,Citation76,Citation115]. For instance, Zhang et al. [Citation116] showed that fusing Zingiber zerumbet CYP71BA1 (α-humulene 8-hydroxylase) to A. thaliana POR did not result in sesquiterpenoid 8-hydroxy-α-humulene formation, in contrast to low-yield separate expressions. In the same way, Sagwan-Barkdoll and Anterola [Citation67] did not obtain an improved product formation when fusing CYP725A4 to POR gene in E. coli. In contrast, Albertsen et al. [Citation113] suggested that enzyme trimming can be used to optimise the active site positioning of enzymes in a fusion construct. A recent study by Haslinger and Prather [Citation117] showed that linking the P450 and reductase partners outperformed their separate expression for caffeic acid production in E. coli. Indeed, the nature has favored mankind with plenty of naturally evolved fusion proteins where the P450 heme domain is covalently bound to a reductive domain [Citation118] and currently, new electron transfer pathways in these self-sufficient P450s are being discovered [Citation119]. Examples include the widely-studied Bacillus megaterium self-sufficient P450 (P450BM3) [Citation120], which has an excellent electron transfer system and the fastest substrate oxidation rate among the P450s owing to the proximity and specific positioning of its domains [Citation9,Citation103], Rhodococcus sp. ECU0066 P450SMO [Citation121], and Rhodococcus sp. NCIMB 9784 CYP116B2 (P450RhF) [Citation122]. These natural systems can be effectively mimicked for constructing the artificial P450-reductase fusion proteins. For instance, to systematically generate the self-sufficient P450s, Neil Bruce’s group [Citation103,Citation123] proposed the ligase- and restriction enzyme-free LICRED cloning method using P450RHF reductase domain (RhFRED).
The first successful, heterologous self-sufficient P450 system was reported by Sibbesen et al. [Citation124], which was composed of a triple fusion of P. putida putidaredoxin reductase (PdR), putidaredoxin (Pdx) and P450cam domains to produce 5-exo-hydroxycamphor and 5-oxocamphor from camphor in E. coli. In another non-terpenoid example, fusion of RhFRED to S. venezuelae P450 PikC with a 16-aa linker improved the enzyme catalytic activity by four-fold towards both 12-membered ring macrolactone YC-17 and 14-membered ring macrolactone narbomycin [Citation125]. In terpenoid research, a self-sufficient P450 system, consisting of Thermus thermophilus CYP175A1, ferredoxin (Fdx), and ferredoxin-NADP+ oxidoreductase (FNR) improved the beta-carotene hydroxylation up to 25-fold through improved electron transfer [Citation126]. Artificial fusion of the P450cam variant (Y96F/V247L)-P450SMO reductase domain using a GS linker also enabled the hydroxylation of the terpenes limonene, α-pinene and camphor in Luan et al. [Citation127] study.
Enhancing the electrostatic interactions between P450 and reductase proteins can improve protein-protein interactions and electron transfer [Citation128], resulting in higher P450 turnover number [Citation129,Citation130]. In fusion proteins, this can potentially occur through perfecting the repositioning of redox center and the reduction potential. Indeed, the shorter distance between the Fdx iron-sulfur cluster and P450 heme-iron center as well as between the FAD components and the Fdx iron-sulfur cluster like in plastidic-type (FAD-containing ferredoxin reductase) FdRs (6–8 Å), can enable more optimal electron transfer as can be seen in plastidic-type FdRs [Citation28,Citation30]. Other studies have also suggested a distance of 6.2 Å based on Pseudomonas aeruginosa NAD(P)H:rubredoxin reductase (RdxR)-rubredoxins (Rdxs) electron transfer system [Citation131] or a distance of 14 Å or less between redox center edges for maximised and robust electron transfer rate [Citation132]. However, current limited knowledge and the lack of protein structure of many P450s with terpenoid biosynthesis activity confine advising on proper distancing between the Fdxs. Despite this, previous studies have emphasised on the key role of Fdx aromatic amino acid (F65) and FNR hydrophobic amino acids (L76, L78, and V136) for Fdx as well as flavodoxin (Fld) binding and orientation [Citation128,Citation133]. Therefore, a proper design must be considered not to interfere with biocatalysis which stress the need for auxiliary structures to facilitate this process.
Linkers influence the P450 productivity
Linkers can properly distance the P450 and its active site from redox gene partners and improve their internal spatial and steric orientations [Citation134]. These artificial amino acid chains aim to mimic the natural linkers found in self-sufficient P450 systems or in NADPH-FAD binding domains that closely connect the FAD and FMN domains for improved electron transfer [Citation135]. However, the scientific community has been skeptical about applying these artificial constructs for creating the P450-POR fusions. Contrary to these disagreements, there has been some success in using linkers as previous works suggest. For instance, Li et al. [Citation125], using a 16-aa linker (covalent linkage) stabilised P450-redox partner interaction in PikC-RhFRED system and positively influenced the electron transfer. Zao et al. [Citation76], using flexible linkers to fuse the truncated cytochrome P450-type protopanaxadiol synthase (PPDS) to a truncated A. thaliana reductase (ATR1), improved the yield by 71% and terpenoid substrate (dammarenediol-II) conversion by 4.5-fold in S. cerevisiae.
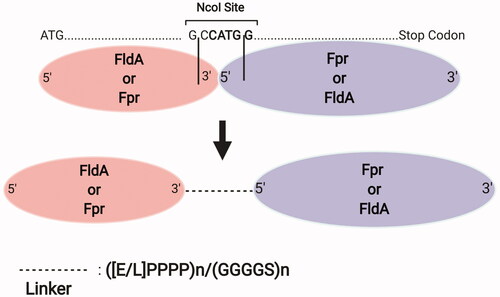
The previous studies have stated a few key parameters for constructing successful P450-redox partner fusions, particularly when using linkers. Beside their lengths and amino acid contents, their position relative to the P450 and the redox gene are important. In this regard, two interesting studies by Vlada B. Urlacher’s group have focused thoroughly on the effect of linkers for constructing P450 fusions. In the first study, DuaLinX method is proposed as a single-cloning fusion methodology which introduces two distinct linkers simultaneously. Here, E. coli flavodoxin (FldA) was fused to its flavodoxin reductase (Fpr) with either a flexible or rigid linker [Citation136] (). Using Bacillus subtilis CYP109B1 and FldA, and Fpr as reductase partners, the best performance was achieved with a linker length of more than 15 residues as well as with rigid linkers. Similar results using a free mixture of the redox partners at 4:4:1 (Fpr:FldA:CYP109B1) ratio were achieved upon using the ([E/L]PPPP)4 linker, both reaching around 62% myristic acid conversion and 2.75-fold cytochrome c reductase activity in vitro. Also, a later study by this research group [Citation137] demonstrated the best electron transfer for B. megaterium CYP106A2 and bovine CYP21A2 using rigid, proline-rich linkers. In the same study, fusing the B. subtilis flavodoxin (YkuN) to N-terminus of E. coli Fpr with ([E/L]PPPP)5 linker increased the myristic acid conversion rate and coupling efficiency of CYP109B1 to 94% and by 1.7-fold, respectively, compared to separate expressions. In addition, the proline-rich linkers were the best in coupling the electron transfer with B. megaterium CYP106A2 and bovine CYP21A2.
Figure 4. DuaLinX method. In this system, E. coli flavodoxin (FldA) and flavodoxin reductase (Fpr) are fused with a proline- or glycine-rich linker in any of N- or C-termini attachment orientations, through insertion of an NcoI restriction enzyme site between these two genes. Figure created with BioRender.com.
The proline-rich, rigid linkers have been shown to enable more dynamic conformational changes due to their disordered loop shape [Citation138,Citation139]. Moreover, they promote the electron transfer due to better control of enzyme spatial arrangements and structural isolation of fusion domains from the surrounding amino acids and redox partners [Citation136,Citation140,Citation141]. Despite this, the electron transfer improvement using glycine-rich, flexible linkers has also been reported [Citation142,Citation143]. Bakkes et al. [Citation137] reported that glycine rich-linker showed the highest coupling efficiency of 81.2% with up to 8.1-fold decrease in NADPH oxidation rate. Some good applications of flexible linkers in terpenoid biosynthesis are described in [Citation143,Citation144] studies in Yarrowia lipolytica. Li et al. [Citation144], using a GSTSSG linker for fusion of M. truncatula CYP716A12 to an N-terminally truncated A. thaliana POR (ATR1), improved the β-amyrin to triterpenoid oleanolic acid conversion to 100% compared to 72.7% with no linker. The same authors had previously engineered Y. lipolytica for production of another triterpenoid, compound K [Citation143]. Here, using the same linker for P450 PPDS and A. thaliana truncated ATR1 reductase fusion improved the substrate, dammarenediol II, conversion and final protopanaxadiol titer by 98% and 1.33-fold, respectively. Interestingly, this linker, however, resulted in around 16-fold lower (+)-nootkatone titer using fusion of nootkatone synthase CYP706M1 to truncated A. thaliana reductase (46AtPOR1) in Y. lipolytica. This denotes the importance of other key parameters in tuning the effect of linker, like linker length. Indeed, the linker length can affect the orientation, folding, correct conformation and flexibility of the enzymes [Citation21,Citation145]. Supportive of this statement, the recent study by Moon et al. [Citation142] showed that the fusion of steviol biosynthesis gene CYP714A2 to A. thaliana POR by a series of flexible (GGGGS)n = 1–3 linkers in E. coli was only improved by 1.7-fold using a linker length of 3 (15 amino acids) in comparison to length of 1. As evident, a more standardised approach for determining suitable linker seems essential. In this regard, a recent study by Gräwe et al. [Citation146] has proposed a new type IIS restriction enzymes- and T4 DNA ligase-assisted iterative functional linker cloning (iFlinkC) strategy, with a variety of rigid, flexible, short and semi-flexible linkers for fusion protein construction.
Linker-mediated P450-reductase self-assembly systems
The protein self-assembly methods and scaffolds are other flexible tools for co-localising the P450 and redox genes and bringing the redox centers to close proximity for an enhanced electron transfer [Citation111,Citation130]. In this regard, Truyuki Nagamune group, introduced a self-assembling system using Sulfolobus solfataricus heterotrimeric proliferating cell nuclear antigen (PCNA) to form a complex known as PUPPET (PCNA-utilized protein complex of P450 and its two-electron transfer-related proteins). Here, P. putida P450cam components were localised through fusing the N-termini of P. putida P450 components (P450cam, putidaredoxin: Pdx, and putidaredoxin reductase: PdR) to C-termini of three PCNAs, which enabled up to two-fold in vitro catalytic activity while achieving 25% of native P450cam activity [Citation147,Citation148] (). Later, the same group, Haga et al. [Citation140], showed that improving the Pdx positioning through optimising the linker between Pdx and PCNA, particularly with a rigid linker (G4SP20G4S), enhanced the P450cam activity by 1.9-fold, despite not achieving a similar enzyme turnover number as in P450cam. With using glycine-rich linkers ((G4S)n, n = 1–6), no improvement was achieved, and this was justified by the short end-to-end distance of the linker that potentially resulted in unfavorable interaction of Pdx with P450cam. In their more recent study, Haga et al. [Citation148] successfully recovered 92% of P450cam activity by assembling it with more reductase moieties; three PdRs and two Pdxs moieties. Haslinger and Pratha [Citation117] were the first to deploy the PUPPET system for whole-cell catalysis to produce caffeic acid polyphenol in E. coli. In this study, up to six- and eight-fold increase in caffeic acid titer was obtained using palustrisredoxin (Pux)/ putidaredoxin reductase (PdR) and putidaredoxin (Pdx)/PdR, where they fused these ferredoxin reductase (FdR), Fdx and Rhodopseudomonas palustris CYP199A2 F185L NΔ7 counterparts to PCNAs 1, 2 and 3, respectively.
Figure 5. PUPPET system. In this system, the N-termini of Pseudomonas putida P450 components are attached to C-termini of Sulfolobus solfataricus heterotrimeric proliferating cell nuclear antigen (PCNA), which enables the colocalisation of the redox and P450 enzymes. Adapted with modifications from [Citation147]. Figure created with BioRender.com.
![Figure 5. PUPPET system. In this system, the N-termini of Pseudomonas putida P450 components are attached to C-termini of Sulfolobus solfataricus heterotrimeric proliferating cell nuclear antigen (PCNA), which enables the colocalisation of the redox and P450 enzymes. Adapted with modifications from [Citation147]. Figure created with BioRender.com.](/cms/asset/97d28c9e-c7c6-4307-83e3-f830403e7efc/ibty_a_1990210_f0005_c.jpg)
Are linkers capable of altering P450 kinetics and product profiles?
Variable study outcomes have made it very difficult to reach a consensus on the type of effect that linkers have on P450 enzyme kinetics. For example, Haslinger and Prather [Citation117] reported that the fusion construct developed using proliferating cell nuclear antigen (PCNA) technology accelerated the P450cam enzymatic activity compared to the free enzymes system. However, the fusion construct (PCNA1–PdR:PCNA2–PdX:PCNA3–P450cam) did not alter its kinetics [Citation147]. Hence, the intermolecular interactions between the redox partner and P450 genes need to be properly investigated to evaluate the efficacy of the linkers. In line with it, Belsare et al. [Citation21] suggested that the heme and FMN sites in P450cin CinA (heme active site)-10aa linker-CinC (FMN-containing flavodoxin) fusion were not fully interacting. This resulted in around 1.7-fold less product formation rate compared to free components mixture. From another viewpoint, the linker effect might be determined by its position in relation to the reductase enzyme. In Jin et al. [Citation149] study, the total triterpenoids concentration decreased by around two-fold or no catalysis occurred when the linker was fused to the N-terminus and C-terminus of POR, respectively. This was regardless of the rigid or flexible linker type for fusing Betula platyphylla CYP716A180 to Lotus japonicus POR or M. truncatula POR for sequential oxidations of lupeol to triterpenoid, betulinic acid. Also, the linkers might only have minimal effect on the activity and stability of the properly-folded P450 as was shown for the endogenous linker of P450 2C2 [Citation150]. It is noteworthy that the linker structure has proved to be a more important parameter than flexibility for determining the distance and orientation of enzyme partners in a fusion construct [Citation151]. This would potentially play a part in tuning the P450-reductase interactions and their overall activity. Therefore, overall, caution should be exercised to carefully design them to eliminate the possible.
Different viewpoints also exist regarding the potential role of linkers in determining the product spectra. For instance, Biggs et al. [Citation152] showed that fusing the N-terminally modified and truncated CYP725A4 and Taxus POR proteins with a GSTGS linker shifted the diterpenoid product: by-product ratio to 1:1, still favoring the by-product (5(12)‐oxa‐3(11)‐cyclotaxane) formation in E. coli. This was similar to an older study where fusing the truncated Taxus POR to N-terminally modified and truncated CYP725A4 with the same linker resulted in almost equal ratio of main product (taxa-4(20),11-dien-5alpha-ol) and by-product (5(12)‐oxa‐3(11)‐cyclotaxane) formation in E. coli. However, these results do not seem sufficient to provide a sound base for claiming that the linkers are the sole players in novel P450 product synthesis. Indeed, the role of evolution for the superior kinetic properties of self-sufficient enzymes shall not be ignored. For instance, the linker-containing, self-sufficient enzyme like P450BM3 with remarkable kinetic properties, has long served as a model for engineering the other P450s, while also being used to catalyse the sesquiterpene valencene oxidation in E. coli, albeit with low selectivity [Citation153]. However, looking at this issue differently, an interesting discovery by Winzer et al. [Citation154] revealed that the presence of a natural fusion protein called ″STORR: [(S)- to (R)-reticuline]” which links P450 (CYP82Y2) to aldo-keto reductase (oxidoreductase in high-reticuline (an alkaloid)-producing mutants of poppy lines), prevented the accumulation of highly reactive or unstable intermediates. Therefore, it remains a question whether the by-products are actually the result of inefficient electron coupling due to inefficient linker design or P450-reductase interactions, and they are the actual transient intermediates (metabolons) in the metabolic pathway [Citation155] that rapidly form and are replaced with more stable compounds, or they are simply formed as a result of underground metabolism in the heterologous host [Citation156], or are formed due to low P450 enzymatic activity [Citation73,Citation157]. Clearly, the eventual decision over fusing or separating P450 and reductases and the linker type depends on the context and the individual enzymes. On this matter, the complementary computational protein modeling tools like I-TASSER have proven to be useful for fusion protein design [Citation158,Citation159] to reduce the possibility of the inefficient electron transfer and formation of by-products.
Regeneration of reducing equivalents for efficient P450 activity
Bioelectrocatalysis and photochemical catalysis: developing cell-free systems for cofactor regeneration
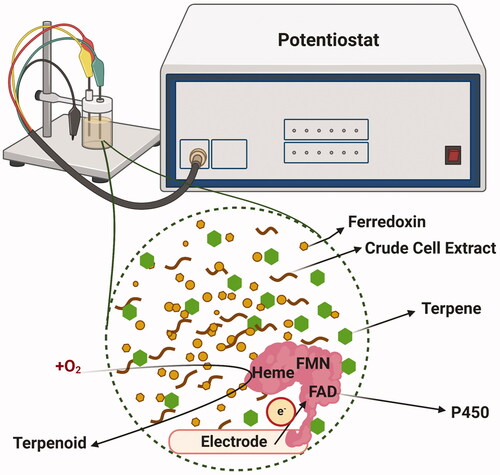
In addition to whole-cell systems, a variety of cell-free techniques such as bioelectrocatalysis have been used in P450 research to provide the required reducing environment. The direct NAD(P)H supplementation in the reaction mixture or P450 immobilisation along with a reductant and electron mediator such as zinc dust and cobalt(II) sepulchrate trichloride have been shown to overcome the self-sufficient P450 enzymes instability and increase the electron transfer rate [Citation110,Citation130,Citation160,Citation161] (). For instance, in a recent ground-breaking study by Frank et al. [Citation162], a three-electrode setup in conjunction with a 96-multipotentiostat and vertical-divided cell architecture and indium tin oxide as the electrode material, preserved up to 80% bioelectrocatalytic activity of the immobilised flavin-deficient variant of P450BM3. In another study, by Ulrich Schwaneberg’s group [Citation163], the 2-β-hydroxy-1,8-cineole formation rate by Citrobacter braakii P450 CinA-10aa-CinC fusion protein [Citation164] was increased by 1.5-fold using either zinc/cobalt (III) sepulchrate or platinum/cobalt (III) sepulchrate, in comparison to equimolar of CinA, CinC and Fpr mixture in presence of NADPH. However, several studies have shown that bioelectrocatalysis is also at the risk of direct O2 reduction by the cathode and reactive oxygen species (ROS) generation, unfavorable enzyme orientation, infeasible electrochemical reaction [Citation165,Citation166], and P450 conformation changes [Citation161]. For instance, in Mie et al. [Citation161] study, using lower molecular weight redox partners like ferredoxin (Fdx) compared to ferredoxin reductase (FdR), optimising the concentration of the purified enzyme and using larger and surface-modified electrode (ITO-AUPA) outperformed the conventional NADPH and catalase supplementation methods in reducing the electron uncoupling and side reactions, while increasing the 25-hydroxyvitamin D3 concentration. However, the P450 activity was decreased by 17% with time in presence of sufficient substrate, which was attributed to some of the most common drawbacks for applying these bioelectrochemical systems. These include the electrode low stability and its deterioration and/or enzyme deactivation and ROS generation. Also, similar to Frank et al. [Citation162], using whole-cell, unpurified E. coli crude extract decreased the product titer by 70%, potentially due to electron loss from mediator or fouling of high molecular weight or competing biopolymers that masked the electron transfer [Citation162].
Figure 6. A hypothetical representation of bioelectrocatalytic, cell-free system for self-sufficient P450-mediated production of terpenoids. Either the self-sufficient P450-expressing crude microbial cell extract with ferredoxin or electrode-bound purified P450 enzyme can be used to potentially hydroxylate the terpenes. The redox potential is simultaneously monitored with a potentiostat device. Figure created with BioRender.com.
Self-assembly of enzymes with nanomaterials is also another bioelectrocatalysis method which can aid in electron delivery and improve the enzyme catalytic activity [Citation130]. For instance, Darimont et al. [Citation167] used carbon nanotube for anchoring the P450BM3, which, together with low pH buffer and high catalase concentration, improved the coupling efficiency and NADPH-dependent activity by 32- and 13-fold, respectively. However, inadequate reduction equivalents reduced the hydroxylation activity, re-emphasising the key role of electron supply pool. Furthermore, apart from being costly for large scale applications, the accumulated NAD(P) can be inhibitive to NAD(P)H utilising enzymes and can lead to formation of enzymatically inactive NAD(P)H isomers (1,2-NAD(P)H and 1,6-NAD(P)H) [Citation88,Citation168]. New studies have shown that it is possible to build more efficient and stable photoelectrochemical systems like nanostructured electrodes from inexpensive materials that are devoid of inactivate NAD(P)H dimers formation [Citation169]. However, it is questionable if they can sustain production of P450-catalysed natural products like terpenoids in long term.
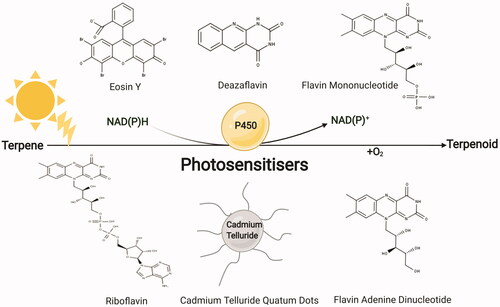
In contrast to bioelectrochemical systems, the light-driven, photochemical reduction systems rely on exogenously supplementing and covalently attaching light-harvesting units (photosensitisers) such as fluorescent eosin Y dye as electron shuttles for rapid electron injection and transfer to P450 heme active site. However, most of the photochemical-based P450 catalysis have so far relied on P450BM3. In Park et al. [Citation170], novel cofactor-free and reductase-independent platform from bacterial P450BM3 and human P450s (CYP1A1, 1B1, 1A2, 2A6, 2E1 and 3A4) were developed using photosensitiser eosin Y which enabled 7‐ethoxycoumarin o-dealkylation and drug bioconversions, respectively. However, eosin Y dye suffers from operating under certain light intensities for providing photons that facilitate P450 turnover and cofactor regeneration and there is a risk of photobleaching at higher light irradiation [Citation171]. However, there are reports using other good photosensitisers such as: riboflavin, FAD and FMN [Citation172], Cadmium telluride (CdTe) quantum dots [Citation173] as well as deazaflavin [Citation174] (). For example, Spradlin et al. [Citation175] studied the attachment of P450BM3 heme domain to ruthenium(II)-diimine photosensitiser via a non-native single cysteine residue and a single point mutation enabled up to 60 and 90% more photocatalytic activity and product conversion, respectively. Overall, better electron coupling and lower electron transfer rate have been reported in photochemical systems [Citation171,Citation175] compared to convenient NADPH-based assays. Therefore, using them in combination with microbial cells can be a new avenue for terpenoid biosynthesis research. However, similar to any other process upscaling, their economic feasibility must be initially assessed.
Figure 7. Schematic of photochemical-mediated P450 catalysis in in vitro systems for terpenoids synthesis. Photosensitisers are light-harvesting units that provide electrons to P450 enzymes for biosynthesis. MarvinSketch 20.17.0 was used for displaying the chemical structures, ChemAxon (https://www.chemaxon.com). Figure created with BioRender.com.
Not going so far, the photosynthetic organisms are natural, in vivo photochemical systems. Thanks to the reducing environment of chloroplast stroma, the P450 stabilisation and activity is promoted together with a great pool of ferredoxin (Fdx) and oxygen [Citation160,Citation176]. On this matter, an emerging technology, although still in its infancy stage, is to couple the P450 to chloroplast or cyanobacterial thylakoids. Here, the photosynthesis is mediated through electron carriers found in thylakoid lumen and stroma and this favors P450 activity with a continuous supply of reducing factors [Citation160] (). Upon water oxidation at photosystem II (PSII) and electron transfer to the photosystem I (PSI), the electrons will reduce the Fdx isomers and they will be majorly spent for NADP reduction through sequential binding of two Fdx molecules to ferredoxin-NADP+ oxidoreductase (FNR) [Citation177,Citation178]. The generated NADPH can potentially be synchronised for P450-mediated terpenoid biosynthesis and the competing electron sinks can also be deleted to further optimise the desired P450 activity [Citation179]. For instance, Berepiki et al. [Citation180] utilised cyanobacterium Synechococcus PCC 7002 photosynthetic electron transport chain to drive heterologous expression of CYP1A1 by using electrons from water splitting and knocking out NDH-1 (a subunit of type I NADH dehydrogenase complex in cyclic photosynthetic electron flow) which doubled the P450 activity.
Figure 8. Photosynthetic electron transport chain reduces NAD(P) to fuel terpenoid biosynthesis by P450 enzymes. PSI: Photosystem I; PSII: Photosystem II; PQ: Plastoquinone; FNR: Ferredoxin-NADP+ oxidoreductase; Fdx: Ferredoxin; NDH-1: NADPH dehydrogenase; SOD: Superoxide dismutase; Cyt b6/f Complex: Cytochrome b6f complex. Adapted from “Light Dependent Reactions of Photosynthesis”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
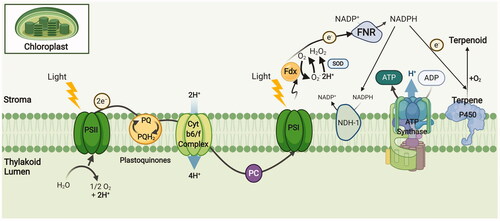
Despite the advantages the natural, light-driven P450 biosynthesis offers, it requires sufficient electron allocations in competition with endogenous processes that rely on Fdx. These include: FNR, lipid and chlorophyll biosynthesis, nitrate and sulfur assimilation, thioredoxin-mediated redox regulation of chloroplast metabolism and cyclic electron flow [Citation160,Citation181,Citation182]. NADP+ and P450 also both compete for reduced Fdx produced from PSI. This is consistent with Nielsen et al. [Citation176] findings, who reported that electron flux is directed toward FNR in the excess of exogenous NADP+, resulting in 64% reduction in CYP79A1 in vitro activity present in isolated thylakoids. In this scenario, a possible way to maximise the heterologous P450 activity would be to either mutate the P450 interacting phase of Fdx or to fuse the P450 to Fdx to manage electron partitioning as also indicated previously [Citation160]. Applying the later strategy, Goñi et al. [Citation183] presented a hybrid electron transfer chain in which B. megaterium ATCC 13368 CYP106A2 was reduced by cyanobacterium (Anabaena sp. PCC 7119) photosynthetic electron transfer flavodoxin (Fld) and Fdx in vivo. However, from another viewpoint, although the photosynthetic organisms can accumulate NADPH and ATP under macronutrient depletion [Citation184,Citation185], the reaction of oxygen with these high potential electrons can lead to reactive oxygen species (ROS) generation. This phenomenon that can even occur under mild light irradiation, is unlikely to be compensated with alternate electron transfer pathways that do not yield NADPH [Citation180].
Enzyme-mediated cofactor recycling
There is a growing interest in in vitro systems and chemo-enzymatic cascade reactions for the synthesis of useful chemicals [Citation186–188]. Here, the reaction depends on exogenously supplementing the NADP(H) cofactor for purified enzyme or crude cell extract. However, this cofactor has proven to be the most expensive component for the generally costly cell-free synthesis [Citation189]. Therefore, unless an NADPH-rich, lyophilised cell is utilised [Citation190], adapting in vitro systems instead of whole-cell biocatalysis must be re-considered economically from the aspect of P450 enzyme overall contribution. It must be noted that interesting cofactor analog such as carba-NADP(H) has also been recently proposed [Citation191]. But, its weaker activity compared to the commonly used NAD(P)H justifies the superiority of whole-cell biocatalysis. There might be also some concerns over the lack of sufficient microbial research infrastructures in chemistry labs [Citation129]. However, the valuable open-source microcontroller-based toolkits and 3D printing tools can effectively assist in constructing relatively low-cost miniature bioreactors from scratch for effective control of the microbial systems [Citation192,Citation193].
Going forward, to promote the whole-cell biocatalysis, the enzymatic methods have been extensively used for NAD(P)H cofactor regeneration (), among which hydrogenases, dehydrogenases and oxidases being the most applied. Hydrogenases have 100% atom efficiency and are by-product free compared to commonly used dehydrogenases [Citation166]. As they can be deactivated in the presence of oxygen, aerobic-tolerant hydrogenases like Ralstonia eutropha [NiFe] hydrogenase, with inhibition tolerance of 20% at 60% oxygen of total gas (0.7 mM of O2) can be used for P450 reaction [Citation194], although this has not been reported for terpenoid biosynthesis so far. However, perhaps, a good example is a study by Lonsdale et al. [Citation195], where it was reported that using R. eutropha soluble NAD+-reducing hydrogenase for Polaromonas sp. JS666 alkane-oxidising CYP153A in P. putida, increased the 1-octanol yield by three-fold. The first use of hydrogenases for oxygen-dependent enzymes was recently shown by Preissler et al. [Citation196] using a mutant variant of R. eutropha O2‐tolerant NAD+‐reducing hydrogenase (E341A/S342R), which supported the activity of P450BM3 for octane oxidation.
Figure 9. Selection of enzymatic routes to NAD(P)H regeneration for P450-mediated terpenoid synthesis. Figure created with BioRender.com.
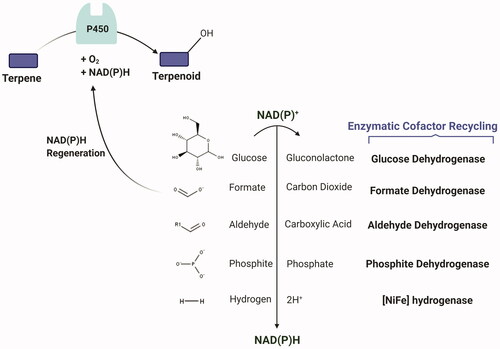
From dehydrogenases, aldehyde, glucose and formate dehydrogenases, have been used for optimising the P450 product yield [Citation197,Citation198]. However, formate dehydrogenase is probably the least optimal due to its low specific activity and stability, preference for NAD+, as well as product loss in the form of CO2 (Citation197). However, formate dehydrogenases from Burkholderia cepasia and acidobacterium Granulicella mallensis with both NAD+ and NADP+ specificity, have been identified [Citation198,Citation199]. While being recognised for in vitro P450 catalysis using bacterial crude cell extracts [Citation200] or for co-immobilisation [Citation201], glucose dehydrogenases are used in microbial terpenoid biosynthesis, a notable example being Luan et al. [Citation127]. Here, the glucose dehydrogenase coexpression improved (−)-limonene conversion to (−)-isopiperitenol by 10-fold in E. coli expressing Rhodococcus sp. ECU0066 P450SMO reductase-GS linker- P. putida PpGl P450cam (Y96F/V247L). Brummund et al. [Citation202] also used a crude, cell-free extract of E. coli K12 derivative to convert sesquiterpene α-ionone to its oxidised form using a mutant variant of P450BM3 (R47L/F87V/L188Q), which relied on commercial glucose dehydrogenase system for NADPH regeneration. Chen et al. [Citation203] also reported up to 14-fold increased artemisinic acid production when coexpressing A. annua aldehyde dehydrogenase with N-terminus truncated A. annua CYP71AV1-POR fusion in S. cerevisiae. Likewise, using B. megaterium glucose dehydrogenase and E. coli transhydrogenase improved the titer up to two- and four-folds, respectively. Thermostable phosphite dehydrogenases have also been used in fusion with self-sufficient Baeyer–Villiger monooxygenases (CRE2 (coenzyme regenerating enzyme)/BVMO) for NADPH regeneration for ketone conversion using E. coli crude cells [Citation129,Citation204]. Although these studies did not use P450s, they lend the opportunity to deploy phosphite dehydrogenases for P450 biotransformation through using phosphite-oxidising microbes in co-culture systems [Citation205,Citation206]. However, using glucose dehydrogenase involves the co-production of acidifying gluconate/gluconic acid in the medium, which although they might be overcome with biphasic culture systems, increased glucose feeding and pH control, they would increase the costs and complicate the identification and purification of substrates from the products [Citation197,Citation200,Citation207].
The dehydrogenases might act distinctly when partnered with P450s for terpenoid biosynthesis as illustrated in Meng et al. study [Citation208]. Here, while using short-chain dehydrogenase/reductase (SDR) superfamily dehydrogenases (Z. zerumbet ZSD1 and Citrus sinensis ABA2 or P. pastoris dehydrogenase (AHD-C3)) yielded between 49 and 60 mg/L of (+)-nootkatone in S. cerevisiae, the endogenous dehydrogenases (ADH2 and AHD6) did not significantly change the product concentration. This was attributed to their interference with P450 and reductase enzymes (H. muticus premnaspirodiene oxygenase (HPO) variant (V482I/A484I) and A. thaliana cytochrome P450 reductase (ATR1)). The other general disadvantage of dehydrogenases is that they can get deactivated by NADPH-mediated product inhibition, lending the opportunity to NAD(P)H oxidases (NOXs), despite not have been reported for P450-catalysed terpenoid biosynthesis so far. NOXs show more specificity towards NADH, even in species with dual (NADH and NADPH) specificity [Citation197]. However, a water-forming Lactobacillus reuteri NOX with equal efficiency for both NADPH and NADH and higher activity with NADPH, has been reported to improve the Gluconobacter oxydans sorbitol dehydrogenase total turnover by around 53-fold [Citation209]. Moreover, dual-specific NOXs can be artificially constructed as was shown with P450BM3 for vanillin production [Citation210].
Excess cofactor regeneration: good or bad?
Despite the great potential of cofactor regeneration systems, the cofactor supply might not be always the rate-limiting step for terpenoid formation. Li et al. [Citation211] demonstrated that increasing the exogenous supplementation of mutated 2,3-butanediol dehydrogenase (mBDH1) substrate, acetoin, although initially improved the betulinic acid concentration by 1.5-fold at 20 mM concentration at the price of decreased growth rate, it did not further increase the product concentration at 100 mM. Similarly, recently, Otto et al. [Citation73] reported that the titer of sesquiterpenoid, abscisic acid (ABA) produced by an engineered S. cerevisiae strain expressing Botrytis cinereal four-step ABA biosynthesis pathway genes (bcaba1234), did not depend on the NADPH supply. Instead, the rate-limiting enzymes were the P450s BcABA1 and BcABA2, which improved the ABA titer by 4.1-fold upon overexpression. Janocha and Bernhardt [Citation105] had also shown that the co-expression of Lactobacillus brevis alcohol dehydrogenase and dissolving the substrate, abietic acid, in 2-propanol as co-substrate for NADPH regeneration only enhanced the 15-hydroxyabietic acid formation rate by 10% from whole-cell E. coli JM109 system, transiently expressing resin acid diterpenoid hydroxylase (Streptomyces griseolus CYP105A1), Schizosaccharomyces pombe adrenodoxin reductase homolog 1 (Arh1) and electron transfer protein 1 and ferredoxin domain (Etp1fd). However, the limited concentration of the reducing cofactor might also not be favorable to P450 catalysis. For instance, Ensari et al. [Citation212] could only recover the efficiency of hydroxy- and keto-fatty acid methyl esters production by using ethanol as a co-substrate for producing NADH cofactor in E. coli expressing Candida parapsilosis alcohol dehydrogenase 5 (cpADH5 (W286A)) and P450BM3 (R47S/Y51W/T235S/N239R/I401 M). Therefore, although excess NAD(P)H might inactivate P450s, the efficient P450 biocatalysis strongly depends on proper redox balance. Such shortcomings in previously explained cell-free or whole-cell enzymatic methodologies can be systematically addressed through systems metabolic engineering and mathematical modeling approaches. As a common practice, the NADPH-consuming central carbon metabolism reactions are downregulated or knocked out and NADPH generating pathways are upregulated, albeit at the risk of reduced strain fitness. Instead, the genome-scale models are becoming more popular to help scientists modulate the microbial growth and productivity. However, not all microbial horse powers are favored to have a good model yet and the models for more widely used microorganisms like S. cerevisiae are still evolving to enhance their prediction power [Citation213]. In recent years, the metabolic and regulatory target predicting algorithms [Citation214–218] have been integrated into programming work benches to address these issues [Citation219–223]. Among relevant applications to the P450 biocatalysis are the cofactor swapping methodology for redox balance [Citation224] and cofactor modification analysis [Citation225]. Also, there has been an emerging interest in improving the microbial biocatalysis through computational approaches in addition to flux balance analysis and multi-omics data integration [Citation226–229]. Hence, it is not ambitious to envisage that mathematical modeling tools will reach a level to be the next players in optimising the microbial terpenoid biosynthesis with the least effect on cell activities as compared to rather blind enzyme selections described above.
Practical considerations for redox manipulation: reductase-associated toxicity
The natural, inevitable uncoupling in P450s is possibly negligible compared to higher reactive oxygen species (ROS) and proton leak caused by mitochondria and oxidative phosphorylation, a price that cells pay for aerobic energy transformation [Citation230,Citation231]. As discussed, the excess reductase expression can lead to electron transfer uncoupling, ROS production and reduced terpenoid titer. This might explain the higher ratio of P450s to PORs in plants, reaching up to 15:1 [Citation33,Citation232]. Despite this, the ROS problem is commonly tackled using scavenging systems and antioxidants like superoxide dismutase and catalase. However, these methods are inefficient for removing the lower hydrogen peroxide levels [Citation233] or are associated with energy-intensive uncoupling [Citation127,Citation231]. Although hydrogen peroxide is blamed for causing cellular death, it has been shown to improve antioxidant carotenoids production by three-fold [Citation234] and it can be effectively controlled to power P450-based catalysis. For instance, Jiang et al. [Citation235] used Streptomyces coelicolor A3(2) alditol oxidase (AldO) and commercial lipase (CRL) for hydrogen peroxide formation for in situ free fatty acids decarboxylation by an artificial three-enzyme cascade of OleTJE P450 peroxygenase and fatty acid decarboxylase. This approach was also found to be FMN- and NADPH-saving and preserved the enzyme from hydrogen peroxide-mediated deactivation and instability, despite loss of activity at higher hydrogen peroxide concentrations. Zhang et al. [Citation106] also observed the partial contribution of superoxide to novel N-demethylation activity of P450 MycG enzyme. An old study by Cirino and Arnold [Citation236] also showed the applicability of hydrogen peroxide for improving the hydroxylase activity of a P450BM3 variant in E. coli. For this to occur, other than developing more stable P450s, the laboratory microbial strains need to be equipped with better oxidative stress resistance through adaptive evolution at adverse conditions [Citation237] to facilitate the large-scale P450-based terpenoid biosynthesis.
Although not tested, it would be interesting to explore microbial cells with superior endogenous hydrocarbon degrading mechanisms such as n-alkane utilising Alcanivorax dieselolei strain B‐5 [Citation238] and alkylguaiacols-utilising Rhodococcus rhodochrous EP4 [Citation239], which might better handle the ROS-caused toxicities for improved P450-based terpenoid production. Despite their potential benefits, the tools and knowledge for genetic manipulation and cultivation of these microorganisms are limited. There are, however, more widely used host microbes like P. putida which has remarkable oxidation capacity for terpenoid production [Citation240]. It also has unique and elaborate oxidative stress response owing to its Entner–Doudoroff pathway [Citation241], adaptive membrane fluidity, chaperone-assisted protein refolding and efflux pumps in some of its strains [Citation242]. The other stress-resistant microorganisms are Bacillus spp. with a network of metal- and thiol-based oxidative stress response mechanisms [Citation243]. The diatom Phaeodactylum tricornutum has also been reported to well-tolerate the P450 and POR enzymes without affecting the cell growth, suggesting its potential for wider applications in industrial P450 biotransformation [Citation244]. Alternatively, increasing the ergosterol and triacylglycerol biosynthesis rate or exogenous ergosterol supplementation seem promising particularly for non-oleaginous microbial chassis [Citation237,Citation245]. This is because the microorganisms with more oxidative stress resistance like two non-conventional yeast strains, Torulaspora delbrueckii and Metschnikowia pulcherrima, have been reported to have higher ergosterol/squalene and phosphatidylcholine/phosphatidylethanolamine content in their plasma membrane, which are postulated to increase their hydrogen peroxide permeability as reported before [Citation246]. Contrary to these findings, studies like Brennan et al. [Citation247] have not reported ergosterol synthesis upregulation in limonene-producing yeast with known toxicity. Hence, this approach might involve preliminary trial and error experiments.
Conclusions and prospects
Cytochrome P450s can be undoubtedly one of those most versatile and popular choices for industrial biotechnology, either for producing natural products or to synthesise new-to-nature chemicals. Aiming to hopefully replace the sophisticated organic chemistry approaches as a cheap and eco-friendly alternative, there is a need to finely tune them to meet industrial requirements for high productivity. Despite the scientific advances and exploratory attempts to reach a comprehensive understanding of redox state and its functional dimensions for P450-catalysed terpenoid biosynthesis, there are moderate industrial achievements.
Among many decisive parameters for their optimal functions, the current paper focuses on the important regulatory role of NAD(P)H electron-deriving reductases in microbial hosts for heterologous production of terpenoids. The optimal construction of P450-reductase fusion proteins and selection of proper linkers can both impact the P450 functionality and product profile through affecting the P450-reductase interaction. However, the P450 research community still needs user-friendly standardised methodologies, synthetic biology and protein modeling toolkits to accelerate the protein fusion design for the biosynthesis of P450-catalysed terpenoids and other natural products.
Beside the fusion proteins, several non-enzymatic and enzymatic NAD(P)H regeneration approaches also exist that could exert strong impact on the activity of whole P450-redox partner systems. Although the course of this paper was based on the whole microbial cell biosynthesis, a number of the latest cutting-edge discoveries in cell-free P450-redox tuning were discussed as potential avenues to be adapted and combined in whole-cell systems. However, as these methodologies increase the risk of redox imbalance and oxidative stress, using more stress-resistant microbial hosts or developing microbial consortia are suggested. While non-model microorganisms from extreme environments could possess superior catalysis functionalities relative to commonly used laboratory strains, their common usage is still far from reality. Instead, it is anticipated that evolving new microbial strains, while not being superior to extremophiles, can remarkably enhance the microorganism characteristics for P450-catalysed terpenoid biosynthesis
With these limitations, there is still room for improvement with regards to cofactor regeneration and modulation of P450 and reductase expressions at whole-cell. Indeed, P450 research can instead benefit from systems biology-guided metabolic engineering to decrease the associated costs and risks, and increase the flux towards terpenoid synthesis, through engineering the cell factory and/or modifying the bioprocessing parameters like growth medium composition.
Having broad selectivity and being favored with available cofactor pool, microorganisms provide ample opportunities to extend our reach to a diverse range of terpenoids with roots in P450-dominated biosynthetic pathways. We therefore close by noting that we have just scratched the surface of whole-cell biocatalysis and to our belief, microorganism-wise modulation of reductase partner and cofactors can systematically shape the next generation of P450 whole-cell catalysis research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Janocha S, Schmitz D, Bernhardt R. Terpene hydroxylation with microbial cytochrome P450 monooxygenases. In: Schrader J, Bohlmann J, editors. BT - biotechnology of isoprenoids. Cham: Springer International Publishing; 2015. p. 215–250. Available from: https://doi.org/10.1007/10_2014_296
- Pankov KV, McArthur AG, Gold DA, et al. The cytochrome P450 (CYP) superfamily in cnidarians. Sci Rep. 2021;11(1):9834.
- Zheng X, Li P, Lu X. Research advances in cytochrome P450-catalysed pharmaceutical terpenoid biosynthesis in plants. J Exp Bot. 2019;70(18):4619–4630.
- Nelson DR. Cytochrome P450 diversity in the tree of life. Biochim Biophys Acta Proteins Proteom. 2018;1866(1):141–154.
- Di Nardo G, Gilardi G. Natural compounds as pharmaceuticals: the key role of cytochromes P450 reactivity. Trends Biochem Sci. 2020;45(6):511–525.
- Li Z, Jiang Y, Guengerich FP, et al. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J Biol Chem. 2020;295(3):833–849.
- Xu L-H, Du Y-L. Rational and semi-rational engineering of cytochrome P450s for biotechnological applications. Synth Syst Biotechnol. 2018;3(4):283–290.
- Huang M-Z, Li J-Y. Physiological regulation of reactive oxygen species in organisms based on their physicochemical properties. Acta Physiol (Oxf). 2020;228(1):e13351.
- Guengerich FP, Munro AW. Unusual cytochrome P450 enzymes and reactions. J Biol Chem. 2013;288(24):17065–17073.
- Munro AW, McLean KJ, Grant JL, et al. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem Soc Trans. 2018;46(1):183–196.
- Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124(1):128–145.
- Aeling KA. WO 2019/014310 Al [Internet]. 2018. Available from: https://patents.google.com/patent/WO2019014310A1/en?oq=WO+2019%2F014310+Al.
- Schalk M, Rocci L. WO 2018/015453 A2 [Internet]. Vol. 7. 2017. Available from: https://patents.google.com/patent/WO2018015453A2/en?oq=WO+2018%2F015453+A2.
- Rice CF, Stonehouse EA, Memmer N, et al. WO 2019/171230 A1 [Internet]. Vol. 1. 2019. Available from: https://patents.google.com/patent/WO2019171230A1/en?oq=WO+2019%2F171230+A1.
- Chappell J, Zhuang X, Wu S. US 10,597,665 B1 [Internet]. Vol. 1. 2013. Available from: https://patents.google.com/patent/US10597665B1/en?oq=US+10%2C597%2C665+B1.
- Paddon CJ, Westfall PJ, Pitera DJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532.
- Wang M, Roberts DL, Paschke R, et al. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA. 1997;94(16):8411–8416.
- Meharenna YT, Li H, Hawkes DB, et al. Crystal structure of P450cin in a complex with its substrate, 1,8-cineole, a close structural homologue to D-camphor, the substrate for P450cam. Biochemistry. 2004;43(29):9487–9494.
- Zhang L, Xie Z, Liu Z, et al. Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase. Nat Commun. 2020;11(1):2676.
- Sevrioukova IF, Li H, Zhang H, et al. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci USA. 1999;96(5):1863–1868.
- Belsare KD, Ruff AJ, Martinez R, et al. Insights on intermolecular FMN-heme domain interaction and the role of linker length in cytochrome P450cin fusion proteins. Biol Chem. 2020;401(11):1249–1255.
- Cook DJ, Finnigan JD, Cook K, et al. Chapter five - cytochromes P450: history, classes, catalytic mechanism, and industrial application. In: Christov CZBT-A, editor. Insights into enzyme mechanisms and functions from experimental and computational methods. Cambridge, MA: Academic Press; 2016. p. 105–126.
- Hannemann F, Bichet A, Ewen KM, et al. Cytochrome P450 systems—biological variations of electron transport chains. Biochim Biophys Acta - Gen Subj. 2007;1770(3):330–344.
- Finnigan JD, Young C, Cook DJ, et al. Chapter nine - cytochromes P450 (P450s): a review of the class system with a focus on prokaryotic P450s. In: Karabencheva-Christova T, Christov CBT-A, editors. Advances in protein chemistry and structural biology. Cambridge, MA: Academic Press; 2020. p. 289–320.
- Li S, Du L, Bernhardt R. Redox partners: function modulators of bacterial P450 enzymes. Trends Microbiol. 2020;28(6):445–454.
- Munro AW, Girvan HM, McVey JP, et al. Cytochrome P450 redox partner systems: biodiversity and biotechnological implications. In: Urlacher VB, Schmid RD, editors. Modern Biooxidation [Internet]. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2007. p. 123–53. (Wiley Online Books).
- Munro AW, Girvan HM, McLean KJ. Cytochrome P450-redox partner fusion enzymes. Biochim Biophys Acta. 2007;1770(3):345–359.
- Chiliza ZE, Martínez-Oyanedel J, Syed K. An overview of the factors playing a role in cytochrome P450 monooxygenase and ferredoxin interactions. Biophys Rev. 2020;12(5):1217–1222.
- Wang Z, Shaik S, Wang B. Conformational motion of ferredoxin enables efficient electron transfer to heme in the full-length P450TT. J Am Chem Soc. 2021;143(2):1005–1016.
- Zhang W, Du L, Li F, et al. Mechanistic insights into interactions between bacterial class I P450 enzymes and redox partners. ACS Catal. 2018;8(11):9992–10003.
- de las Heras B, Rodríguez B, Boscá L, et al. Terpenoids: sources, structure elucidation and therapeutic potential in inflammation. Curr Top Med Chem. 2003;3(2):171–185.
- Pattanaik B, Lindberg P. Terpenoids and their biosynthesis in cyanobacteria. Life (Basel). 2015;5(1):269–293.
- Jensen K, Møller BL. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry. 2010;71(2–3):132–141.
- Nadler SG, Strobel HW. Role of electrostatic interactions in the reaction of NADPH-cytochrome P-450 reductase with cytochromes P-450. Arch Biochem Biophys. 1988;261(2):418–429.
- Ravichandran KG, Boddupalli SS, Hasermann CA, et al. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450’s. Science. 1993;261(5122):731–736.
- Esteves F, Campelo D, Gomes BC, et al. The role of the FMN-Domain of human cytochrome P450 oxidoreductase in its promiscuous interactions with structurally diverse redox partners. Front Pharmacol. 2020;11:299.
- Mukherjee G, Nandekar PP, Wade RC. An electron transfer competent structural ensemble of membrane-bound cytochrome P450 1A1 and cytochrome P450 oxidoreductase. Commun Biol. 2021;4(1):55.
- Kenaan C, Zhang H, Shea EV, et al. Uncovering the role of hydrophobic residues in cytochrome P450-cytochrome P450 reductase interactions. Biochemistry. 2011;50(19):3957–3967.
- Shen AL, Kasper CB. Role of acidic residues in the interaction of NADPH-Cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem. 1995;270(46):27475–27480.
- Laursen T, Jensen K, Møller BL. Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. Biochim Biophys Acta. 2011;1814(1):132–138.
- Sugishima M, Taira J, Sagara T, et al. Conformational equilibrium of NADPH–cytochrome P450 oxidoreductase is essential for heme oxygenase reaction. Antioxidants. 2020;9(8):673.
- Iijima M, Ohnuki J, Sato T, et al. Coupling of redox and structural states in cytochrome P450 reductase studied by molecular dynamics simulation. Sci Rep. 2019;9(1):9341.
- Gutierrez A, Munro AW, Grunau A, Wolf CR, et al. Interflavin electron transfer in human cytochrome P450 reductase is enhanced by coenzyme binding. Relaxation kinetic studies with coenzyme analogues. Eur J Biochem. 2003;270(12):2612–2621.
- Freeman SL, Martel A, Raven EL, et al. Orchestrated domain movement in catalysis by cytochrome P450 reductase. Sci Rep. 2017;7(1):9741.
- Barnaba C, Ramamoorthy A. Picturing the membrane-assisted choreography of cytochrome P450 with lipid nanodiscs. Chemphyschem. 2018;19(20):2603–2613.
- DeJong JM, Liu Y, Bollon AP, et al. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93(2):212–224.
- Walls LE, Malcı K, Nowrouzi B, et al. Optimizing the biosynthesis of oxygenated and acetylated taxol precursors in Saccharomyces cerevisiae using advanced bioprocessing strategies. Biotechnol Bioeng. 2021;118(1):279–293.
- Renault H, Bassard J-E, Hamberger B, et al. Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr Opin Plant Biol. 2014;19:27–34.
- Thesseling FA, Hutter MC, Wiek C, et al. Novel insights into oxidation of fatty acids and fatty alcohols by cytochrome P450 monooxygenase CYP4B1. Arch Biochem Biophys. 2020;679:108216.
- Vergères G, Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 1995;77(7–8):604–620.
- Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003;97(2):139–152.
- Storbeck K-H, Swart AC, Goosen P, et al. Cytochrome b5: novel roles in steroidogenesis. Mol Cell Endocrinol. 2013;371(1–2):87–99.
- Elahian F, Sepehrizadeh Z, Moghimi B, et al. Human cytochrome b5 reductase: structure, function, and potential applications. Crit Rev Biotechnol. 2014;34(2):134–143.
- Shirabe K, Fujimoto Y, Yubisui T, et al. An in-frame deletion of codon 298 of the NADH-cytochrome b5 reductase gene results in hereditary methemoglobinemia type II (generalized type). a functional implication for the role of the COOH-terminal region of the enzyme. J Biol Chem. 1994;269(8):5952–5957.
- McLean KJ, Sabri M, Marshall KR, et al. Biodiversity of cytochrome P450 redox systems. Biochem Soc Trans. 2005;33(Pt 4):796–801.
- Porter TD. The roles of cytochrome b5 in cytochrome P450 reactions. J Biochem Mol Toxicol. 2002;16(6):311–316.
- Stiborová M, Indra R, Moserová M, et al. Comparison of human cytochrome P450 1A1-catalysed oxidation of benzo[a]pyrene in prokaryotic and eukaryotic expression systems. Monatsh Chem. 2017;148(11):1959–1969.
- Liu J, Zhang X, Wu H, et al. Characteristics and roles of cytochrome b5 in cytochrome P450-mediated oxidative reactions in Locusta migratoria. J Integr Agric. 2020;19(6):1512–1521.
- Noble MA, Girvan HM, Smith SJ, et al. Analysis of the interactions of cytochrome b5 with flavocytochrome P450 BM3 and its domains. Drug Metab Rev. 2007;39(2–3):599–617.
- Hayashi K, Sakaki T, Kominami S, et al. Coexpression of genetically engineered fused enzyme between yeast NADPH-P450 reductase and human cytochrome P450 3A4 and human cytochrome b5 in yeast. Arch Biochem Biophys. 2000;381(1):164–170.
- Zhang H, Gruenke L, Arscott D, et al. Determination of the rate of reduction of oxyferrous cytochrome P450 2B4 by 5-deazariboflavin adenine dinucleotide T491V cytochrome P450 reductase. Biochemistry. 2003;42(40):11594–11603.
- Im S-C, Waskell L. The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch Biochem Biophys. 2011;507(1):144–153.
- Shet MS, Faulkner KM, Holmans PL, et al. The effects of cytochrome b5, NADPH-P450 reductase, and lipid on the rate of 6 beta-hydroxylation of testosterone as catalyzed by a human P450 3A4 fusion protein. Arch Biochem Biophys. 1995;318(2):314–321.
- Bart AG, Scott EE. Structural and functional effects of cytochrome b5 interactions with human cytochrome P450 enzymes. J Biol Chem. 2017;292(51):20818–20833.
- Estrada DF, Laurence JS, Scott EE. Substrate-modulated cytochrome P450 17A1 and cytochrome b5 interactions revealed by NMR. J Biol Chem. 2013;288(23):17008–17018.
- Wang C, Su X, Sun M, et al. Efficient production of glycyrrhetinic acid in metabolically engineered Saccharomyces cerevisiae via an integrated strategy. Microb Cell Fact. 2019;18(1):95.
- Sagwan-Barkdoll L, Anterola AM. Taxadiene-5α-ol is a minor product of CYP725A4 when expressed in Escherichia coli. Biotechnol Appl Biochem. 2018;65(3):294–305.
- Gao Q, Doneanu CE, Shaffer SA, et al. Identification of the interactions between cytochrome P450 2E1 and cytochrome b5 by mass spectrometry and site-directed mutagenesis*. J Biol Chem. 2006;281(29):20404–20417.
- Zhang T, Rao J, Li W, et al. Mechanism-based inactivation of cytochrome P450 enzymes by natural products based on metabolic activation. Drug Metab Rev. 2020;52(4):501–530.
- Emmerstorfer A, Wimmer-Teubenbacher M, Wriessnegger T, et al. Over-expression of ICE2 stabilizes cytochrome P450 reductase in Saccharomyces cerevisiae and Pichia pastoris. Biotechnol J. 2015;10(4):623–635.
- Chen J, Fan F, Qu G, et al. Identification of absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone. Metab Eng. 2020;57:31–42.
- Szczebara FM, Chandelier C, Villeret C, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat Biotechnol. 2003;21(2):143–149.
- Otto M, Teixeira PG, Vizcaino MI, et al. Integration of a multi-step heterologous pathway in Saccharomyces cerevisiae for the production of abscisic acid. Microb Cell Fact. 2019;18(1):205.
- Biggs BW, Lim CG, Sagliani K, et al. Overcoming heterologous protein interdependency to optimize P450-mediated taxol precursor synthesis in Escherichia coli. Proc Natl Acad Sci USA. 2016;113(12):3209–3214.
- Sarrade-Loucheur A, Ro D-K, Fauré R, et al. Synthetic derivatives of (+)-epi-α-Bisabolol are formed by mammalian cytochromes P450 expressed in a yeast reconstituted pathway. ACS Synth Biol. 2020;9(2):368–380.
- Zhao F, Bai P, Liu T, et al. Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnol Bioeng. 2016;113(8):1787–1795.
- Glick BR. Metabolic load and heterologous gene expression. Biotechnol Adv. 1995;13(2):247–261.
- Moser S, Pichler H. Identifying and engineering the ideal microbial terpenoid production host. Appl Microbiol Biotechnol. 2019;103(14):5501–5516.
- Thirumurthy MA, Jones AK. Geobacter cytochrome OmcZs binds riboflavin: implications for extracellular electron transfer. Nanotechnology. 2020;31(12):124001.
- Huang L, Liu X, Ye Y, et al. Evidence for the coexistence of direct and riboflavin-mediated interspecies electron transfer in geobacter co-culture. Environ Microbiol. 2020;22(1):243–254.
- Zhou K, Qiao K, Edgar S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33(4):377–383.
- Ouyang X, Cha Y, Li W, et al. Stepwise engineering of Saccharomyces cerevisiae to produce (+)-valencene and its related sesquiterpenes. RSC Adv. 2019;9(52):30171–30181.
- Zelcbuch L, Antonovsky N, Bar-Even A, et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013;41(9):e98–e98.
- Ye L, Zhang C, Bi C, et al. Combinatory optimization of chromosomal integrated mevalonate pathway for β-carotene production in Escherichia coli. Microb Cell Fact. 2016;15(1):202.
- Mao W, Berenbaum MR, Schuler MA. Modifications in the N-terminus of an insect cytochrome P450 enhance production of catalytically active protein in baculovirus-Sf9 cell expression systems. Insect Biochem Mol Biol. 2008;38(1):66–75.
- Ajikumar PK, Xiao W-H, Tyo KEJ, et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74.
- Kudla G, Murray AW, Tollervey D, et al. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924):255–258.
- Morrison CS, Paskaleva EE, Rios MA, et al. Improved soluble expression and use of recombinant human renalase. PLoS One. 2020;15(11):e0242109.
- Fox JD, Routzahn KM, Bucher MH, et al. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 2003;537(1–3):53–57.
- Christensen U, Vazquez-Albacete D, Søgaard KM, et al. De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP fusions. Appl Microbiol Biotechnol. 2017;101(10):4103–4113.
- Nørholm MHH, Toddo S, Virkki MTI, et al. Improved production of membrane proteins in Escherichia coli by selective codon substitutions. FEBS Lett. 2013;587(15):2352–2358.
- Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88(13):5597–5601.
- Vazquez-Albacete D, Cavaleiro AM, Christensen U, et al. An expression tag toolbox for microbial production of membrane bound plant cytochromes P450. Biotechnol Bioeng. 2017;114(4):751–760.
- Hausjell J, Halbwirth H, Spadiut O. Recombinant production of eukaryotic cytochrome P450s in microbial cell factories. Biosci Rep. 2018;38(2):BSR20171290.
- Zelasko S, Palaria A, Das A. Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expr Purif. 2013;92(1):77–87.
- Denisov IG, Sligar SG. Cytochromes P450 in nanodiscs. Biochim Biophys Acta. 2011;1814(1):223–229.
- Durairaj P, Hur J-S, Yun H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb Cell Fact. 2016;15(1):125.
- Luthra A, Gregory M, Grinkova YV, et al. Nanodiscs in the studies of membrane-bound cytochrome P450 enzymes. Methods Mol Biol. 2013;987:115–127.
- Lee JH, Nam DH, Lee SH, et al. New platform for cytochrome P450 reaction combining in situ immobilization on biopolymer. Bioconjugate Chem. 2014;25(12):2101–2104.
- Rouck JE, Biggs BW, Kambalyal A, et al. Heterologous expression and characterization of plant taxadiene-5α-hydroxylase (CYP725A4) in Escherichia coli. Protein Expr Purif. 2017;132:60–67.
- Girvan HM, Munro AW. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr Opin Chem Biol. 2016;31:136–145.
- Schempp FM, Drummond L, Buchhaupt M, et al. Microbial cell factories for the production of terpenoid flavor and fragrance compounds. J Agric Food Chem. 2018;66(10):2247–2258.
- Sabbadin F, Hyde R, Robin A, et al. LICRED: a versatile drop-in vector for rapid generation of redox-self-sufficient cytochrome P450s. Chembiochem. 2010;11(7):987–994.
- Wong J, d’Espaux L, Dev I, et al. De novo synthesis of the sedative valerenic acid in Saccharomyces cerevisiae. Metab Eng. 2018;47:94–101.
- Janocha S, Bernhardt R. Design and characterization of an efficient CYP105A1-based whole-cell biocatalyst for the conversion of resin acid diterpenoids in permeabilized Escherichia coli. Appl Microbiol Biotechnol. 2013;97(17):7639–7649.
- Zhang W, Liu Y, Yan J, et al. New reactions and products resulting from alternative interactions between the P450 enzyme and redox partners. J Am Chem Soc. 2014;136(9):3640–3646.
- Lim Y-R, Eun C-Y, Park H-G, et al. Regioselective oxidation of lauric acid by CYP119, an orphan cytochrome P450 from Sulfolobus acidocaldarius. J Microbiol Biotechnol. 2010;20(3):574–578.
- Klenk JM, Dubiel P, Sharma M, et al. Characterization and structure-guided engineering of the novel versatile terpene monooxygenase CYP109Q5 from Chondromyces apiculatus DSM436. Microb Biotechnol. 2019;12(2):377–391.
- Sun W, Xue H, Liu H, et al. Controlling chemo- and regioselectivity of a plant P450 in yeast cell toward rare licorice triterpenoid biosynthesis. ACS Catal. 2020;10(7):4253–4260.
- O'Reilly E, Köhler V, Flitsch SL, et al. Cytochromes P450 as useful biocatalysts: addressing the limitations. Chem Commun. 2011;47(9):2490–2501.
- Behrendorff JBYH, Gillam EMJ. Prospects for applying synthetic biology to toxicology: future opportunities and current limitations for the repurposing of cytochrome P450 systems. Chem Res Toxicol. 2017;30(1):453–468.
- Zhang K, Shafer BM, Demars MD, et al. Controlled oxidation of remote sp3 C–H bonds in artemisinin via P450 catalysts with fine-tuned regio- and stereoselectivity. J Am Chem Soc. 2012;134(45):18695–18704.
- Albertsen L, Chen Y, Bach LS, et al. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol. 2011;77(3):1033–1040.
- Wang X, Pereira JH, Tsutakawa S, et al. Efficient production of oxidized terpenoids via engineering fusion proteins of terpene synthase and cytochrome P450. Metab Eng. 2021;64:41–51.
- Degregorio D, Sadeghi SJ, Di Nardo G, et al. Understanding uncoupling in the multiredox Centre P450 3A4-BMR model system. J Biol Inorg Chem. 2011;16(1):109–116.
- Zhang C, Liu J, Zhao F, et al. Production of sesquiterpenoid zerumbone from metabolic engineered Saccharomyces cerevisiae. Metab Eng. 2018;49:28–35.
- Haslinger K, Prather KLJ. Heterologous caffeic acid biosynthesis in Escherichia coli is affected by choice of tyrosine ammonia lyase and redox partners for bacterial cytochrome P450. Microb Cell Fact. 2020;19(1):26.
- Kim BS, Kim SY, Park J, et al. Sequence-based screening for self-sufficient P450 monooxygenase from a metagenome library. J Appl Microbiol. 2007;102(5):1392–1400.
- Zhang C, Lu M, Lin L, et al. Riboflavin is directly involved in N-dealkylation catalyzed by bacterial cytochrome P450 monooxygenases. Chembiochem. 2020;21(16):2297–2305.
- Munro AW, Leys DG, McLean KJ, et al. P450 BM3: the very model of a modern flavocytochrome. Trends Biochem Sci. 2002;27(5):250–257.
- Zhang J-D, Li A-T, Yang Y, et al. Sequence analysis and heterologous expression of a new cytochrome P450 monooxygenase from Rhodococcus sp. for asymmetric sulfoxidation. Appl Microbiol Biotechnol. 2010;85(3):615–624.
- Roberts GA, Grogan G, Greter A, et al. Identification of a new class of cytochrome P450 from a Rhodococcus sp. J Bacteriol. 2002;184(14):3898–3908.
- Schückel J, Rylott EL, Grogan G, et al. A gene-fusion approach to enabling plant cytochromes p450 for biocatalysis. Chembiochem. 2012;13(18):2758–2763.
- Sibbesen O, De Voss JJ, de Montellano PRO. Putidaredoxin reductase-putidaredoxin-cytochrome P450cam triple fusion protein. Construction of a self-sufficient Escherichia coli catalytic system. J Biol Chem. 1996;271(37):22462–22469.
- Li S, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J Am Chem Soc. 2007;129(43):12940–12941.
- Mandai T, Fujiwara S, Imaoka S. Construction and engineering of a thermostable self-sufficient cytochrome P450. Biochem Biophys Res Commun. 2009;384(1):61–65.
- Luan Z-J, Yin Y-C, Li A-T, et al. Monoterpene hydroxylation with an artificial self-sufficient P450 utilizing a P450SMO reductase domain for the electron transfer. J Mol Catal B Enzym. 2015;116:78–82.
- Nogués I, Martínez-Júlvez M, Navarro JA, et al. Role of hydrophobic interactions in the flavodoxin mediated electron transfer from photosystem I to ferredoxin-NADP + reductase in Anabaena PCC 7119. Biochemistry. 2003;42(7):2036–2045.
- Dong J, Fernández-Fueyo E, Hollmann F, et al. Biocatalytic oxidation reactions: a chemist’s perspective. Angew Chem Int Ed. 2018;57(30):9238–9261.
- Yu Y, Liu X, Wang J. Expansion of redox chemistry in designer metalloenzymes. Acc Chem Res. 2019;52(3):557–565.
- Hagelueken G, Wiehlmann L, Adams TM, et al. Crystal structure of the electron transfer complex rubredoxin–rubredoxin reductase of Pseudomonas aeruginosa. Proc Natl Acad Sci. 2007;104(30):12276–12281.
- Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402(6757):47–52.
- Martínez-Júlvez M, Nogués I, Faro M, et al. Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP + reductase for optimal complex formation and electron transfer to ferredoxin. J Biol Chem. 2001;276(29):27498–27510.
- Quehl P, Schüürmann J, Hollender J, et al. Improving the activity of surface displayed cytochrome P450 enzymes by optimizing the outer membrane linker. Biochim Biophys Acta Biomembr. 2017;1859(1):104–116.
- Kandel SE, Lampe JN. Role of protein-protein interactions in cytochrome P450-mediated drug metabolism and toxicity. Chem Res Toxicol. 2014;27(9):1474–1486.
- Bakkes PJ, Biemann S, Bokel A, et al. Design and improvement of artificial redox modules by molecular fusion of flavodoxin and flavodoxin reductase from Escherichia coli. Sci Rep. 2015;5(1):12158.
- Bakkes PJ, Riehm JL, Sagadin T, et al. Engineering of versatile redox partner fusions that support monooxygenase activity of functionally diverse cytochrome P450s. Sci Rep. 2017;7(1):9570.
- Aalbers FS, Fraaije MW. Enzyme fusions in biocatalysis: coupling reactions by pairing enzymes. Chembiochem. 2019;20(1):20–28.
- Papaleo E, Saladino G, Lambrughi M, et al. The role of protein loops and linkers in conformational dynamics and allostery. Chem Rev. 2016;116(11):6391–6423.
- Haga T, Hirakawa H, Nagamune T. Fine tuning of spatial arrangement of enzymes in a PCNA-mediated multienzyme complex using a rigid Poly-L-Proline linker. PLoS One. 2013;8(9):e75114.
- George RA, Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng Des Sel. 2002;15(11):871–879.
- Moon JH, Lee K, Lee JH, et al. Redesign and reconstruction of a steviol-biosynthetic pathway for enhanced production of steviol in Escherichia coli. Microb Cell Fact. 2020;19(1):20.
- Li D, Wu Y, Zhang C, et al. Production of triterpene ginsenoside compound K in the non-conventional yeast Yarrowia lipolytica. J Agric Food Chem. 2019;67(9):81–88.
- Li D, Wu Y, Wei P, et al. Metabolic engineering of Yarrowia lipolytica for heterologous oleanolic acid production. Chem Eng Sci. 2020;218:115529.
- Guo X, Sun J, Li D, et al. Heterologous biosynthesis of (+)-nootkatone in unconventional yeast Yarrowia lipolytica. Biochem Eng J. 2018;137:125–131.
- Gräwe A, Ranglack J, Weyrich A, et al. iFLinkC: an iterative functional linker cloning strategy for the combinatorial assembly and recombination of linker peptides with functional domains. Nucleic Acids Res. 2020;48(4):e24–e24.
- Hirakawa H, Nagamune T. Molecular assembly of P450 with ferredoxin and ferredoxin reductase by fusion to PCNA. Chembiochem. 2010;11(11):1517–1520.
- Haga T, Hirakawa H, Nagamune T. Artificial self-sufficient cytochrome P450 containing multiple auxiliary proteins demonstrates improved monooxygenase activity. Biotechnol J. 2018;13(12):1800088.
- Jin C-C, Zhang J-L, Song H, et al. Boosting the biosynthesis of betulinic acid and related triterpenoids in Yarrowia lipolytica via multimodular metabolic engineering. Microb Cell Fact. 2019;18(1):77.
- Chen C-D, Doray B, Kemper B. Efficient assembly of functional cytochrome P450 2C2 requires a spacer sequence between the N-terminal signal anchor and catalytic domains*. J Biol Chem. 1997;272(36):22891–22897.
- Li G, Huang Z, Zhang C, et al. Construction of a linker library with widely controllable flexibility for fusion protein design. Appl Microbiol Biotechnol. 2016;100(1):215–225.
- Biggs BW, Rouck JE, Kambalyal A, et al. Orthogonal assays clarify the oxidative biochemistry of taxol P450 CYP725A4. ACS Chem Biol. 2016;11(5):1445–1451.
- Sowden RJ, Yasmin S, Rees NH, et al. Biotransformation of the sesquiterpene (+)-valencene by cytochrome P450cam and P450BM-3. Org Biomol Chem. 2005;3(1):57–64.
- Winzer T, Kern M, King AJ, et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science. 2015;349(6245):309–312.
- Laursen T, Borch J, Knudsen C, et al. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science. 2016;354(6314):890–893.
- D'Ari R, Casadesús J. Underground metabolism. Bioessays. 1998;20(2):181–186.
- Gold ND, Fossati E, Hansen CC, et al. A combinatorial approach to study cytochrome P450 enzymes for De novo production of steviol glucosides in baker’s yeast. ACS Synth Biol. 2018;7(12):2918–2929.
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40.
- Zhang Y, Wang Y, Wang S, et al. Engineering bi-functional enzyme complex of formate dehydrogenase and leucine dehydrogenase by peptide linker mediated fusion for accelerating cofactor regeneration. Eng Life Sci. 2017;17(9):989–996.
- Lassen LM, Nielsen AZ, Ziersen B, et al. Redirecting photosynthetic electron flow into light-driven synthesis of alternative products including high-value bioactive natural compounds. ACS Synth Biol. 2014;3(1):1–12.
- Mie Y, Yasutake Y, Takayama H, et al. Electrochemically boosted cytochrome P450 reaction that efficiently produces 25-hydroxyvitamin D3. J Catal. 2020;384:30–36.
- Frank R, Prönnecke C, Azendorf R, et al. Advanced 96-microtiter plate based bioelectrochemical platform reveals molecular short cut of electron flow in cytochrome P450 enzyme. Lab Chip. 2020;20(8):1449–1460.
- Belsare KD, Horn T, Ruff AJ, et al. Directed evolution of P450cin for mediated electron transfer. Protein Eng Des Sel. 2017;30(2):119–127.
- Belsare KD, Ruff AJ, Martinez R, et al. P-LinK: a method for generating multicomponent cytochrome P450 fusions with variable linker length. Biotechniques. 2014;57(1):13–20.
- Hollmann F, Arends IWCE, Buehler K. Biocatalytic redox reactions for organic synthesis: nonconventional regeneration methods. ChemCatChem. 2010;2(7):762–782.
- Lauterbach L, Lenz O, Vincent KA. H2-driven cofactor regeneration with NAD(P)+-reducing hydrogenases. Febs J. 2013;280(13):3058–3068.
- Darimont D, Weissenborn MJ, Nebel BA, et al. Modulating proposed electron transfer pathways in P450BM3 led to improved activity and coupling efficiency. Bioelectrochemistry. 2018;119:119–123.
- Wang X, Saba T, Yiu HHP, et al. Cofactor NAD(P)H regeneration inspired by heterogeneous pathways. Chem. 2017;2(5):621–654.
- Kadowaki JT, Jones TH, Sengupta A, et al. Copper oxide-based cathode for direct NADPH regeneration. Sci Rep. 2021;11(1):180.
- Park JH, Lee SH, Cha GS, et al. Cofactor-free light-driven whole-cell cytochrome P450 catalysis. Angew Chem Int Ed Engl. 2015;54(3):969–973.
- Lee SH, Kwon Y-C, Kim D-M, et al. Cytochrome P450-catalyzed O-dealkylation coupled with photochemical NADPH regeneration. Biotechnol Bioeng. 2013;110(2):383–390.
- Girhard M, Kunigk E, Tihovsky S, et al. Light-driven biocatalysis with cytochrome P450 peroxygenases. Biotechnol Appl Biochem. 2013;60(1):111–118.
- Jiang L, Li Y, Wei W, et al. Confining nanohybrid of CdTe quantum dots and cytochrome P450 2D6 in macroporous ordered siliceous foam for drug metabolism. J Electroanal Chem. 2016;781:345–350.
- Zilly FE, Taglieber A, Schulz F, et al. Deazaflavins as mediators in light-driven cytochrome P450 catalyzed hydroxylations. Chem Commun. 2009;46:7152–7154.
- Spradlin J, Lee D, Mahadevan S, et al. Insights into an efficient light-driven hybrid P450 BM3 enzyme from crystallographic, spectroscopic and biochemical studies. Biochim Biophys Acta. 2016;1864(12):1732–1738.
- Nielsen AZ, Ziersen B, Jensen K, et al. Redirecting photosynthetic reducing power toward bioactive natural product synthesis. ACS Synth Biol. 2013;2(6):308–315.
- Mellor SB, Vavitsas K, Nielsen AZ, et al. Photosynthetic fuel for heterologous enzymes: the role of electron carrier proteins. Photosynth Res. 2017;134(3):329–342.
- Hanke GUY, Mulo P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 2013;36(6):1071–1084.
- Büchsenschütz HC, Solymosi D, Dyczmons NG, et al. Engineering of NADPH supply boosts photosynthesis-driven biotransformations. ACS Catal. 2020;10:11864–11877.
- Berepiki A, Gittins JR, Moore CM, et al. Rational engineering of photosynthetic electron flux enhances light-powered cytochrome P450 activity. Synth Biol (Oxf). 2018;3(1):ysy009.
- Benz JP, Lintala M, Soll J, et al. A new concept for ferredoxin-NADP(H) oxidoreductase binding to plant thylakoids. Trends Plant Sci. 2010;15(11):608–613.
- Chitnis PR. PHOTOSYSTEM I: function and physiology. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:593–626.
- Goñi G, Zöllner A, Lisurek M, et al. Cyanobacterial electron carrier proteins as electron donors to CYP106A2 from Bacillus megaterium ATCC 13368. Biochim Biophys Acta. 2009;1794(11):1635–1642.
- Grossman A. Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist. 2000;151(3):201–224.
- Tripathy BC, Oelmüller R. Reactive oxygen species generation and signaling in plants. Plant Signal Behav. 2012;7(12):1621–1633.
- Ilie A, Harms K, Reetz MT. P450-catalyzed regio- and stereoselective oxidative hydroxylation of 6-Iodotetralone: preparative-scale synthesis of a key intermediate for Pd-catalyzed transformations. J Org Chem. 2018;83(14):7504–7508.
- Alwaseem H, Frisch BJ, Fasan R. Anticancer activity profiling of parthenolide analogs generated via P450-mediated chemoenzymatic synthesis. Bioorg Med Chem. 2018;26(7):1365–1373.
- Hernandez-Ortega A, Vinaixa M, Zebec Z, et al. A toolbox for diverse oxyfunctionalisation of monoterpenes. Sci Rep. 2018;8(1):14396.
- Leuchs S, Lima-Ramos J, Greiner L, et al. Reaction engineering of biocatalytic enantioselective reduction: a case study for aliphatic ketones. Org Process Res Dev. 2013;17(8):1027–1035.
- Jakoblinnert A, Rother D. A two-step biocatalytic Cascade in micro-aqueous medium: using whole cells to obtain high concentrations of a vicinal diol. Green Chem. 2014;16(7):3472–3482.
- Zachos I, Döring M, Tafertshofer G, et al. carba Nicotinamide adenine dinucleotide phosphate: robust cofactor for redox biocatalysis. Angew Chemie Int Ed. 2021;60:14701–14706.
- Wong BG, Mancuso CP, Kiriakov S, et al. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat Biotechnol. 2018;36(7):614–623.
- Steel H, Habgood R, Kelly CL, et al. In situ characterisation and manipulation of biological systems with Chi.Bio. PLoS Biol. 2020;18(7):e3000794.
- Schneider K, Schlegel HG. Production of superoxide radicals by soluble hydrogenase from Alcaligenes eutrophus H16. Biochem J. 1981;193(1):99–107.
- Lonsdale TH, Lauterbach L, Honda Malca S, et al. H2-driven biotransformation of n-octane to 1-octanol by a recombinant Pseudomonas putida strain co-synthesizing an O2-tolerant hydrogenase and a P450 monooxygenase. Chem Commun (Camb). 2015;51(90):16173–16175.
- Preissler J, Reeve HA, Zhu T, et al. Dihydrogen-Driven NADPH recycling in imine reduction and P450-catalyzed oxidations mediated by an engineered O2-tolerant hydrogenase. ChemCatChem. 2020;12(19):4853–4861.
- Ferrandi EE, Monti D, Riva S. New trends in the in situ enzymatic recycling of NAD(P)(H) cofactors. In: Cascade biocatalysis. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2014. p. 23–42.
- Robescu MS, Rubini R, Beneventi E, et al. From the amelioration of a NADP+-dependent formate dehydrogenase to the discovery of a new enzyme: round trip from theory to practice. ChemCatChem. 2020;12(9):2478–2487.
- Hatrongjit R, Packdibamrung K. A novel NADP+-dependent formate dehydrogenase from Burkholderia stabilis 15516: screening, purification and characterization. Enzyme Microb Technol. 2010;46(7):557–561.
- Kaluzna I, Schmitges T, Straatman H, et al. Enabling selective and sustainable P450 oxygenation technology. Production of 4-Hydroxy-α-isophorone on kilogram scale. Org Process Res Dev. 2016;20(4):814–819.
- Garny S, Beeton-Kempen N, Gerber I, et al. The co-immobilization of P450-type nitric oxide reductase and glucose dehydrogenase for the continuous reduction of nitric oxide via cofactor recycling. Enzyme Microb Technol. 2016;85:71–81.
- Brummund J, Müller M, Schmitges T, et al. Process development for oxidations of hydrophobic compounds applying cytochrome P450 monooxygenases in-vitro. J Biotechnol. 2016;233:143–150.
- Chen X, Zhang C, Too H-P. Multienzyme biosynthesis of dihydroartemisinic acid. Molecules. 2017;22(9):1422.
- Torres Pazmiño DE, Riebel A, de Lange J, et al. Efficient biooxidations catalyzed by a new generation of self-sufficient Baeyer-Villiger monooxygenases. Chembiochem. 2009;10(16):2595–2598.
- Figueroa IA, Barnum TP, Somasekhar PY, et al. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc Natl Acad Sci USA. 2018;115(1):E92–E101.
- Schink B, Thiemann V, Laue H, et al. Desulfotignum phosphitoxidans sp. nov., a new marine sulfate reducer that oxidizes phosphite to phosphate. Arch Microbiol. 2002;177(5):381–391.
- Nowrouzi B, Li RA, Walls LE, et al. Enhanced production of taxadiene in Saccharomyces cerevisiae. Microb Cell Fact. 2020;19(1):200.
- Meng X, Liu H, Xu W, et al. Metabolic engineering Saccharomyces cerevisiae for de novo production of the sesquiterpenoid (+)-nootkatone. Microb Cell Fact. 2020;19(1):21.
- Gao H, Li J, Sivakumar D, et al. NADH oxidase from Lactobacillus reuteri: a versatile enzyme for oxidized cofactor regeneration. Int J Biol Macromol. 2019;123:629–636.
- Holec C, Neufeld K, Pietruszka J. P450 BM3 monooxygenase as an efficient NAD(P)H-oxidase for regeneration of nicotinamide cofactors in ADH-catalysed preparative scale biotransformations. Adv Synth Catal. 2016;358(11):1810–1819.
- Li J, Zhang Y. Modulating betulinic acid production in Saccharomyces cerevisiae by managing the intracellular supplies of the co-factor NADPH and oxygen. J Biosci Bioeng. 2015;119(1):77–81.
- Ensari Y, de Almeida Santos G, Ruff AJ, et al. Engineered P450 BM3 and cpADH5 coupled Cascade reaction for β-oxo fatty acid methyl ester production in whole cells. Enzyme Microb Technol. 2020;138:109555.
- Lu H, Li F, Sánchez BJ, et al. A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat Commun. 2019;10(1):3586.
- Jensen K, Broeken V, Hansen ASL, et al. OptCouple: joint simulation of gene knockouts, insertions and medium modifications for prediction of growth-coupled strain designs. Metab Eng Commun. 2019;8:e00087.
- von Kamp A, Klamt S. Enumeration of smallest intervention strategies in genome-scale metabolic networks. PLoS Comput Biol. 2014;10(1):e1003378.
- Kim J, Reed JL. OptORF: optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC Syst Biol. 2010;4(1):53.
- Stanford NJ, Millard P, Swainston N. RobOKoD: microbial strain design for (over)production of target compounds. Front Cell Dev Biol. 2015;3:17.
- Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84(6):647–657.
- Cardoso JGR, Jensen K, Lieven C, et al. Cameo: a python library for computer aided metabolic engineering and optimization of cell factories. ACS Synth Biol. 2018;7(4):1163–1166.
- Ebrahim A, Lerman JA, Palsson BO, et al. COBRApy: COnstraints-Based reconstruction and analysis for python. BMC Syst Biol. 2013;7(1):74.
- Heirendt L, Arreckx S, Pfau T, et al. Creation and analysis of biochemical constraint-based models using the COBRA toolbox v.3.0. Nat Protoc. 2019;14(3):639–702.
- von Kamp A, Thiele S, Hädicke O, et al. Use of CellNetAnalyzer in biotechnology and metabolic engineering. J Biotechnol. 2017;261:221–228.
- Becker SA, Feist AM, Mo ML, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox. Nat Protoc. 2007;2(3):727–738.
- King ZA, Feist AM. Optimal cofactor swapping can increase the theoretical yield for chemical production in Escherichia coli and Saccharomyces cerevisiae. Metab Eng. 2014;24:117–128.
- Chung BK-S, Lakshmanan M, Klement M, et al. Genome-scale in silico modeling and analysis for designing synthetic terpenoid-producing microbial cell factories. Chem Eng Sci. 2013;103:100–108.
- Zeng H, Yang A. Modelling overflow metabolism in Escherichia coli with flux balance analysis incorporating differential proteomic efficiencies of energy pathways. BMC Syst Biol. 2019;13(1):3.
- Fondi M, Pinatel E, Talà A, et al. Time-resolved transcriptomics and constraint-based modeling identify system-level metabolic features and overexpression targets to increase spiramycin production in Streptomyces ambofaciens. Front Microbiol. 2017;8:835.
- Hendry JI, Prasannan CB, Joshi A, et al. Metabolic model of synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour Technol. 2016;213:190–197.
- Costello Z, Martin HG. A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. Npj Syst Biol Appl. 2018;4(1):19.
- Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35(6–7):811–820.
- Holtmann D, Hollmann F. The oxygen dilemma: a severe challenge for the application of monooxygenases? Chembiochem. 2016;17(15):1391–1398.
- Shephard EA, Phillips IR, Bayney RM, et al. Quantification of NADPH: cytochrome P-450 reductase in liver microsomes by a specific radioimmunoassay technique. Biochem J. 1983;211(2):333–340.
- Riebel BR, Gibbs PR, Wellborn WB, et al. Cofactor regeneration of NAD+ from NADH: novel water-forming NADH oxidases. Adv Synth Catal. 2002;344(10):1156–1168.
- Reyes LH, Gomez JM, Kao KC. Improving carotenoids production in yeast via adaptive laboratory evolution. Metab Eng. 2014;21:26–33.
- Jiang Y, Li Z, Zheng S, et al. Establishing an enzyme cascade for one-pot production of α-olefins from low-cost triglycerides and oils without exogenous H2O2 addition. Biotechnol Biofuels. 2020;13(1):52.
- Cirino PC, Arnold FH. A self-sufficient peroxide-driven hydroxylation biocatalyst. Angew Chem Int Ed Engl. 2003;42(28):3299–3301.
- Vázquez J, Grillitsch K, Daum G, et al. The role of the membrane lipid composition in the oxidative stress tolerance of different wine yeasts. Food Microbiol. 2019;78:143–154.
- Liu C, Wang W, Wu Y, et al. Multiple alkane hydroxylase systems in a marine alkane degrader, alcanivorax dieselolei B-5. Environ Microbiol. 2011;13(5):1168–1178.
- Fetherolf MM, Levy-Booth DJ, Navas LE, et al. Characterization of alkylguaiacol-degrading cytochromes P450 for the biocatalytic valorization of lignin. Proc Natl Acad Sci USA. 2020;117(41):25771–25778.
- Mi J, Becher D, Lubuta P, et al. De novo production of the monoterpenoid geranic acid by metabolically engineered Pseudomonas putida. Microb Cell Fact. 2014;13(1):170.
- Kim J, Park W. Oxidative stress response in Pseudomonas putida. Appl Microbiol Biotechnol. 2014;98(16):6933–6946.
- Udaondo Z, Duque E, Fernández M, et al. Analysis of solvent tolerance in Pseudomonas putida DOT-T1E based on its genome sequence and a collection of mutants. FEBS Lett. 2012;586(18):2932–2938.
- Zuber P. Management of oxidative stress in Bacillus. Annu Rev Microbiol. 2009;63:575–597.
- D’Adamo S, Schiano di Visconte G, Lowe G, et al. Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol J. 2019;17(1):75–87.
- Liu J, Zhu Y, Du G, et al. Exogenous ergosterol protects Saccharomyces cerevisiae from d-limonene stress. J Appl Microbiol. 2013;114(2):482–491.
- Landolfo S, Zara G, Zara S, et al. Oleic acid and ergosterol supplementation mitigates oxidative stress in wine strains of Saccharomyces cerevisiae. Int J Food Microbiol. 2010;141(3):229–235.
- Brennan TCR, Krömer JO, Nielsen LK. Physiological and transcriptional responses of Saccharomyces cerevisiae to d-limonene show changes to the cell wall but not to the plasma membrane. Appl Environ Microbiol. 2013;79(12):3590–3600.
