Abstract
Alignment of three nucleic acids strands, in which the third strand is identical to one of the DNA duplex strands, occurs in various cellular systems. In the case of telomeric t-loops, recognition between the DNA duplex and the homologous single strand is likely to be mediated by proteins through formation of the transient recombination-type R-triplex. Earlier, using 2-aminopurine as a fluorescent reporting base, we evaluated the thermodynamic characteristics of intramolecular R-triplex formed by a mixed nucleotide sequence. Here, we used this approach to explore a propensity of the telomeric TTAGGG repeat to form the R-triplex. The circular dichroism spectral changes detected upon formation of the R-triplex suggest that this process is accompanied by specific conformational changes in DNA, including a local destabilization of the target duplex next to a GGG run revealed by the fluorescence of the reporting 2-aminopurine base. Surprisingly, stability of the R-triplex formed by telomeric sequence depends strikingly on the counter ion, being higher for Na+ than for Li+. Taken together these findings indicate a significant conformational variability of telomeric DNA in the context of recombination-type R-triplex, a phenomenon of possible biological relevance.
Introduction
The ends of mammalian and other telomeres are organized in t-loops formed by a backward folding of the 3′ telomeric overhang along the duplex telomeric repeat (Cesare, Quinney, Willcox, Subramanian, & Griffith, Citation2003; Griffith et al., Citation1999; Tomaska, Makhov, Griffith, & Nosek, Citation2002). It has been suggested that t-loops represent a general structural characteristic of eukaryotic nuclear chromosomes (Fouche et al., Citation2006; Munoz-Jordan, Cross, de Lange, & Griffith, Citation2001; Murti & Prescott, Citation1999), reviewed in Amiard et al. (Citation2007). The t-loop structure, however, exists in a dynamic state and must be destabilized in order to be replicated and re-established through numerous cell divisions.
TRF2 protein binds to the junction between the telomeric ds DNA and the three-stranded DNA alignment (Griffith et al., Citation1999). The junction-specific binding may allow TRF2 to mediate invasion of the single-strand overhang completing t-loop formation (Amiard et al., Citation2007; Fouche et al., Citation2006). There is evidence that t-loops play a role in a capping function of telomeres, protecting the ends of chromosomes against degrading enzymes and DNA repair machinery (Tomaska et al., Citation2002). On the other hand, the single-stranded repeating sequence d(TTAGGG)n is able to form self-folded G-quadruplexes of several conformations depending on specific sequence and the type of counter ion (Neidle & Parkinson, Citation2008). The replication-dependent formation of G-quadruplex structures in the genome of mammalian cells modulated during cell cycle progression was demonstrated with engineered, structure-specific antibody (Biffi, Tannahill, McCafferty, & Balasubramanian, Citation2013). G-quadruplex preferentially forms at the very 3′ end of vertebrate telomeric DNA rather than at internal positions of 3′ overhang (Tang et al., Citation2008). This property provides a molecular basis for telomerase inhibition by G-quadruplex formation. Moreover, it may also regulate those processes that depend on the structure of the very 3′ telomere end, for instance, telomere t-loop formation. Thus, formation of G-quadruplex during replication may compete with new t-loop formation after replication in the late S-G2 phase of the cell cycle (Vannier, Pavicic-Kaltenbrunner, Petalcorin, Ding, & Boulton, Citation2012).
The shelterin proteins assemble the three DNA strands in the junction enabling sequence-specific DNA–DNA interactions that contribute to the functioning of the telomeres (de Lange, Citation2005; Takai, Hooper, Blackwood, Gandhi, & de Lange, Citation2010). The ss overhang is aligned parallel to the identical strand in the telomeric DNA duplex, which provides a possibility of R-triplex serving as a transient intermediate leading to the exchange of identical strands and resulting in formation of the D-loop within t-loop. R-type triplexes comprising DNA duplex and a homologous third DNA/RNA strand oriented parallel to the identical duplex strand, have been proposed to participate in DNA replication (Lestienne, Citation2009, 2011; Lestienne, Boudsocq, & Bonnet, Citation2008) and in mediating de novo CpG methylation of rRNA genes (Schmitz, Mayer, Postepska, & Grummt, Citation2010). Earlier, we presented thermodynamic characteristics of the R-triplex formed by a mixed nucleotide sequence indicating that its stability is extremely sensitive to the presence of a mismatch base in the third strand (Shchyolkina, Kaluzhny, Arndt-Jovin, Jovin, & Zhurkin, Citation2006). This feature is consistent with the R-triplex propensity for the ss strand alignment to the homologous strand of duplex.
Here, we explore a potential for the R-triplex formation by the mammalian telomere G-rich sequence using a special oligonucleotide construct and solution conditions facilitating the protein-independent interaction of 5′-d(GGTTAGGGTTAG) strand with the homologous duplex.
Materials and methods
Oligonucleotides
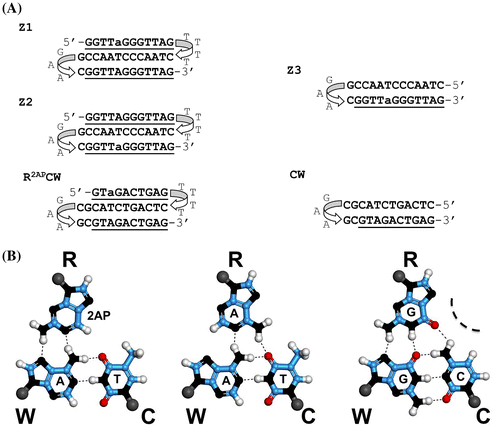
Oligonucleotides Z1, Z2, and Z3 were synthesized and purified with HPLC by Syntol (Moscow) or Midland Certified Reagent Co. Inc. (TX). GAA loop in Z1, Z2, and Z3 together with the adjacent GC base pair stabilized the duplex part of the folds, whereas the 5′-d(GGTTAGGGTTAG) strand was bound to the duplex part with the more flexible TTTT loop. Such a design minimized displacement of the 3′-terminal nucleotide sequence with the identical 5′-d(GGTTAGGGTTAG) and provided a possibility to monitor a conformational state of the third 5′ strand (Shchyolkina et al., Citation2004). The solution conditions, in particular, couterions, were specially chosen to facilitate the formation of intramolecular species but not intermolecular oligonucleotide associates (Shchyolkina et al., Citation2004). Also, the presence of Li+ excludes formation of G-quadruplexes or diminish probability of their formation (e.g. Włodarczyk, Grzybowski, Patkowski, & Dobek, Citation2005).
Fluorescence measurements
The steady-state fluorescence emission and excitation spectra as well as temperature dependence of 2AP fluorescence intensity were registered with Cary Eclipse fluorescence spectrophotometer. Upon excitation at 310-nm emission at 370 nm was recorded in the temperature range −2°C to 70°C using a temperature ramp of 1°C per minute.
The fluorescence anisotropy of EtBr was recorded using a spectrophotometer (Photon Technology International) with a temperature-controlled cell compartment.
Steady-state fluorescence anisotropy measurements and evaluation of relative hydrodynamic volumes of oligonucleotides using EtBr as a probe
To distinguish between intra and intermolecular oligonucleotide structures, fluorescence anisotropy measurements of intercalated EtBr were carried out. This method has been described in detail in Shchyolkina et al. (Citation2004). The extension of the double or triple helix on binding one intercalator molecule was assumed to be 3.4 Å (Jain, Tsai, & Sobell, Citation1977; Tuite & Nordén, Citation1995), introducing a uniform systematic error of the estimation of the oligonucleotide hydrodynamic volume using EtBr as probe.
The hydrodynamic volume (V) of molecules with low asymmetry can be estimated by the equation:(1)
where τ is the measured fluorescence lifetime of bound EtBr, r is the measured fluorescence anisotropy, and r0 is the limiting anisotropy at infinite viscosity (T/η → 0), where η is the viscosity of the solution, T is the absolute temperature, and k is the Boltzmann constant (Jablonski, Citation1960). The mean fluorescence lifetimes τ of EtBr intercalated in the parallel (recombinant) triplex, conventional antiparallel triplex, and calf thymus DNA were found to be practically the same (Mergny, Collier, Rougée, Montenay-Garestier, & Hélène, Citation1991; Shchyolkina & Borisova, Citation1997). This permitted the calculation of a relative hydrodynamic volume (V) of the oligonucleotides Z1, Z2, and Z3 with respect to the hydrodynamic volume (VCW) of the CW hairpin (see sequence in Figure ):(2)
Figure 1. (A) Oligonucleotide sequences with the reporting 2AP base, used in the experiments. a, 2-aminopurine (2AP); the triplex-forming oligonucleotides: Z1 contains 2AP in the third R-strand; Z2 contains 2AP in the W-strand. The hairpin-forming oligonucleotide, Z3, represents the duplex part of the triplex Z2. CW represents a 12-bp hairpin with the mixed base sequence. (B) Schemes of the base triplets 2AP*(T:A), A*(T:A), and G*(C:G).
for the fluorescence anisotropy values determined under the same solution conditions and temperature. The evaluation of the oligonucleotide hydrodynamic volume in solution provides a sensitive detection of intermolecular species present in a sample (Shchyolkina et al., Citation1994, 2004). Using EtBr as a fluorescent probe to study the oligonucleotide stoichiometry is convenient, as EtBr does not destabilize DNA duplexes or triplexes at low occupancy (Shchyolkina & Borisova, Citation1997; Shchyolkina et al., Citation2004). Importantly, this approach reveals even relatively weak oligonucleotide associations in solution, which may disintegrate during polyacrylamide gel electrophoresis.
Thermodynamic analysis of the intramolecular triplex formation
The detailed analysis of the thermal denaturation profiles, recorded by fluorescence of the 2AP reporter, has been described elsewhere (Shchyolkina et al., Citation2004). In essence, we analyzed the intramolecular binding of the dangling third strand to its double-helical CW part by fitting a theoretical curve to the experimental fluorescence melting curve. From these data, we deduced the basic thermodynamic parameters of the intramolecular transition (denoted below as the ‘triple helix formation’).
Circular dichroism
Circular dichroism (CD) spectra were recorded with a Jasco 715 spectropolarimeter using 1-cm cell in a thermostatted cell holder. All CD spectra were collected at 50 nm/min, with five scans averaged and were corrected for baselines.
Results and discussion
Z1, Z2, and Z3 oligonucleotides form intramolecular folds under the experimental conditions
We designed oligonucleotides Z1 and Z2 so that the intramolecular folds represented a simple model of three DNA strands alignment in D-loop of t-loop. The flexible loop TTTT sequence attaches the third strand and decreases the entropy of mixing of the duplex and the third strand. The duplex part of the oligonucleotide is reinforced by the structured GAA sequence.
It is well known that G-rich oligonucleotides including the telomeric sequences, can form diverse intra and intermolecular structures (Adrian, Heddi, & Phan, Citation2012; Neidle & Parkinson, Citation2008). As a first step in specifying the studied oligonucleotide conformations, we established whether they form intramolecular folds or, alternatively, intermolecular associates under the experimental conditions.
We estimated hydrodynamic volumes of oligonucleotides Z1, Z2, and Z3 (Figure ) using the EtBr probe (Equation (Equation1(1) )). The intensity of EtBr fluorescence in the presence of Z1, Z2, and Z3 increased 30-fold, indicative of the intercalation binding type, similar to that observed in double-stranded DNA (LePecq & Paoletti, Citation1967; Shchyolkina & Borisova, Citation1997; Shchyolkina et al., Citation2000).
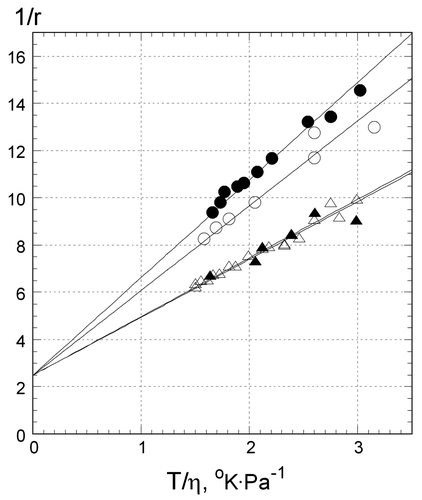
Figure shows the dependence of reciprocal fluorescence anisotropy (1/r) on T/η. In a broad temperature range, from 1°C to 20°C, it was linear up to 20°C (T/η ≈ 3oK Pa−1) for Z1 (open triangles), Z2 (filled triangles), and Z3 (open circles), as well as for CW (filled circles), demonstrating the lack of abrupt changes in their hydrodynamic volumes. Both hairpins, CW and Z3, can be modeled as ellipsoids with low asymmetry and Equation (Equation1(1) ) (Methods) was used for the calculation of their apparent hydrodynamic volumes. The ratio of the slopes of CW and Z3 is ~(1.10 ± .02) and corresponds to their hydrodynamic volumes and molecular masses (27–29 nt) within experimental error, in correspondence to Equation (Equation2
(2) ). Similarly, the ratio of the slopes of both Z3 to Z1 and Z3 to Z2, yields hydrodynamic volumes of Z1 and Z2 ~1.5 times that of Z3. Such a ratio is consistent with the hydrodynamic volume and molecular mass of a single, self-folded triplex with third strand tightly attached to the duplex part of Z1 and Z2. A dissociation of the third strand from the duplex part at higher temperatures would increase the hydrodynamic volume nonlinearly (Shchyolkina et al., Citation2004). We conclude that under our experimental conditions oligonucleotides Z1, Z2, and Z3 are monomolecular structures.
Figure 2. Reciprocal fluorescence anisotropy of EtBr bound to oligonucleotides at different temperatures: Z1 in .5 M LiCl (open triangles), Z2 in .5 M NaCl (filled triangles), Z3 in .5 M LiCl (open circles), and CW in .5 M LiCl (filled circles). Y axis: reciprocal anisotropy 1/r, X axis: ratio of the absolute temperature to viscosity of solution. Value of T/η = 3 corresponds to 24°C (see Methods). Samples contained 1 μM oligonucleotides, 1 μM EtBr,10 mM Tris–HCl buffer, pH 7.6.
R-triplex formation of 5′-d(GGTTAGGGTTAG) monitored by the fluorescence of 2-aminopurine
We performed strand-specific labeling of the oligonucleotide folds by replacing an adenine with the fluorescent 2-aminoprine (2AP) (Figure ). Such a substitution provides a 2AP:T base pair or a 2AP*T:A base triplet isomorphous to the corresponding structures with adenines and do not affect R-triplex stability (Shchyolkina et al., Citation2004). The steady-state fluorescence intensity of 2AP in place of adenine in the 5′-terminal strand of an intramolecular fold was shown earlier to be a suitable reporter for monitoring the strand conformational state (Shchyolkina et al., Citation2006). Figure shows the temperature dependence of the fluorescence intensities of Z1, Z2, and Z3 depending on temperature in the presence of Li+ (Figure (A)) and Na+ (Figure (B)).
Figure 3. Temperature dependence of the fluorescence emission of 2AP, incorporated in the oligonucleotides in .5 M LiCl (A) and .5 M NaCl (B): Z1 (filled circles), Z2 (open circles), and Z3 (triangles). Every 20th experimental point is marked. The excitation wavelength was 310 nm, emission was measured at 370 nm. Samples contained 1 μM oligonucleotides, 10 mM Tris–HCl buffer, pH 7.6.
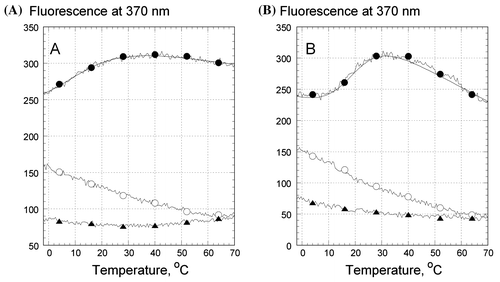
We now compare the fluorescence of the reporting base 2AP placed in the duplex Z3, the duplex part of the triplex fold Z2, and in the third strand of Z1 at temperature 0°C (Figure (A) and (B), triangles, open circles, and filled circles, respectively). In the presence of as Li+ (Figure (A)), or Na+ (Figure (B)), there was a significant quenching of Z3 fluorescence, indicative of a profound stacking of 2AP in the double-stranded Z3 hairpin. The higher fluorescence intensity of 2AP in Z1 at 0°C is consistent with a higher solvent accessibility of the base analog positioned in the third strand of the triplex fold (Figure (A) and (B), filled circles), in agreement with model proposed previously (Zhurkin, Raghunathan, Ulyanov, Camerini-Otero, & Jernigan, Citation1994) and corresponding to the other experimental data (Shchyolkina et al., Citation2004, 2006).
The enhancement of Z1 fluorescence in the temperature range from −5°C to 40°C is indicative of an increased exposure of 2AP to solvent upon its unstacking from the adjacent bases during the course of the triplex melting (Figure (A) and (B), filled circles) (Shchyolkina et al., Citation2004, 2006). At temperatures exceeding 40–50°C the temperature dependence of 2AP fluorescence in Z1 was similar to that of 2AP in a single strand (Shchyolkina et al., Citation2004, 2006), constituting evidence for a complete detachment of the third strand 5′-d(GGTTaGGGTTAG) from the duplex part of Z1 fold. The shallow temperature dependence of Z3 fluorescence revealed that the double-stranded hairpin did not melt in the entire temperature range studied (Figure (A) and (B), triangles).
The positions of the fluorescence emission maxima of 2AP were identical in Z2 and Z3 (not shown), supporting a duplex structure in both oligonucleotide folds (Xu, Evans, & Nordlund, Citation1994). However, at 0°C the 2AP fluorescence placed in the duplex Z3 was lower than that in the similar position in the duplex part of the Z2 triplex (Figure (A) and (B), filled triangles and open circles). The more intense 2AP fluorescence in Z2 testifies to a higher dynamical solvent accessibility of the minor groove in Z2 possibly reflecting a structural perturbation, at least locally, in the homologous duplex upon attachment of the 5′-d(GGTTAGGGTTAG) strand (see below). We note also, that a similar fluorescence enhancement took place at lower temperatures in Z2 both in the presence of Li+, and in the presence of Na+ (Figure (A) and (B), open circles), from which we conclude that the same type triplex conformation formed in the presence of each of the counter ions.
An additional finding was that the temperature profile of Z2 was dissimilar to that of Z3 in contrast to previously studied triplexes with a mixed nucleotide content (Shchyolkina et al., Citation2006). That is, 2AP in the mixed nucleotide sequence 5′-d(GTaGACTGAG) fluoresced similarly when placed in the isolated duplex hairpin or in the duplex part of the R-triplex (Shchyolkina et al., Citation2004, 2006). In principle, the monotonous decrease in Z2 fluorescence with rising temperature is typical for 2AP fluorescence in duplexes. The enhanced fluorescence at 0°C of 2AP placed in the duplex part of the triplex will be discussed in the next section.
We derived thermodynamic parameters for the intramolecular R-triplex formation by the 5′-d(GGTTAGGGTTAG) sequence from the Z1 melting curves (Figure (A) and (B), filled circles) using a fitting procedure described in Shchyolkina et al. (Citation2004, 2006), the values of which are presented in Table . The theoretical fits are shown as the solid curves in Figure . Formation of a triplex conformation by the telomeric sequence 5′-d(GGTTAGGGTTAG) appeared to be more favorable in the presence of NaCl, than of LiCl (compare the curves marked with filled circles in Figure (A) and (B)). This was not the case for the mixed sequence 5′-d(GTAGACTGAG) (R2apCW model), with which R-triplex formation was clearly preferable in LiCl rather than in NaCl solutions (Table ). However, in both cases, the most stable structures, RCW in LiCl and Z1 in NaCl, displayed similar average free energies and formation enthalpies and entropies per triplet. It is remarkable that the Li+ counter ion is strongly disadvantageous for the triplex formation of the telomeric sequence.
Table 1. Thermodynamic parameters of 2AP*T: a triplet formation.
We note that the fluorescence anisotropy of EtBr and the fluorescence emission of the reporting 2AP highlight different conformational features of the triplex. The hydrodynamic volume of the triplex was invariant in the temperature range from 0° to ~20°C, indicating that the third strand remained attached to the duplex part either in the presence of LiCl or in the presence of NaCl as revealed by EtBr probing (Figure , open and filled triangles, respectively). On the other hand, the H-bonds between the third strand and the duplex are largely disrupted in this temperature range (Figure (A) and (B), filled circles). We have shown earlier that EtBr intercalation in the triplex slightly increases its melting temperature (appr. for 5°C) (Shchyolkina et al., Citation2004). Additionally, the intercalated EtBr may delay dissociation of the third strand even up to 20–25°C, a temperature at which the majority of H-bonds are melted. The dependence of the intramolecular triplex hydrodynamic volume on temperature and successive dissociation of the third strand from the duplex part were described in Shchyolkina et al. (Citation2004). Here, we applied the evaluation of the triplex relative hydrodynamic volume in order to testify to the intramolecular character of the triplex studied.
Conformational features of the G-run-containing R-triplex in the presence of Na+ and Li+ ions
What accounts for the low stability of Z1 in Li+? We carried out CD experiments to assess the conformational features of the Z1 oligonucleotide fold. The CD spectra of Z1 are shown for 0º and 50°C in Li+ and Na+ in Figure (A) and (B), respectively, indicating somewhat different conformations of the three-stranded folds in the presence of the two counter ions. The calculated differential CD spectra are shown in the solid unmarked curves and reflect the difference between two conformations: R-triplex with the third 5′-d(GGTTaGGGTTAG) sequence bound to the duplex part and a conformation with this stand dangling away from the intact double-stranded part of Z1 at the higher temperature (Figure (A) and (B)). The spectral signatures are very different in the presence of the two different counter ions. Here, we note that the differential CD spectra are quite reproducible and specific, despite the low intensity of the bands. The shape of the differential CD spectrum in Li+ (Figure (A)) is a sign of a different stacking interaction of guanine bases (for example, see Gray et al., Citation2008) suggesting an A-type conformation of the double helix (Ivanov, Minchenkova, Schyolkina, & Poletayev, Citation1973; Trantı́rek et al., Citation2000). We assume that the conformational change occurs in the duplex part of Z1. FTIR data support this assumption, as the N-type sugars were observed mainly in the oligoG strand of oligoG:oligoC duplex upon binding of the third oligoG strand to the duplex (Ouali et al., Citation1993). Nonetheless, we cannot state whether the overall duplex geometry shifts to A-form and/or the changes involve a sugar puckering switch.
Figure 4. CD spectra of Z1 oligonucleotide in (A) .5 M LiCl and in (B) .5 M NaCl. Temperature was 0°C (filled circles) or 50°C (open circles). The unmarked curves are the difference between CD spectra at 0°C and at 50°C. Samples contained 1 μM oligonucleotides, 10 mM Tris–HCl buffer, pH 7.6. CD is presented as molar CD per nucleotide.
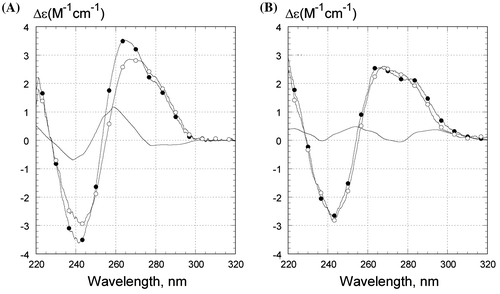
The differential CD spectrum of Z1 in the presence of Na+ has a substantially smaller amplitude than that in the presence of Li+ (compare Figure (A) and (B)), which is indicative of relatively modest changes in base stacking induced by the intramolecular transition in NaCl.
The following mechanism can account for the A-shift in the duplex part of the triplex in the presence of Li+ ions. Counter ions tend binding to the RC groove (Figure (B), G*(C:G) triplet, dashed curve) at GGG site, as there are N7 and O6 of guanines accessible from this groove (Zhurkin et al., Citation1994). The Li+ ion is smaller than its Na+ counterpart, facilitating the compression of the R and C strands together and thereby narrowing of the major groove of the duplex. This conformational change would correspondingly widen the duplex minor groove, which, in turn, leads to an A-type shift in the duplex conformation. Earlier, Minchenkova et al. observed a bias to an A-form for a DNA duplex with a homo-G sequence (Minchenkova, Schyolkina, Chernov, & Ivanov, Citation1986). Three or more subsequent guanines in a sequence were found to account for the A-type stacking or A-type helix geometry in the duplex (Trantı́rek et al., Citation2000).
Several conclusions from the CD experiments may be drawn. Upon triplex formation in Li+, the duplex part of Z1 acquires certain A-like conformational features which involve the G3:C3 block and probably several other adjacent base pairs. This B-to-A-like transition may be responsible for the low stability of the telomeric R-triplex in LiCl (Table ). In the presence of Na+ ions, the conformational changes accompanying the triplex formation are smaller. These local deformations evidently do not diminish or undermine the stability of the telomeric R-triplex in NaCl (Table ).
Increased fluorescence of the reporting base 2AP in Z2 (at 0°C), in comparison with that in Z3, substantiates the inference of a conformational change in DNA duplex as a result of the triplex formation (Figure (A) and (B), open circles and triangles, correspondingly). The position of 2AP next to G3 run (whose conformation is likely to be shifted to an A-like form) may lead to an enhanced 2AP fluorescence in Z2, both in LiCl and in NaCl (Figure (A) and (B), open circles) (O’Neil & Barton, Citation2002). We see that the base next to GGG tract is conformationally flexible (as follows from its increased exposure to solvent). Potentially, this flexibility would facilitate invasion of the telomeric single strand into the homologous duplex at the some point next to a GGG block. Interestingly, the presence of a specific solvent-accessible base in the duplex part of R-triplex is an exclusive feature of the telomeric sequence having GGG blocks. Such a feature was absent in a mixed nucleotide sequence RCW (Shchyolkina et al., Citation2006).
The results described above are consistent with the two important observations made by Amiard et al. (Citation2007). First, the uptake of single strand by homologous supercoiled DNA occurring without shelterin proteins implies that the telomeric 5′-T2AG3 repeats are particularly good substrates for strand invasion (at least, in the presence of Na+ ions, when the R-triplex is rather stable according to our data). Second, binding of the telomeric single strand to the corresponding duplex brings up an enhanced sensitivity of the duplex DNA to single strand-specific nuclease, detected next to the telomeric repeat. The latter observation may be related to the effect reported above, namely, an increased conformational flexibility of the reporting base 2AP located next to G3 run, which is clearly visible upon formation of the telomeric R-triplex (Figure (B), open circles).
Concluding remarks
We have extended a library of oligonucleotide structures that sequences containing G runs can adopt and found that the mammalian telomere sequences can form either a less or a more stable R-triplex depending on the ionic environment. In the presence of Na+ ions, the telomeric R-triplex is more energetically advantageous than the R-triplex of a mixed nucleotide sequence (Table ), while the Li+ ions produce an opposite effect (Table ). The observed plasticity of the telomeric R-triplex may affect the predilection of a sequence for homologous strand exchange in cell, as formation of a relatively flexible (and mobile) R-triplex would not offer a kinetic trap on the way to strand exchange. We propose that the R-triplex serves as a putative intermediate in the formation of D-loop within t-loop which, together with the shelterin proteins, could provide correct initial phasing of the single-strand overhang along telomeric DNA.
According to our molecular modeling (Zhurkin et al., Citation1994), the three homologous triplets, A*(T·A), G*(C·G) and T*(A·T), have practically the same geometry (as shown in Figure (B)), whereas the triplet C*(G·C) has somewhat different conformation. Therefore, the telomeric 5′-T2AG3 repeats (missing cytosines in the 3′ overhang) are expected to be especially favorable for the R-triplex formation. Interestingly, the telomeric sequence was found to bind to the homologous double helix and presumably invade it even in the absence of proteins (Amiard et al., Citation2007). At the same time, the strand association was not observed when a probe of identical length but hybridizing outside the telomeric tract was used.
We note that stability of the observed structure is as yet relatively low in our model system. This may be related to the fact that in order to acquire homogeneous intramolecular structures, we used Li+ or Na+ counter ions but not K+ or Mg++ and Mn++. The latter counter ions promote intermolecular association of oligonucleotides (and thus would obscure detection of the intra-molecular duplex-to-triplex transitions). In other systems (e.g. superhelical plasmids in cellular environment), the formation of telomeric R-triplex may be facilitated by small ligands and certain proteins (in particular TRF2). Further displacement of homologous strand followed by D-loop formation may proceed under the action of other shelterin proteins.
Funding
This work was supported by Russian Academy of Sciences, Molecular and Cellular Biology Program.
Acknowledgements
The authors appreciate helpful discussions with V. L. Florent’ev and M.A. Livshits. The study was supported by Molecular and Cellular Biology Program of the Russian Academy of Sciences. Funding to pay the Open Access publication charges for the article was provided by the National Institutes of Health (USA).
References
- Adrian, M., Heddi, B., & Phan, A. T. (2012). NMR spectroscopy of G-quadruplexes. Methods, 57, 11–24.
- Amiard, S., Doudeau, M., Pinte, S., Poulet, A., Lenain, C., Faivre-Moskalenko, C., … Giraud-Panis, M. J. (2007). A topological mechanism for TRF2-enhanced strand invasion. Nature Structural and Molecular Biology, 14, 147–154.10.1038/nsmb1192
- Biffi, G., Tannahill, D., McCafferty, J., & Balasubramanian, S. (2013). Quantitative visualization of DNA G-quadruplex structures in human cells. Nature Chemistry, 5, 182–186.10.1038/nchem.1548
- Cesare, A. J., Quinney, N., Willcox, S., Subramanian, D., & Griffith, J. D. (2003). Telomere looping in P. sativum (common garden pea). The Plant Journal, 36, 271–279.10.1046/j.1365-313X.2003.01882.x
- Fouche, N., Cesare, A. J., Willcox, S., Ozgur, S., Compton, S. A., & Griffith, J. D. (2006). The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. Journal of Biological Chemistry, 281, 37486–37495.10.1074/jbc.M608778200
- Gray, D. M., Wen, J. D., Gray, C. W., Repges, R., Repges, C., Raabe, G., & Fleischhauer, J. (2008). Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality, 20, 431–440.10.1002/chir.v20:3/4
- Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., & de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514.10.1016/S0092-8674(00)80760-6
- Ivanov, V. I., Minchenkova, L. E., Schyolkina, A. K., & Poletayev, A. I. (1973). Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers, 12, 89–110.10.1002/(ISSN)1097-0282
- Jablonski, A. (1960). On the notion of emission anisotropy. Bulletin de l’Academie Polonaise des Sciences, Serie A, 8, 259–264.
- Jain, S. C., Tsai, C. C., & Sobell, H. M. (1977). Visualization of drug-nucleic acid interactions at atomic resolution. II. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodocytidylyl (3′-5′) guanosine. Journal of Molecular Biology, 114, 317–331.10.1016/0022-2836(77)90253-4
- de Lange, T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Development, 19, 2100–2110.10.1101/gad.1346005
- LePecq, J. B., & Paoletti, C. (1967). A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. Journal of Molecular Biology, 27, 87–106.
- Lestienne, P. P. (2009). Are there three polynucleotide strands in the catalytic centre of DNA polymerases? Biochimie, 91, 1523–1530.10.1016/j.biochi.2009.07.008
- Lestienne, P. P. (2011). Priming DNA replication from triple helix oligonucleotides: Possible threestranded DNA in DNA polymerases. Molecular Biology International, 2011. 562849.
- Lestienne, P. P., Boudsocq, F., & Bonnet, J. E. (2008). Initiation of DNA replication by a third parallel DNA strand bound in a triple-helix manner leads to strand invasion. Biochemistry, 47, 5689–5698.10.1021/bi702318g
- Mergny, J. L., Collier, D., Rougée, M., Montenay-Garestier, T., & Hélène, C. (1991). Intercalation of ethidium bromide into a triple-stranded oligonucleotide. Nucleic Acids Research, 19, 1521–1526.10.1093/nar/19.7.1521
- Minchenkova, L. E., Schyolkina, A. K., Chernov, B. K., & Ivanov, V. I. (1986). CC/GG contacts facilitate the B to A transition of DNA in solution. Journal of Biomolecular Structure and Dynamics, 4, 463–476.10.1080/07391102.1986.10506362
- Munoz-Jordan, J. L., Cross, G. A., de Lange, T., & Griffith, J. D. (2001). t-Loops at trypanosome telomeres. The EMBO Journal, 20, 579–588.10.1093/emboj/20.3.579
- Murti, K. G., & Prescott, D. M. (1999). Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proceedings of the National Academy of Sciences of the United States of America, 96, 14436–14439.10.1073/pnas.96.25.14436
- Neidle, S., & Parkinson, G. N. (2008). Quadruplex DNA crystal structures and drug design. Biochimie, 90, 1184–1196.10.1016/j.biochi.2008.03.003
- O’Neil, M. A., & Barton, J. K. (2002). 2-Aminopurine: A probe of structural dynamics and charge transfer in DNA and DNA:RNA hybrids. Journal of the American Chemical Society, 124, 13053–13066.10.1021/ja0208198
- Ouali, M., Letellier, R., Adnet, F., Liquier, J., Sun, J. S., Lavery, R., & Taillandier, E. (1993). A possible family of B-like triple helix structures: Comparison with the Arnott A-like triple helix. Biochemistry, 32, 2098–2103.10.1021/bi00059a030
- Schmitz, K. M., Mayer, C., Postepska, A., & Grummt, I. (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes & Development, 24, 2264–2269.10.1101/gad.590910
- Shchyolkina, A. K., & Borisova, O. F. (1997). Stabilizing and destabilizing effects of intercalators on DNA triplexes. FEBS Letters, 419, 27–31.10.1016/S0014-5793(97)01417-8
- Shchyolkina, A. K., Borisova, O. F., Livshits, M. A., Pozmogova, G. E., Chernov, B. K., Klement, R., & Jovin, T. M. (2000). Parallel-stranded DNA with mixed AT/GC composition: Role of trans G.C base pairs in sequence dependent helical stability. Biochemistry, 39, 10034–10044.10.1021/bi9913909
- Shchyolkina, A. K., Kaluzhny, D. N., Arndt-Jovin, D. J., Jovin, T. M., & Zhurkin, V. B. (2006). Recombination R-triplex: H-bonds contribution to stability as revealed with minor base substitutions for adenine. Nucleic Acids Research, 34, 3239–3245.10.1093/nar/gkl431
- Shchyolkina, A. K., Kaluzhny, D. N., Borisova, O. F., Hawkins, M. E., Jernigan, R. L., Jovin, T. M., … Zhurkin, V. B. (2004). Formation of an intramolecular triple-stranded DNA structure monitored by fluorescence of 2-aminopurine or 6-methylisoxanthopterin. Nucleic Acids Research, 32, 432–440.10.1093/nar/gkh158
- Shchyolkina, A. K., Timofeev, E. N., Borisova, O. F., Il’icheva, I. A., Minyat, E. E., Khomyakova, E. B., & Florentiev, V. L. (1994). The R-form of DNA does exist. FEBS Letters, 339, 113–118.10.1016/0014-5793(94)80396-X
- Takai, K. K., Hooper, S., Blackwood, S., Gandhi, R., & de Lange, T. (2010). In vivo stoichiometry of shelterin components. Journal of Biological Chemistry, 285, 1457–1467.10.1074/jbc.M109.038026
- Tang, J., Kan, Z. Y., Yao, Y., Wang, Q., Hao, Y. H., & Tan, Z. (2008). G-quadruplex preferentially forms at the very 3′ end of vertebrate telomeric DNA. Nucleic Acids Research, 36, 1200–1208.
- Tomaska, L., Makhov, A. M., Griffith, J. D., & Nosek, J. (2002). t-Loops in yeast mitochondria. Mitochondrion, 1, 455–459.10.1016/S1567-7249(02)00009-0
- Trantı́rek, L., Štefl, R., Vorlı́čková, M., Koča, J., Sklenar, V., & Kypr, J. (2000). An A-type double helix of DNA having B-type puckering of the deoxyribose rings. Journal of Molecular Biology, 297, 907–922.10.1006/jmbi.2000.3592
- Tuite, E., & Nordén, B. (1995). Intercalative interactions of ethidium dyes with triplex structures. Bioorganic & Medicinal Chemistry, 3, 701–711.10.1016/0968-0896(95)00061-K
- Vannier, J. B., Pavicic-Kaltenbrunner, V., Petalcorin, M. I., Ding, H., & Boulton, S. J. (2012). RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell, 149, 795–806.10.1016/j.cell.2012.03.030
- Włodarczyk, A., Grzybowski, P., Patkowski, A., & Dobek, A. (2005). Effect of ions on the polymorphism, effective charge, and stability of human telomeric DNA. Photon correlation spectroscopy and circular dichroism studies. The Journal of Physical Chemistry B, 109, 3594–3605.10.1021/jp045274d
- Xu, D., Evans, K. O., & Nordlund, T. M. (1994). Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry, 33, 9592–9599.10.1021/bi00198a027
- Zhurkin, V. B., Raghunathan, G., Ulyanov, N. B., Camerini-Otero, R. D., & Jernigan, R. L. (1994). A parallel DNA triplex as a model for the intermediate in homologous recombination. Journal of Molecular Biology, 239, 181–200.10.1006/jmbi.1994.1362
