Abstract
COVID-19 is the disease caused by SARS-CoV-2 which has led to 2,643,000 deaths worldwide, a number which is rapidly increasing. Urgent studies to identify new antiviral drugs, repurpose existing drugs, or identify drugs that can target the overactive immune response are ongoing. Antiretroviral drugs (ARVs) have been tested in past human coronavirus infections, and also against SARS-CoV-2, but a trial of lopinavir and ritonavir failed to show any clinical benefit in COVID-19. However, there is limited data as to the course of COVID-19 in people living with HIV, with some studies showing a decreased mortality for those taking certain ARV regimens. We hypothesized that ARVs other than lopinavir and ritonavir might be responsible for some protection against the progression of COVID-19. Here, we used chemoinformatic analyses to predict which ARVs would bind and potentially inhibit the SARS-CoV-2 main protease (Mpro) or RNA-dependent-RNA-polymerase (RdRp) enzymes in silico. The drugs predicted to bind the SARS-CoV-2 Mpro included the protease inhibitors atazanavir and indinavir. The ARVs predicted to bind the catalytic site of the RdRp included Nucleoside Reverse Transcriptase Inhibitors, abacavir, emtricitabine, zidovudine, and tenofovir. Existing or new combinations of antiretroviral drugs could potentially prevent or ameliorate the course of COVID-19 if shown to inhibit SARS-CoV-2 in vitro and in clinical trials. Further studies are needed to establish the activity of ARVs for treatment or prevention of SARS-CoV-2 infection .
Communicated by Ramaswamy H. Sarma
1. Introduction
The novel coronavirus, SARS-CoV-2, was first reported as a new viral infection in humans in late 2019, and over the course of 2020 the viral infection has become a pandemic. Coronavirus disease, COVID-19, has in turn taken the lives of more than 2,643,000 people by March 2021, with hundreds of thousands of more deaths expected in the absence of effective treatments or additional control measures. COVID-19 presents a major worldwide public health emergency. Only three drugs have been given emergency use authorization for use against COVID-19 in the USA, and intense research efforts are underway to find effective antiviral treatments via novel drug design or drug repurposing (Duarte et al., Citation2020; Elmezayen et al., Citation2020; Sanders et al., Citation2020). Many in silico methods have already been employed for their potential to discover repurposed drugs with activity towards SARS-CoV-2 (Aanouz et al., Citation2020; Boopathi et al., Citation2020; Elmezayen et al., Citation2020; Enayatkhani et al., Citation2020; Enmozhi et al., Citation2020; Gupta et al., Citation2020; Hasan et al., Citation2020; Joshi et al., Citation2020; Sarma et al., Citation2020), including ARVs used in HIV infection, with variable results (Muralidharan et al., Citation2020; Pant et al., Citation2020) (Khan et al., Citation2020). Some studies found no change in morbidity or mortality (Byrd et al., Citation2020; Härter et al., Citation2020; Karmen-Tuohy et al., Citation2020), but others have observed lower SARS-CoV-2 infection rates, or lower hospitalizations in PLWH who take certain Antiretroviral drugs (ARV) regimens, like tenofovir/emtricitabine, (TDF/FTC) or Truvada (Del Amo et al., 2020). In addition, a South African study also observed a lower mortality rate for those taking tenofovir/emtricitabine although potential confounders such as healthier people taking tenofovir/emtricitabine create a channeling bias which needs to be further investigated (Boulle et al., Citation2020).
Antiretroviral drugs (ARVs), normally used to treat HIV infection, have also been tested in Hepatitis B Virus (HBV) infection (Boettiger et al., Citation2016), and in Amyotrophic Lateral Sclerosis (ALS) [NCT02437110]. ARVs have been studied in silico, in vitro and in vivo for their activity against Severe Acute Respiratory Syndrome (SARS) (Chu et al., Citation2004; Elfiky & Azzam, Citation2020; Yamamoto et al., Citation2004), and Middle East Respiratory Syndrome (MERS) (de Wilde et al., Citation2014), which led to adoption of two ARVs, lopinavir and ritonavir, as putative antiviral drugs against SARS-CoV-2. However, a small randomized controlled clinical trial of lopinavir and ritonavir co-administered to hospitalized adults with severe COVID-19 showed no clinical benefit over the standard of care (Cao et al., Citation2020), and a large randomized, controlled, open-label trial (RECOVERY), showed no clinical benefit for COVID-19 (Recovery-Collaborative-Group, Citation2020). For other ARVs, a preprint reported that tenofovir and emtricitabine acted as chain terminators in the replication of viral RNA by the SARS-CoV-2 RdRp (Jockusch et al., Citation2020). Other in vitro studies have also shown that ARVs such as tenofovir and abacavir are capable of terminating RNA synthesis catalyzed by the RdRp of SARS-Cov-2 as well, whereas lamivudine and emtricitabine were poor substrates for the RdRp (Chien et al., Citation2020). Remdesivir is approved for use by the FDA for its ability to act as a chain terminator of the growing RNA strands catalyzed by the SARS-CoV-2 RdRp. In this context, in silico studies can help to gauge projected mechanisms and likely molecules which bind to key viral targets, in a cost effective, high throughput manner, whereas in vitro studies provide necessary details for effective inhibitory concentrations of compounds tested, and clinical studies would provide evidence for effectiveness in people.
Here, we describe a comprehensive in silico analysis of the binding of known ARVs to the catalytically active sites of the Main protease (Mpro) (PDB ID: 6Y2E) (Zhang et al., Citation2020) and RNA dependent RNA polymerase (RdRp) (PDB ID: 6M71) (Gao et al., Citation2020) of SARS-CoV-2 at atomic resolution. We used the Schrodinger’s induced fit docking algorithm and molecular dynamics (MD) to identify specific HIV ARVs that could inhibit the viral replication cycle of SARS-CoV-2 by binding to these essential proteins. Results from these analyses suggest the SARS-CoV-2 Mpro is stably bound by both protease inhibitors (PI) atazanavir and indinavir. The ARVs predicted from the same protocol to bind the catalytic site of the RdRp include NRTIs, abacavir, emtricitabine, tenofovir and zidovudine.
Our results suggest that some commonly used ARVs have binding potential in silico to SARS-CoV-2 including ARVs commonly given to people without HIV as PrEP, or also used in the treatment of chronic HBV infection. Further studies should explore the activity of ARVs in vitro, in animal models, and in clinical trials, to assess whether the ARVs we identified could be used either in COVID-19 treatment regimens or as pre-exposure prophylaxis for SARS-CoV-2 infection.
2. Materials and methods
2.1. Methods overview
Maestro (v12.2.012, release 2019-4, Schrödinger) software was used to facilitate small molecule docking calculations involving high throughput virtual screening (HTVS), including flexible Induced Fit Docking (IFD), and scoring with the Glide XP (extra precision) (Friesner et al., Citation2006) docking score (Schrödinger, 2020, USA). Select HIV ARVs were downloaded in their active forms from the ZINC database, and PubChem when not available via ZINC. These structures were then processed using LigPrep. All defaults parameters were used, including retaining specified chiralities while varying other chiral centers.
2.2. Protein preparation and ligand preparation
The main protease (Mpro) (Protein Data Bank [PDB] ID: 6Y2E) Cryo-EM structure, and the RNA-dependent RNA polymerase (RdRp) (PDB ID: 6M71) crystal structure, of SARS-Cov-2 were used for the entirety of this work. The ligand docking sites were specified as the catalytically active sites by Zhang et al. (Zhang et al., Citation2020) and Gao et al. (Gao et al., Citation2020), using an inner box distance of 10 Å around the catalytically active sites, and outer box was set to automatic determination. 6Y2E’s waters were separated and were not included in further analysis. The protein preparation wizard (Schrödinger) was used on its default settings to prepare both proteins. Hydrogen bonds were assigned and optimized, and a restrained minimization was carried out, using the OPLS3e forcefield (Roos et al., Citation2019).
Ligand Preparation was carried out using LigPrep in Maestro interface. As stated above, all HIV ARVs were downloaded as SDF files in their active forms from the ZINC database (Irwin & Shoichet, Citation2005), and PubChem (Kim et al., Citation2021) when not available via ZINC. The OPLS3e force field was applied. Ionization was carried out using Epik (Shelley et al., Citation2007), at a target of pH 7.0 +/- 2.0. Desalting was allowed, and tautomers were generated. The stereoisomers were set to retain specified chiralities while varying unspecified chiral centers.
2.3. Protein-ligand small molecule docking using Schrodinger’s Maestro interface
In order to identify potential binders of HIV ARVs to the catalytic sites of the SARS-COV-2 Mpro and RdRp enzymes, we performed in silico molecular docking analyses using Schrodinger’s induced fit docking (IFD) followed by Glide XP docking. In the molecular structure of the Mpro (PDB ID: 6Y2E) the catalytic site is marked by residues Gln189, His41 and Cys145 (Zhang et al., Citation2020). For the RdRp structure (PDB ID: 6M71) the substrate binding site is marked by residues 753-FSMMILSDDAVVCFN-767 that includes the motif 759-SDD-761, which is conserved amongst polymerases (Gao et al., Citation2020).
2.4. Induced fit docking
The enzymes’ catalytic sites were selected based on the residues cited above and ligands were allowed to dock within 20 Å of these sites. The side chains were refined and then optimized if docked within 5 Å of the ligands pose. Redocking was done for structures within 30 kcal/mol of the best structure and within the top 20 structures overall with extra precision. Ligands were docked flexibly. The top 20 poses for each ligand were allowed, but only the top 5 poses for each were used for ranking according to their emodel score. The Glide XP docking score was then used to rank the top 5 dock for each ligand. Following selection of the molecule with the most negative emodel score, ligands were ranked according to their Glide XP scores. There was an exception to this ranking system. Indinavir had an emodel score difference of 0.8, but a docking score 2 points more negative for the pose with the slightly more positive emodel score. No other ligands fit this scoring discrepancy. Because of this, and because it was the highest scoring ligand both poses were used for MD analysis, and the one with the superior docking score was far more stable over time in the binding pocket, and is the dock pose we discuss further in the paper.
2.5. Molecular dynamics
The unliganded Mpro and RdRp were used as references for MD analysis. In addition, the top 3 scoring ligands for the Mpro, and the top 3 ligands for the RdRp were chosen based on their IFD docking scores, and by their known mechanism of action in HIV. We also added tenofovir to this analysis, despite being the 5th ranked according to docking score, due to its reported in vitro activity in this paper, and proposed potential in several clinical trials (Ayerdi et al., 2020; Chien et al., Citation2020; Del Amo et al., 2020). These 7 top docked structures were used for further analyses due to these known properties and their simulated docking scores. This includes the RdRp docked to abacavir, emtricitabine and zidovudine and the Mpro docked to indinavir, amprenavir, and atazanavir. A 250 ns MD simulation was performed with each docked structure with Desmond (D. E. Shaw Research, New York) to assess the stability of bound structures to their target enzymes. The system was solved in a TIP3P water model and 0.15 M NaCl. Neutralization of the protein-ligand complex system was achieved by adding Na + or Cl − counterions that balanced the net charge of the system. The full system energy minimization step was completed. The MD simulation was run for 250 ns at 300 K temperature and standard pressure (1.01325), within an orthorhombic box with buffer dimensions of 10 Å × 10 Å × 10 Å and the NPT ensemble class. The energy of 1.2 (kcal/mol) was recorded at intervals of every 100 ps resulting in 2,501 frames for each simulation. The simulations were then loaded into Maestro, for visualization of the protein ligands structures over time. The Nose-Hoover chain was used to sustain temperature at 300 K. The Martyna-Tobias-Klein dynamic algorithm was used to sustain the pressure at 1.01325 bar. The simulation interaction diagram in Maestro was utilized to depict results captured in MD simulations. The final docked structures were always the reference frame used across all MD simulations and are the structures represented at time 0.
2.6. Prime MM-GBSA free energy calculations
Schrodinger’s Prime MM-GBSA version 3 (Shivakumar et al., Citation2010) was used to calculate the free energy of binding for the 7 protein-ligand models. Specifically, the free energy of binding was calculated at 1-ns resolution, for each of the 250 ns simulations. The average free energy of binding and related standard deviations are reported in and .
Table 3. MM-GBSA results for the top Mpro-ligand hits calculated from MD trajectories of 250 ns.
Table 4. MM-GBSA results for the top RdRp-ligand hits calculated from MD trajectories of 250 ns.
2.7. Data controls and interpretation
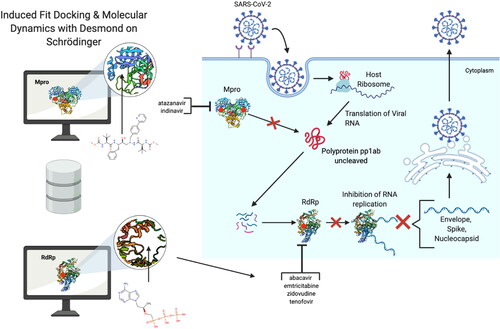
Because remdesivir is approved for COVID-19 treatment and has been shown in in vitro and in vivo to inhibit the SARS-CoV-2 RdRp, we used its docking score as the positive control for reporting our results (Beigel et al., 2020; Gordon et al., Citation2020). Remdesivir has an EC50 of 0.77 μM against SARS-CoV-2 viral replication in vitro (M. Wang et al., Citation2020). We also used boceprevir as a positive control, as it has been shown to have in vitro activity against the Mpro. Boceprevir has an IC50 of 4.13 µM, and an EC50 of 1.90 µM against the SARS-CoV-2 virus (Ma et al., Citation2020). The image portrayed in was created using BioRender.
Figure 1. Overall schematic for the methods used in this paper and our significant results. Molecules are docked to key residues of the viral enzymes. The HIV ARVs that are predicted to bind to designated catalytic sites of the viral enzymes are those listed next to either bracket. The red dots indicate the designated catalytic sites for the purpose of this study. The binding and potential inhibition of these enzymes would disrupt the replication machinery of this virus, as shown by the replication schematic.
3. Results
3.1. Estimates of COVID-19 and HIV infection
There are several important issues that complicate the issue of COVID-19 and HIV co-infection, and in the USA, PLWH of color are disproportionally impacted by COVID-19. In addition, half of PLWH in the USA are over 50 years of age, and co-morbidities are more common in this group. We estimated the number of PLWH at risk for death from COVID-19 in the USA by August 4th 2020, based upon projected estimates (, supplementary material). These estimates are confounded by many variables, and since the mortality rates are fluctuating with changing testing algorithms, and data availability, they are limited in utility, yet in order to make sense of data in any observational study, such numbers need to be estimated. An observational study to determine the incidence or disease course from SARS-CoV-2 infection in PLWH who take ARV therapy, compared to those that do not is technically challenging, and problematic because ARV therapy is the standard of care for HIV worldwide. A prospective, randomized study of anti-HIV PrEP medications to prevent COVID-19, would be more appropriate. In addition, linking COVID-19-related deaths to HIV registries could provide some of these insights as well.
Table 1. Docking scores for HIV drugs tested against SARS-CoV-2 Mpro.
3.2. Induced fit docking
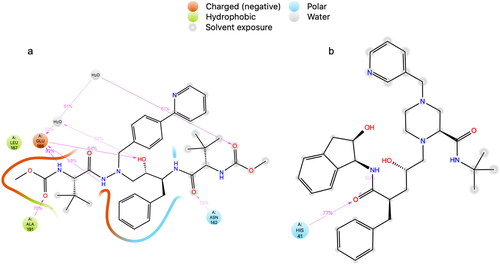
The protocol described in was used to analyze the ability of HIV ARV drugs to bind to the catalytic sites of the SARS-CoV-2 main protease (Mpro) (Zhang et al., Citation2020) and RNA-dependent RNA polymerase (RdRp) enzyme (Gao et al., Citation2020). The standard deviation of results presented in and , is 1.867 for the Mpro, and 2.32 for the RdRp. See the ligand interaction diagrams in atazanavir and 2(b)—indinavir, each docked to the Mpro. See also ligand interaction diagrams for abacavir, emtricitabine, tenofovir and zidovudine in . We chose the top scoring docked ligands; in addition, we considered their known mechanism in HIV. Further, we included tenofovir in the MD analysis, due to its effect in vitro and in vivo against SARS-CoV-2 (Ayerdi et al., 2020; Chien et al., Citation2020; Del Amo et al., 2020). and show the top scored ligands and their Glide XP docking scores when docked to the Mpro and RdRp respectively. The mechanism of each inhibitor against HIV is listed. Controls and other antivirals are included for comparison.
Figure 2. Ligand interaction diagrams in 2D from induced fit docking of (a) atazanavir, and (b) indinavir to the Mpro.
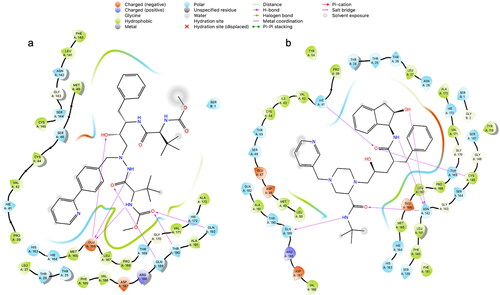
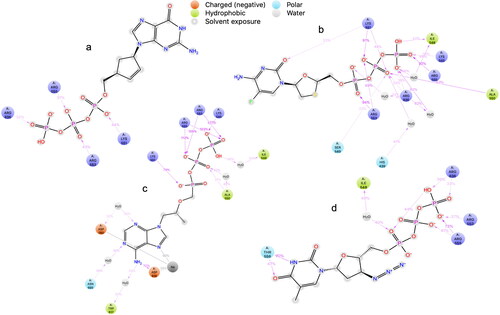
Figure 3. Ligand interaction diagrams in 2D from induced fit docking of (a) abacavir, (b) emtricitabine, (c) tenofovir, and (d) zidovudine.
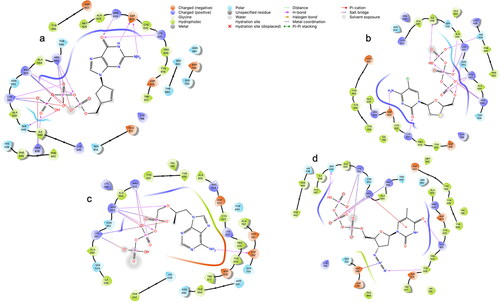
Table 2. Docking scores for HIV drugs tested against SARS-CoV-2 RdRp.
3.3. Molecular dynamics
The unliganded Mpro was stable over time only fluctuating between an RMSD of 1.2 Å and 2.4 Å. The RMSF of all residues was between 0.4 Å and 3.2 Å. However only 3 residues have an RMSF greater than 2 Å. The unliganded RdRp has an RMSD between 1.6 Å and 3.2 Å for the entire simulation. The RMSF of the unliganded RdRp is between 0.6 Å and 5 Å. The high number in some residues, stems from the multiple subunits of the RdRp which are not connected by a single amino acid chain. The binding site residues of the RdRp, at residues near 100, and 1,000, and between 210 and 800, are stable with an RMSF ranging from 0.6 Å to 2.4 Å. See supplemental material for unliganded material. The Mpro docked with either, amprenavir, atazanavir, or indinavir, were the systems used for further MD analysis. Atazanavir was the most stable protein ligand structure over time. The Cα RMSD is very stable over time for all tested structures in . The ligand RMSD for the Mpro-atazanavir structure is relatively stable over the entire simulation fluctuating primarily between 1.2 Å and 3.6 Å. Atazanavir is stable inside the binding pocket throughout the entire simulation. Both the Mpro-amprenavir and Mpro-indinavir models exhibit stable Cα RMSD over time as well. But upon review of the ligand RMSD for amprenavir leads to a sharp rise in RMSD which appears to be in conjunction amprenavir’s instability in the binding pocket and subsequent conformational changes which leave amprenavir outside of the catalytic site (see supplemental material). The indinavir ligand RMSD is quite variable until about 120 ns when a stable conformation is achieved. for atazanavir and indinavir respectively, shows that the ligands are bound to residues with RMSFs mainly around 1 Å to 2 Å. The protein-ligand interactions are seen in , for atazanavir and indinavir respectively. Atazanavir has a majority of its interactions with Glu 166, which has been shown to be a key residue in the proper function of the Mpro in site directed mutagenesis experiments (Cheng et al., Citation2010). Atazanavir also interacts with His 41, Asn 142 in addition to others seen in . Atazanavir forms interactions for more than 30% of the simulation with sidechains and waters shown in . Indinavir forms less interactions than atazanavir and the interaction fraction scale is smaller for that reason in . Indinavir has a majority of its interactions with the key residue His 41, in addition to Thr 26, Gln 189, Asn 142, Gly 143, Met 49, and others. Indinavir forms interactions for more than 30% of the simulation with sidechains and waters shown in . All MD results not presented here are found in the supplemental material, including the ligand RMSF, radius of gyration, intramolecular hydrogen bonding, and SASA, MolSA and PSA.
Figure 4. Protein-ligand RMSD plots for the Mpro, (a) atazanavir, and (c) indinavir complexes. For Cα atoms of the Mpro the RMSD is represented by the blue line (scale on left). The RMSD of the ligands are represented by the red line with the scale to the right of the figures in the top row. Please note that all graphs have various scales. The x-axis scale is in nanoseconds, the y-axis is in Angstroms. Protein RMSF plots in bottom row of (b) atazanavir, and (d) indinavir complexed with Mpro. Secondary structural elements of alpha helices and beta strands are represented by highlighted red and blue backgrounds, respectively. Protein ligand contacts are marked with green vertical bars. Please note the varying scales used in each graph.
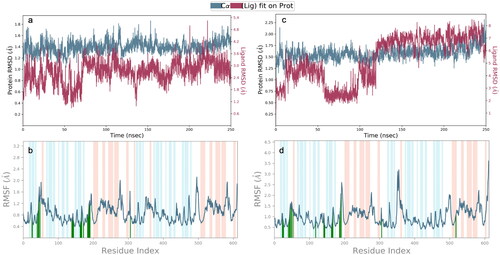
Figure 5. Protein ligand contacts of the Mpro with the (a) atazanavir, and (b) indinavir ligands, respectively. Hydrogen bonds represented in green, purple representing hydrophobic interactions, pink for ionic, and blue for water bridges.
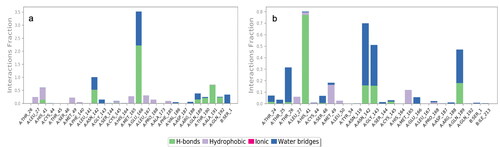
Figure 6. Detailed protein ligand interactions which occur over time in the MD simulation are shown for (a) atazanavir, and (b) indinavir. Only interactions which occur for more than 30% of the simulation are shown in each.
The RdRp docked with either abacavir, emtricitabine, tenofovir, or zidovudine was the selected structures used in further MD analyses based on their docking scores and reported in vitro, and in vivo (Ayerdi et al., 2020; Chien et al., Citation2020; Del Amo et al., 2020). The protein RMSD of all four were completely stable over the 250 ns simulation as determined by the Cα represented blue lines in . Looking at each MD trajectory over time makes clear that each ligand is stable within the binding pocket and does not ever leave the binding site during the 250 ns simulation. The ligand RMSD of each structure was variable, with tenofovir having the most stable ligand RMSD throughout the simulation shown in . Abacavir was stable until 100 ns, when a sharp conformational change occurs, and a stable conformer is achieved. Emtricitabine’s ligand RMSD in increases for 25 ns, followed by an overall relatively stable conformation throughout the remainder of the time. The zidovudine ligand RMSD has a slow increase until about 75 ns when a stable conformation is achieved, and which remains for the remainder of the 250 ns. The RMSF of each protein-ligand structure is shown in . Interacting residues form interactions with residues which consistently have lower RMSFs; interacting residues can be seen in detail in and . Interactions which occur for more than 30% of the simulation are seen in . All MD results are found in the supplemental material, and include the ligand RMSF, radius of gyration, intramolecular hydrogen bonding, and SASA, MolSA and PSA.
Figure 7. RMSD plots in left column, for the Cα (blue, scale left) from MD analysis of RdRp (a) abacavir, (b) emtricitabine, (c) tenofovir, and (d) zidovudine complexes. The ligand RMSD plot (magenta, scale right). Protein RMSF plots in right column, of (e) abacavir, (f) emtricitabine, (g) tenofovir, and (h) zidovudine complexed with RdRp. Secondary structural elements of alpha helices and beta strands are represented by highlighted red and blue backgrounds, respectively. Protein ligand contacts are marked with green vertical bars. Please note the varying scales in each graph.
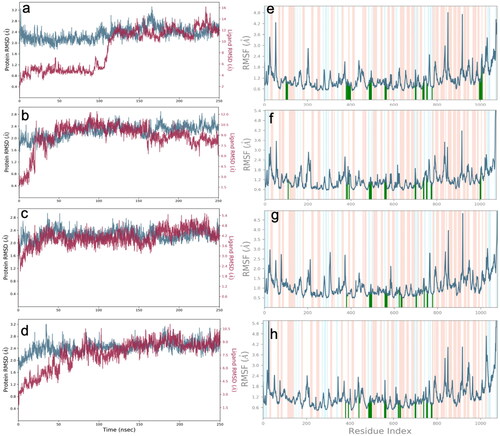
Figure 8. Protein ligand contacts of the RdRp with the (a) abacavir, (b) emtricitabine, (c) tenofovir, and (d) zidovudine ligands, respectively. Hydrogen bonds represented in green, purple representing hydrophobic interactions, pink for ionic, and blue for water bridges.
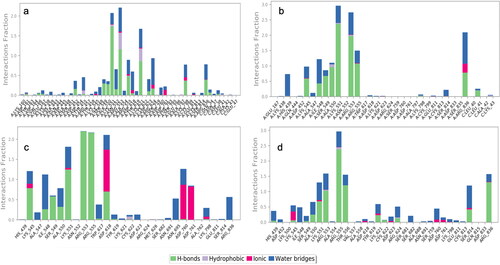
Figure 9. Detailed protein ligand interactions which occur over time in the MD simulation are shown for the RdRp- (a) abacavir, (b) emtricitabine, (c) tenofovir, and (d) zidovudine complexes. Only interactions which occur for at least 30% of the simulation are shown in each. For more detailed interactions across each please see the supplemental material. The Glide XP docking scores utilize an empirical scoring function which approximates the ligand binding free energy, when the ligands bind to the designated catalytic sites of either the Mpro or RdRp. More negative numbers suggest more free energy associated with the predicted binding event ( and ).
3.4. MM-GBSA
Molecular mechanics with generalized born surface area and solvent accessibility (MM-GBSA), was used to assess the binding affinity of each structure assessed in the MD analysis, across the 250 ns simulation. The Mpro-atazanavir structure was by far the strongest binder according to its ΔGbinding of −86.19 kcal/mol, followed by indinavir −51.44 kcal/mol (). The strongest binder for the RdRp was abacavir with ΔGbinding −105.02 kcal/mol, followed closely by emtricitabine at −102.23 kcal/mol, then tenofovir at −96.70 kcal/mol, and zidovudine −93.48 kcal/mol ().
3.5. Data availability
Remaining structural files, of docked ligands relating to the lower Glide XP scored conformers, and their subsequent poses will be made available upon request to the authors of this paper.
4. Discussion
The data presented here, along with anecdotal evidence and recent in vitro studies, present a case for the possible prevention and treatment of COVID-19 with repurposed HIV ARV drugs. HIV ARVs were studied in past human coronavirus infections of SARS-CoV and MERS (Ford et al., Citation2020), and are being used in SARS-CoV-2 clinical trials as listed in and . Recent publications assessing COVID-19 and HIV coinfection have suggested a decreased mortality amongst some PLWH taking certain ARVs such as TDF/FTC, while others show very little difference in PLWH with no other comorbidities (Ayerdi et al., 2020; Boulle et al., Citation2020; Del Amo et al., 2020). In addition, two clinical trials assessing lopinavir and ritonavir failed to show any clinical benefit in people with COVID-19 (Cao et al., Citation2020; Recovery-Collaborative-Group, Citation2020). In order to better determine potential mechanistic reasons for such associations we explored the potential binding of these HIV ARVs to the known active sites of the Mpro and RdRp of SARS-CoV-2. We used chemoinformatic analyses to predict which ARVs would bind to the SARS-CoV-2 Mpro or RdRp enzymes and identified a number of compounds with predicted binding ability. Future in vitro work should be guided by the in-silico data present here. While further studies are needed to establish the value of ARVs in COVID-19, existing or new combinations of ARV regimens for preventing or treating HIV infection could potentially prevent or ameliorate the course of COVID-19.
Table 5. Ongoing and planned clinical trials that use HIV antiretrovirals (ARVs) for the prevention and/or treatment of SARS-CoV-2.
Table 6. Clinical trials and observational studies of PLWH or PrEP users, and the prevalence of COVID-19.
The rationale supporting our work lies in previously published work describing similarities between the RNA-dependent RNA polymerase’s from previous coronaviruses, including SARS, and the HIV reverse transcriptase (RT) (Oberg, Citation2006). Because of that similarity, ARVs were tested in the setting of SARS infection (Chu et al., Citation2004). SARS-CoV and SARS-CoV-2 Mpro are 96% identical (Chen et al., Citation2020), and SARS-CoV and the SARS-CoV-2 RdRp 98% (Shannon et al., Citation2020). The similarity between the enzymes of SARS-CoV and SARS-CoV-2 is marked and provided the rationale for testing drugs which were effective in vitro against SARS-CoV, in the setting of SARS-CoV-2 as experimental therapies.
Both the RdRp and the Mpro were analyzed by MD in their unliganded forms. Both structures had very stable RMSDs over the 250 ns MD simulations. The RdRp has a few small chains which become detached, but the overall structure is stable with no changes in conformation observed. In our IFD study presented here, indinavir was the most efficient binder and surpassed all controls with known binding affinity by the IFD analysis. However, atazanavir was the most stable in the MD analysis over the 250 ns simulation with very little variation (, and 4a). Atazanavir was also the most efficient binder according to MM-GBSA, with a ΔGbinding of −86.19. In comparison indinavir’s ΔGbinding is −51.44, see . We hope that our results will encourage in vitro studies to test if indinavir or atazanavir are active against the SARS-CoV-2 Mpro. In our study amprenavir was also predicted to be a good binder by IFD, but according to the MD analysis, was very unstable in the catalytic site of the Mpro and made few interactions which sustained the length of the MD simulation. Despite this, both drugs could still be tested further in vitro, although widespread use of protease inhibitors would be challenging to implement because of adverse drug-drug interactions and tolerability issues.
We used boceprevir as a positive control in the IFD analysis, due to reported in vitro activity against the Mpro. Boceprevir has an IC50 of 4.13 µM, and an EC50 of 1.90 µM against the SARS-CoV-2 virus (Ma et al., Citation2020). Remdesivir, an adenosine nucleotide analogue currently with emergency FDA-approval for COVID-19 treatment, has activity against the RdRp of SARS-CoV-2 (Gordon et al., Citation2020) by incorporation into the growing RNA strand and inhibiting further viral transcription. Remdesivir was the first agent to show clinical efficacy in a randomized (Beigel et al., 2020) placebo-controlled study. However, more accessible oral drugs would help expand the antiviral drug range. Tenofovir and emtricitabine can inhibit the RdRp of SARS-CoV-2 in vitro (Jockusch et al., Citation2020), as expected from our in silico analyses, and another study using a different method found both tenofovir and abacavir to be better suited than lamivudine or emtricitabine against the RdRp. Interestingly nucleotide reverse transcriptase inhibitors like emtricitabine and lamivudine are –(-) nucleotides and are the mirror image of the D-ribose nucleotides (Hung et al., Citation2019). They are incorporated “backwards” into growing HIV DNA primer strands in their intended HIV inhibition mechanism (Hung et al., Citation2019). Both nucleotide analogs and nucleoside analogs can be incorporated into growing RNA strands and thus may act as chain terminators of growing viral RNA strands under replication by the RdRp, likely due to the low fidelity of the RdRp to its substrate (Jockusch et al., Citation2020; McKenna et al., Citation2010). It should be noted that according to their work, NRTIs like tenofovir and abacavir, seem superior to others like emtricitabine and lamivudine. Our work suggests that people on a regimen containing tenofovir, emtricitabine, abacavir, lamivudine, or zidovudine, could potentially be partially protected from SARS-CoV-2 infection or COVID-19, although only additional in vitro studies like the one mentioned above, and clinical trials can determine this concretely. Considering the data here and the data discussed next, tenofovir seems the most likely candidate for further study.
According to our MD results, tenofovir had one of the lowest docking scores amongst the NRTIs and yet was clearly more stable than the other ligands examined by MD. Zidovudine and emtricitabine were also stable over the entire MD analysis, with only small conformational changes in the ligand conformation occurring until about 50 ns, as seen by the ligand RMSD plots in . The MM-GBSA results show all of the ligands have good, predicted binding to the nucleoside/nucleotide (NT) binding site in . Of course, using this data to determine if these nucleotides/nucleosides will be incorporated into a growing primer RNA strand is not possible with current in silico models. Interestingly only tenofovir exhibited significant binding to the residue Asp760, which is the nucleotide binding site on the RdRp as stated above. It is the most stable of the ligands tested according to ligand RMSD in . Considering the clinical trials, in vitro work, and now this work suggesting the stability of tenofovir in the NT binding site, tenofovir is an important candidate for further study against the SARS-CoV-2 RdRp.
It is not unexpected that NNRTIs as a class did not bind well to the catalytic site for the SARS-CoV-2 RdRp, which is the NT binding site, where elongation of the RNA strand occurs (Gao et al., Citation2020). When NRTIs are metabolized and form triphosphate structures, they are capable of being added to the growing RNA/DNA strand and chain termination will follow (Gordon et al., Citation2020; Jockusch et al., Citation2020; Y. Wang et al., Citation2020). However, HIV reverse transcriptase is inhibited by NNRTIs at a site different from that which the NRTIs bind. Instead, NNRTIs inhibit HIV RT in a non-competitive fashion, binding at a site distant from the polymerase active site, usually stopping key nucleic acid-protein interactions from occurring, or changing the active site structure (Sluis-Cremer & Tachedjian, Citation2008). Past studies have found no potential homologous hydrophobic NNRTI binding site on the previous SARS-CoV RdRp structure (Xu et al., Citation2003), although we identified potential hydrophobic pockets on the surface of the RdRp (data not shown). This will remain an area for further research to address, though COVID-19 research should certainly include second generation NNRTIs with more rotatable bonds and flexibility. If HIV NNRTIs are observed to offer potential protection it should become clear in observational or retrospective studies.
In order to better understand the impact of ARVs on SARS-CoV-2 infection or COVID-19, PLWH or people at risk of HIV infection on pre-exposure prophylaxis (PrEP) could provide critical insights into the course of COVID-19. An early report first showed that PLWH on ARVs can be infected by SARS-CoV-2 (Blanco et al., 2020), but of the first 543 people admitted to a hospital in Barcelona with COVID-19, only five were PLWH. New data has also been presented which compared over 30,000 PLWH and 76,000 matched controls and described 189 PLWH and their COVID-19 outcomes (Park et al., Citation2020). Additionally, registries (Dandachi et al., 2020) can also provide greater insight into the difference or lack of difference in COVID-19 outcomes amongst PLWH.
Our data suggests that select ARVs could also be tested as pre-exposure prophylaxis (PrEP) for COVID-19, if in vitro efficacy was shown. Drugs used for HIV PrEP (tenofovir/emtricitabine) are well tolerated (Mayer et al., 2020), and, if effective against SARS-CoV-2 in vitro, trials would be more easily justified given the excellent tolerability, lack of drug-drug interactions, and well characterized safety profile, when compared to PIs. We also look forward to results from planned research such as the placebo-controlled study of SARS-CoV-2 prophylaxis with tenofovir disoproxil fumarate/emtricitabine vs. hydroxychloroquine vs. both (vs. placebo) in health care workers in Spain (N = 4000) NCT04334928.
5. Conclusions
At this time the authors of this study would like to caution that this report has not made any conclusion or recommendation to change any treatment or prevention regimen. However, our studies suggest that further investigations of the role of ARVs in SARS-CoV-2 prevention or amelioration of COVID-19 are warranted.
| Abbreviations | ||
| AI | = | Attachment Inhibitor |
| ALS | = | Amyotrophic Lateral Sclerosis |
| ARVs | = | Antiretrovirals |
| COVID-19 | = | Coronavirus disease |
| CYP3A | = | Cytochrome P450, family 3, subfamily A |
| Glide XP | = | extra precision |
| EI | = | Entry Inhibitor |
| HBV | = | Hepatitis B virus |
| HTVS | = | High Throughput Virtual Screening |
| INSTI | = | Integrase Strand Transfer Inhibitor |
| MERS | = | Middle East Respiratory Syndrome |
| MM-GBSA | = | Molecular Mechanics with Generalized Born Surface Area and Solvent Accessibility |
| Mpro | = | SARS-CoV-2 Main Protease |
| NNRTI | = | Non-Nucleoside Reverse Transcriptase Inhibitor |
| NRTI | = | Nucleoside/Nucleotide Reverse Transcriptase Inhibitor |
| NT | = | Nucleoside/Nucleotide |
| PDB | = | Protein Data Bank |
| PI | = | Protease Inhibitor |
| PK | = | Pharmacokinetics |
| PLWH | = | People Living With HIV |
| PrEP | = | Pre-Exposure Prophylaxis |
| RdRp | = | SARS-CoV-2 RNA Dependent RNA Polymerase |
| RT | = | Reverse Transcriptase |
| SARS | = | Severe Acute Respiratory Syndrome |
| SARS-CoV-2 | = | Severe Acute Respiratory Syndrome Coronavirus type 2 |
Supplemental_Figures_Final_Feb_25_2021...docx
Download MS Word (227.9 MB)Disclosure statement
All authors declare no conflict of interest
Additional information
Funding
References
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. https://doi.org/10.1080/07391102.2020.1758790
- Ayerdi, O., Puerta, T., Clavo, P., Vera, M., Ballesteros, J., Fuentes, M. E., Estrada, V., Rodríguez, C., Del Romero, J., & Sandoval Study Group. (2020). Preventive efficacy of tenofovir/emtricitabine against severe acute respiratory syndrome coronavirus 2 among pre-exposure prophylaxis users. Open Forum Infectious Diseases, 7(11), ofaa455. https://doi.org/10.1093/ofid/ofaa455
- Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., Hohmann, E., Chu, H. Y., Luetkemeyer, A., Kline, S., Lopez de Castilla, D., Finberg, R. W., Dierberg, K., Tapson, V., Hsieh, L., Patterson, T. F., Paredes, R., Sweeney, D. A., Short, W. R., Touloumi, G., … ACTT-1 Study Group Members. (2020). Remdesivir for the treatment of Covid-19 - final report. New England Journal of Medicine, 383(19), 1813–1826. https://doi.org/10.1056/NEJMoa2007764
- Blanco, J. L., Ambrosioni, J., Garcia, F., Martinez, E., Soriano, A., Mallolas, J., Miro, J. M., & COVID-19 in HIV Investigators. (2020). COVID-19 in patients with HIV: Clinical case series. Lancet HIV, 7(5), e314–e316. https://doi.org/10.1016/S2352-3018(20)30111-9
- Boettiger, D. C., Kerr, S., Ditangco, R., Chaiwarith, R., Li, P. C., Merati, T. P., Thi Thanh Pham, T., Kiertiburanakul, S., Kumarasamy, N., Vonthanak, S., Lee, C. K. C., Van Kinh, N., Pujari, S., Wong, W. W., Kamarulzaman, A., Zhang, F., Yunihastuti, E., Choi, J. Y., Oka, S., Ng, O. T., … TREAT Asia HIV Observational Database. (2016). Tenofovir-based antiretroviral therapy in HBV-HIV coinfection: Results from the TREAT Asia HIV Observational Database. Antiviral Therapy, 21(1), 27–35. https://doi.org/10.3851/IMP2972
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–10. https://doi.org/10.1080/07391102.2020.1758788
- Boulle, A., Davies, M. A., Hussey, H., Ismail, M., Morden, E., Vundle, Z., … Tamuhla, T. (2020). Risk factors for COVID-19 death in a population cohort study from the Western Cape Province, South Africa. Clinical Infectious Diseases. https://doi.org/10.1093/cid/ciaa1198
- Byrd, K. M., Beckwith, C. G., Garland, J. M., Johnson, J. E., Aung, S., Cu-Uvin, S., Farmakiotis, D., Flanigan, T., Gillani, F. S., Macias-Gil, R., Mileno, M., Ramratnam, B., Rybak, N. R., Sanchez, M., Tashima, K., Mylonakis, E., & Kantor, R. (2020). SARS-CoV-2 and HIV coinfection: Clinical experience from Rhode Island, United States. Journal of the International AIDS Society, 23(7), e25573. https://doi.org/10.1002/jia2.25573
- Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., Ruan, L., Song, B., Cai, Y., Wei, M., Li, X., Xia, J., Chen, N., Xiang, J., Yu, T., Bai, T., Xie, X., Zhang, L., Li, C., … Wang, C. (2020). A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. New England Journal of Medicine, 382(19), 1787–1799. https://doi.org/10.1056/NEJMoa2001282
- Chen, Y. W., Yiu, C. B., & Wong, K. Y. (2020). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9, 129. https://doi.org/10.12688/f1000research.22457.2
- Cheng, S. C., Chang, G. G., & Chou, C. Y. (2010). Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophysical Journal, 98(7), 1327–1336. https://doi.org/10.1016/j.bpj.2009.12.4272
- Chien, M., Anderson, T. K., Jockusch, S., Tao, C., Li, X., Kumar, S., Russo, J. J., Kirchdoerfer, R. N., & Ju, J. (2020). Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. Journal of Proteome Research, 19(11), 4690–4697. https://doi.org/10.1021/acs.jproteome.0c00392
- Chu, C. M., Cheng, V. C., Hung, I. F., Wong, M. M., Chan, K. H., Chan, K. S., … Group, H. U. S. S. (2004). Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax, 59(3), 252–256. https://doi.org/10.1136/thorax.2003.012658
- Dandachi, D., Geiger, G., Montgomery, M. W., Karmen-Tuohy, S., Golzy, M., Antar, A., Llibre, J. M., Camazine, M., Díaz-De Santiago, A., Carlucci, P. M., Zacharioudakis, I. M., Rahimian, J., Wanjalla, C. N., Slim, J., Arinze, F., Kratz, A., Jones, J. L., Patel, S. M., Kitchell, E., Francis, A., … HIV-COVID-19 consortium. (2020). Characteristics, comorbidities, and outcomes in a multicenter registry of patients with HIV and coronavirus disease-19. Clinical Infectious Diseases. https://doi.org/10.1093/cid/ciaa1339
- de Wilde, A. H., Jochmans, D., Posthuma, C. C., Zevenhoven-Dobbe, J. C., van Nieuwkoop, S., Bestebroer, T. M., van den Hoogen, B. G., Neyts, J., & Snijder, E. J. (2014). Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrobial Agents and Chemotherapy, 58(8), 4875–4884. https://doi.org/10.1128/AAC.03011-14
- Del Amo, J., Polo, R., Moreno, S., Díaz, A., Martínez, E., Arribas, J. R., Jarrín, I., Hernán, M. A., & The Spanish HIV/COVID-19 Collaboration. (2020). Incidence and severity of COVID-19 in HIV-positive persons receiving antiretroviral therapy: A cohort study. Annals of Internal Medicine, 173(7), 536–541. https://doi.org/10.7326/M20-3689
- Duarte, R. R. R., Copertino, D. C., Jr., Iñiguez, L. P., Marston, J. L., Nixon, D. F., & Powell, T. R. (2020). Repurposing FDA-approved drugs for COVID-19 using a data-driven approach. ChemRxiv. https://doi.org/10.26434/chemrxiv.12148764.v1
- Elfiky, A. A., & Azzam, E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. https://doi.org/10.1080/07391102.2020.1758789
- Elmezayen, A. D., Al-Obaidi, A., Sahin, A. T., & Yelekci, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. https://doi.org/10.1080/07391102.2020.1758791
- Enayatkhani, M., Hasaniazad, M., Faezi, S., Guklani, H., Davoodian, P., Ahmadi, N., … Ahmadi, K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–16. https://doi.org/10.1080/07391102.2020.1756411
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. https://doi.org/10.1080/07391102.2020.1760136
- Ford, N., Vitoria, M., Rangaraj, A., Norris, S. L., Calmy, A., & Doherty, M. (2020). Systematic review of the efficacy and safety of antiretroviral drugs against SARS, MERS or COVID-19: Initial assessment. Journal of the International AIDS Society, 23(4), e25489. https://doi.org/10.1002/jia2.25489
- Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. https://doi.org/10.1021/jm051256o
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.), 368(6492), 779–782. https://doi.org/10.1126/science.abb7498
- Gordon, C. J., Tchesnokov, E. P., Woolner, E., Perry, J. K., Feng, J. Y., Porter, D. P., & Gotte, M. (2020). Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. Journal of Biological Chemistry, 295(20), 6785-6797. https://doi.org/10.1074/jbc.RA120.013679
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. https://doi.org/10.1080/07391102.2020.1751300
- Härter, G., Spinner, C. D., Roider, J., Bickel, M., Krznaric, I., Grunwald, S., Schabaz, F., Gillor, D., Postel, N., Mueller, M. C., Müller, M., Römer, K., Schewe, K., & Hoffmann, C. (2020). COVID-19 in people living with human immunodeficiency virus: A case series of 33 patients. Infection, 48(5), 681–686. https://doi.org/10.1007/s15010-020-01438-z
- Hasan, A., Paray, B. A., Hussain, A., Qadir, F. A., Attar, F., Aziz, F. M., Sharifi, M., Derakhshankhah, H., Rasti, B., Mehrabi, M., Shahpasand, K., Saboury, A. A., & Falahati, M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. https://doi.org/10.1080/07391102.2020.1754293
- Hung, M., Tokarsky, E. J., Lagpacan, L., Zhang, L., Suo, Z., & Lansdon, E. B. (2019). Elucidating molecular interactions of L-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance. Communications Biology, 2, 469. https://doi.org/10.1038/s42003-019-0706-x
- Irwin, J. J., & Shoichet, B. K. (2005). ZINC-a free database of commercially available compounds for virtual screening. Journal of Chemical Information and Modeling, 45(1), 177–182. https://doi.org/10.1021/ci049714+
- Jockusch, S., Tao, C., Li, X., Anderson, T. K., Chien, M., Kumar, S., Russo, J. J., Kirchdoerfer, R. N., & Ju, J. (2020). Triphosphates of the two components in DESCOVY and TRUVADA are inhibitors of the SARS-CoV-2 polymerase. BioRxiv. https://doi.org/10.1101/2020.04.03.022939
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., … Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. https://doi.org/10.1080/07391102.2020.1760137
- Karmen-Tuohy, S., Carlucci, P. M., Zervou, F. N., Zacharioudakis, I. M., Rebick, G., Klein, E., Reich, J., Jones, S., & Rahimian, J. (2020). Outcomes among HIV-positive patients hospitalized with COVID-19. Journal of Acquired Immune Deficiency Syndromes (1999), 85(1), 6–10. https://doi.org/10.1097/QAI.0000000000002423
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. https://doi.org/10.1080/07391102.2020.1751298
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., & Bolton, E. E. (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research, 49(D1), D1388–D1395. https://doi.org/10.1093/nar/gkaa971
- Ma, C., Sacco, M. D., Hurst, B., Townsend, J. A., Hu, Y., Szeto, T., Zhang, X., Tarbet, B., Marty, M. T., Chen, Y., & Wang, J. (2020). Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Research, 30, 678–692. https://doi.org/10.1038/s41422-020-0356-z
- Mayer, K. H., Molina, J.-M., Thompson, M. A., Anderson, P. L., Mounzer, K. C., De Wet, J. J., DeJesus, E., Jessen, H., Grant, R. M., Ruane, P. J., Wong, P., Ebrahimi, R., Zhong, L., Mathias, A., Callebaut, C., Collins, S. E., Das, M., McCallister, S., Brainard, D. M., … Hare, C. B. (2020). Emtricitabine and tenofovir alafenamide vs emtricitabine and tenofovir disoproxil fumarate for HIV pre-exposure prophylaxis (DISCOVER): Primary results from a randomised, double-blind, multicentre, active-controlled, phase 3, non-inferiority trial. Lancet (London, England), 396(10246), 239–254. https://doi.org/10.1016/S0140-6736(20)31065-5
- McKenna, C. E., Kashemirov, B. A., Peterson, L. W., & Goodman, M. F. (2010). Modifications to the dNTP triphosphate moiety: From mechanistic probes for DNA polymerases to antiviral and anti-cancer drug design. Biochimica et Biophysica Acta, 1804(5), 1223–1230. https://doi.org/10.1016/j.bbapap.2010.01.005
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. https://doi.org/10.1080/07391102.2020.1752802
- Oberg, B. (2006). Rational design of polymerase inhibitors as antiviral drugs. Antiviral Research, 71(2-3), 90–95. https://doi.org/10.1016/j.antiviral.2006.05.012
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–10. https://doi.org/10.1080/07391102.2020.1757510
- Park, L. S., Rentsch, C., Sigel, K., Rodriguez-Barradas, M., & Brown, S. T. (2020). COVID-19 in the largest US HIV cohort. Late-breaking poster LBPE023 presented at the AIDS 2020: 23rd International AIDS Conference Virtual. AIDS 2020, 6 – 10 July.
- Recovery-Collaborative-Group. (2020). Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet, 396(10259), 1345-1352. https://doi.org/10.1016/S0140-6736(20)32013-4
- Roos, K., Wu, C., Damm, W., Reboul, M., Stevenson, J. M., Lu, C., Dahlgren, M. K., Mondal, S., Chen, W., Wang, L., Abel, R., Friesner, R. A., & Harder, E. D. (2019). OPLS3e: Extending force field coverage for drug-like small molecules. Journal of Chemical Theory and Computation, 15(3), 1863–1874. https://doi.org/10.1021/acs.jctc.8b01026
- Sanders, J. M., Monogue, M. L., Jodlowski, T. Z., & Cutrell, J. B. (2020). Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA, 323(18), 1824-1836. https://doi.org/10.1001/jama.2020.6019
- Sarma, P., Shekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–9. https://doi.org/10.1080/07391102.2020.1753580
- Shannon, A., Le, N. T.-T., Selisko, B., Eydoux, C., Alvarez, K., Guillemot, J.-C., Decroly, E., Peersen, O., Ferron, F., & Canard, B. (2020). Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Research, 178, 104793. https://doi.org/10.1016/j.antiviral.2020.104793
- Shelley, J. C., Cholleti, A., Frye, L. L., Greenwood, J. R., Timlin, M. R., & Uchimaya, M. (2007). Epik: A software program for pK(a) prediction and protonation state generation for drug-like molecules. Journal of Computer-Aided Molecular Design, 21(12), 681–691. https://doi.org/10.1007/s10822-007-9133-z
- Shivakumar, D., Williams, J., Wu, Y., Damm, W., Shelley, J., & Sherman, W. (2010). Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. Journal of Chemical Theory and Computation, 6(5), 1509–1519. https://doi.org/10.1021/ct900587b
- Sluis-Cremer, N., & Tachedjian, G. (2008). Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Research, 134(1–2), 147–156. https://doi.org/10.1016/j.virusres.2008.01.002
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. https://doi.org/10.1038/s41422-020-0282-0
- Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jin, Y., Fu, S., Gao, L., Cheng, Z., Lu, Q., Hu, Y., Luo, G., Wang, K., Lu, Y., Li, H., Wang, S., Ruan, S., Yang, C., Mei, C., … Wang, C. (2020). Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. The Lancet, 395(10236), 1569–1578. https://doi.org/10.1016/S0140-6736(20)31022-9
- Xu, X., Liu, Y., Weiss, S., Arnold, E., Sarafianos, S. G., & Ding, J. (2003). Molecular model of SARS coronavirus polymerase: Implications for biochemical functions and drug design. Nucleic Acids Research, 31(24), 7117–7130. https://doi.org/10.1093/nar/gkg916
- Yamamoto, N., Yang, R., Yoshinaka, Y., Amari, S., Nakano, T., Cinatl, J., Rabenau, H., Doerr, H. W., Hunsmann, G., Otaka, A., Tamamura, H., Fujii, N., & Yamamoto, N. (2004). HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochemical and Biophysical Research Communications, 318(3), 719–725. https://doi.org/10.1016/j.bbrc.2004.04.083
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science, 368(6489), 409–412. https://doi.org/10.1126/science.abb3405
