ABSTRACT
Circadian rhythm disturbance (CRD) increases the risk of disease, e.g. metabolic syndrome, cardiovascular disease, and cancer. In the present study, we investigated later life adverse health effects triggered by repeated jet lag during gestation. Pregnant mice were subjected to a regular light-dark cycle (CTRL) or to a repeated delay (DEL) or advance (ADV) jet lag protocol. Both DEL and ADV offspring showed reduced weight gain. ADV offspring had an increased circadian period, and an altered response to a jet lag was observed in both DEL and ADV offspring. Analysis of the bones of adult male ADV offspring revealed reduced cortical bone mass and strength. Strikingly, analysis of the heart identified structural abnormalities and impaired heart function. Finally, DNA methylation analysis revealed hypermethylation of miR17-92 cluster and differential methylation within circadian clock genes, which correlated with altered gene expression. We show that developmental CRD affects the circadian system and predisposes to non-communicable disease in adult life.
Introduction
The circadian clock is present in most organisms and generates rhythms in metabolism, physiology, and behavior. Increasing evidence suggests that the circadian clock significantly impacts on health during the life course. Genetic disruption of the circadian clock has been linked to non-communicable disease. Per2 is considered as tumor suppressor gene (Fu et al. Citation2002), as shown by an increased tumor susceptibility of Per2 mutant mice. Also, knockout of Clock leads to features of metabolic syndrome (Turek et al. Citation1995), and Bmal1 knockout mice show features of premature aging and increased tumor incidence (Kondratov et al. Citation2006). Finally, CRY deficiency has been associated with hypertension (Doi et al. Citation2010). Adding to the effect of genetic disruption of the circadian clock on health outcomes, epidemiological and animal studies have associated chronic Circadian Rhythm Disturbance (CRD) in adults (as encountered during shift work and jet lag) with a variety of diseases, including cancer, metabolic syndrome and cardiovascular disease (Barclay et al. Citation2012; Foster and Wulff Citation2005; Haus and Smolensky Citation2013; Roenneberg and Merrow Citation2016). Recently causal evidence was obtained indicating that CRD by chronic jet lag triggers early onset of breast cancer in mice (van Dycke et al. Citation2015).
According to the Developmental Origins of Health and Disease hypothesis (DOHaD), fetal and early-life programming permanently shapes body structure and function. This suggests that environmental factors (e.g. nutritional insults, stress), may (epigenetically) contribute to later life disease (Barker, Citation2007; Godfrey et al. Citation2015; Ladd-Acosta and Fallin Citation2016). As such, gestational CRD is also a candidate for later-life health risk by affecting the epigenetic landscape and predisposing to later-life adverse health effects. Indeed, disruption of the light/dark cycle during pregnancy has been associated with adverse outcomes, such as a higher risk of pre-term birth, low birth weight babies, spontaneous abortion and subfertility (Knutsson Citation2003; Mahoney Citation2010). Recent animal studies have shown that exposure to continuous light during pregnancy is a good model for development of neuropsychiatric disorders and developmental alterations such as cardiovascular dysfunction and skeletal defects in the offspring (Fontanetti et al. Citation2014; Galdames et al. Citation2014; Voiculescu et al. Citation2016). However, under continuous light, the circadian clock is free running, and the question remains open to what extent the circadian system is disturbed by chronic stress, rather than CRD. We, therefore, consider a frequently shifting light-dark schedule (mimicking chronic jet lag) a better model for CRD. This type of protocol was successfully used to induce developmental CRD (Mendez et al. Citation2016; Varcoe et al. Citation2018, Citation2011).
In the present study, we used gestational jet lag in mice as a model for CRD during early development. Pregnant female mice were subjected to one of three experimental conditions (i.e. a regular light-dark cycle, or a repeated phase advanced or phase delayed light-dark cycle). Behavioral and pathological analysis of the jet lag offspring revealed that gestational jet lag affects circadian development and predisposes to disease in adult life. Genome-wide DNA methylation analysis further suggests that the observed changes are mediated by altered methylation of micro-RNA host genes and of circadian clock genes.
Materials and methods
Experimental animals
The current study was performed with C57BL/6J mice. Animals were housed under standard conditions and fed ad libitum. Animals were housed at the Animal Resource Centre (Erasmus University Medical Centre), which operates in compliance with the “Animal Welfare Act” of the Dutch government, using the “Guide for the Care and Use of Laboratory Animals” as its standard. This animal study was approved by DEC Consult, an independent Animal Ethical Committee (Dutch equivalent of the IACUC) under permit numbers EMC3359, in accordance with national and international guidelines.
Experimental setup
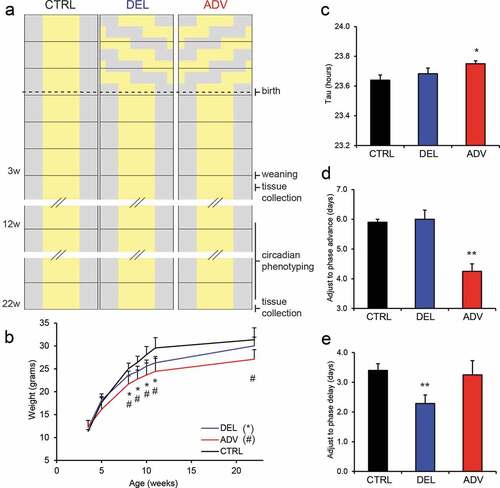
Mating was started under a regular 12:12 hr light-dark cycle, and after determining the presence of a copulatory plug (indicative of successful mating and marking the start of pregnancy), pregnant mice were housed under one of following three experimental conditions: a regular 12:12 hr light-dark-cycle (CTRL), a serial 8 hr delayed light-dark cycle, mimicking a chronic west-bound jet lag (DEL) or a serial 8 hr advanced light-dark cycle, mimicking a chronic east-bound jet lag (ADV). From the moment of delivery (p0), all groups were housed under a regular 12:12 hr light-dark cycle. The jet lag was achieved by advancing or delaying the light period by 8 hr every 3 days, resulting in a total of six shifts. After weaning (p26), the offspring was monitored for growth (body weight) and health status (fur, gait, behavior). This study was focused around the male offspring. From 12 weeks of age, circadian behavior of the male offspring was monitored using a running-wheel set-up (chrono-phenotyping). At 40 weeks of age, the cardiac function of the female offspring of analyzed using micro-computed tomography. Animals were sacrificed at 3 age time points: 4 weeks, 20 weeks and 40 weeks (see ).
Figure 1. Gestational jet lag delays growth and affects circadian performance of the offspring.
Chrono-phenotyping (running-wheel)
At the age of 12 weeks, male offspring was individually housed in cages (30 × 45 cm) equipped with a running wheel (14 cm in diameter) and a sensor to detect wheel revolutions. Animals were maintained in a cycle of 12 hr light (150 lux) and 12 hr darkness (LD) or in continuous darkness (DD), in constant ambient temperature with water and food ad libitum. To determine the capacity to adjust to a jet lag, animals were sequentially exposed to a 6-h phase advanced light-dark cycle and a 6-h phase delayed light-dark cycle (there was a period of 14 days between de two phase shifts). Voluntary wheel running (wheel turns per unit of time) was continuously recorded by an online computer using the ERS program (Groningen University). Activity records were plotted as actograms and the period of locomotor activity was determined by the chi-square method. Unpaired Student’s t-tests were used to make statistical comparisons between the different genotypes.
Microcomputed tomography (µCT) analysis of the femur
Femurs of 20 week-old mice were scanned at a resolution of 9 μm, using a SkyScan 1076 system (SkyScan, Kontich, Belgium). According to guidelines recently published (Bouxsein et al. Citation2010), the following settings were used: X-Ray power and tube current were 40 kV and 250 μA, respectively. Beam hardening was reduced using a 1 mm aluminum filter, exposure time was 2.3 s, and an average of three pictures was taken at each angle with steps of 0.8° to generate final images. Using different software packages from SkyScan (NRecon, CtAn, and Dataviewer), bone microarchitecture parameters were assessed in trabecular and cortical bones of all mice (n = 4–7 for the three groups). The trabecular bone parameters trabecular bone volume fraction (BV/TV), trabecular thickness, trabecular number, trabecular patterning factor (connectivity of trabeculae) and structure model index (0 = plate-like, 3 = rod-like) were determined in the distal metaphysis of the femur (scan area of 1.35 mm from distal growth plate towards middle of femur). In the mid-diaphysis (scan area of 0.45 mm), cortical volume, cortical thickness, endocortical volume, polar moment of inertia (proxy for bone strength) and bone perimeter were analyzed.
Bone mechanical testing (three-point bending)
Femurs were stored in phosphate-buffered saline at −20°C until further use. Before the three-point bending test, femurs were scanned according to the settings mentioned above. The procedure was carried out as previously described in detail (Bouxsein et al. Citation2010). Briefly, femurs were placed in a custom-made three-point bending device, with the lower loading posts 10 mm apart. Mechanical testing was performed, using a Single Column Lloyd LRX System (Lloyd Instruments). Displacement (mm) and force (N) were registered. From the resulting displacement to force graphs ultimate force (N), stiffness (N/mm) and energy (Nmm) were determined as described before (van der Eerden et al. Citation2013).
Bone marrow cultures
Tibial and femoral bone marrow cells were collected using a centrifugation method. From each mouse, 1 femur and 2 tibia were dissected and cut just above the distal growth plate (femur) or at the junction with the fibula (tibiae). Bones were each placed into a 1.5 ml Eppendorf tube containing a handmade plastic adaptor with 250 μl culture medium added. Bones were centrifuged at 1000 g and cell pellets were resuspended in medium containing 50% serum and 10% DMSO and stored in liquid N2 until further use. After thawing, cells were resuspended in 10 ml of α-minimal essential medium (α-MEM; Life Technologies). Next, cells were washed twice with 10 ml of culture medium by centrifugation followed by counting the cells using a Countess cell counter (Thermo Fischer Scientific). Osteoblast cultures were started by seeding 0.6 × 106 (ALP) or 1 × 106 (alizarin) cells per well in 700 μl phenol-red free α-MEM in a 12-well plate, supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Life Technologies), 250 ng/ml amphotericin B (Sigma), 20 mM Hepes, 1.8 mM CaCl2, and 15% (v/v) heat-inactivated FCS (Life Technologies), pH 7.5. Two days after seeding cultures were refreshed with medium supplemented with 50 μM vitamin C (Sigma) and 10 mM β-glycerophosphate (Sigma) twice a week. At days 7 and 21 of culture cells were fixed in 70% ethanol and stained for alkaline phosphatase (ALP) and alizarin red, respectively. For ALP staining, cells were incubated in Tris-HCL (pH 9.5) containing 50 mM MgCl2, 0.6 mg/ml bromo-chloro-indolyl phosphate (Sigma), and 150 μg/ml nitro blue tetrazolium (Sigma) for 20 min and washed with PBS. Alizarin red staining was performed by incubating the cells for 10 min in a saturated alizarin red solution in distilled water (pH 4.2), after which the cells were washed with distilled water. The number of ALP- and alizarin red-positive colonies were counted, using Bioquant software (Bioquant, Nashville, TN, USA).
Osteoclast cultures were started by seeding 1 × 105 cells per well of a 96-well plate. Cells were cultured in the presence of 30 ng/ml recombinant M-CSF (R&D systems) and 20 ng/ml recombinant murine RANKL-TEC (R&D systems) for 6 days and media were refreshed at day 3. At the end of the cultures, cells were washed with PBS, fixed in formalin (10% v/v) and stored at 4°C for tartrate-resistant acid phosphatase (TRAP) staining. To do so, a leucocyte kit (Sigma) was used according to manufacturer’s protocol except for using a higher tartrate concentration of 40 mM to stain specifically for osteoclasts. TRAP-positive multinuclear osteoclasts were counted using ImageJ (https://imagej.nih.gov/ij/). In addition, murine osteoclasts were cultured on OsteoAssay Stripwell plates (Corning) for 6 days and then lysed in distilled water. The hydroxyapatite coating of the OsteoAssay wells was stained with a von Kossa staining. Unstained areas representing actual resorption were quantified, using ImageJ.
Micro-computed tomography (μCT) analysis of the heart
For this study, we used intravenous administration of the iodinated contrast agent eXIA160, which produces strong blood-pool contrast. Micro-CT was done with the Quantum FX MicroCT system to quantitatively assess heart size and function (van Deel et al. Citation2016). The animals received light anesthesia to immobilize them during scanning. Specifically, they were anesthetized with isoflurane (2.5–3% to minimize motion artifacts from respiration and heartbeats). Constant delivery of isoflurane was achieved via a nose cone connected to the scan platform. μCT scans were performed and reconstructed at the Applied Molecular Imaging Erasmus MC facility using the Quantum FX (PerkinElmer). In vivo scans were acquired using a field of view of 20 mm (90 kV/160 mA, 4,5 min). Scans were quantified using Analyze 11.0 software (AnalyzeDirect). Contrast-enhanced blood-pool was quantified by converting original linear attenuation coefficient measurements, by linear transformation, to Hounsfield units (HU). Global thresholding was applied to all scans and determined by visual inspection.
Cardiac histology
Formalin-fixed paraffin embedded heart tissue was serially sectioned into 4-µm slices and stained with Hematoxylin and Eosin (H&E) staining for determination of heart structure. Picrosirius Red dye was used to determine collagen content. Gomorri silver staining was used to identify cell boundaries and determine cardiomyocyte area. Stained slices were scanned using the Nanozoomer (Hamamatsu), and analyzed with Nanozoomer Digital Pathology Imaging software. To determine wall thickness of either the left (LV) of right (RV) ventricle, the mean of 5 measurements along the wall was used per heart. To determine the area of the LV wall, the inner area of the LV was subtracted from the outer area of the LV. To calculate cardiomyocyte area, the mean area of 25 cardiomyocytes per hart was used.
Epigenetic analysis: MeD-Seq
DNA methylation was analyzed in livers of 4 and 20-week old male mice of all three experimental groups. The method used was MeD-seq (Boers et al. Citation2018). In short, DNA from liver samples was extracted using phenol/chloroform (Sigma-Aldrich) extraction. After isopropanol (Sigma-Aldrich) precipitation, DNA was washed with 70% ethanol and dissolved in 20 mM Tris pH8.0. LpnPI, and MspJI (New England Biolabs) digestions were carried out according to manufacturer’s protocol. Reactions contained 1000 ng in a 30 uL volume and digestion took place overnight in the absence of enzyme activators. Digests of genomic DNA with LpnPI results in snippets of 32 bp around the fully methylated recognition site that contains CpG. The 32 bp DNA fragments were prepared for sequencing following manufacturer’s instructions using a Rubicon Genomics Thruplex DNA-SEQ kit. Stem-loop adapters were blunt-end ligated to repaired input DNA and amplified (14 cycles) to include dual-indexed barcodes using a high fidelity polymerase to yield an indexed illumina NGS library. Short DNA fragments were isolated using Pippin system gel purification, the DNA concentration was determined by the Quant-iT™ High-Sensitivity assay (Life technologies; Q33120) and 50 ng ds DNA were prepared using the ThruPlex DNA – seq 96D kit (Rubicon Genomics cat#R400407). Twenty microliter of amplified end product was purified on Pippin HT system with 3% agarose gel cassettes (Sage Science). Multiplexed samples were sequenced on Illumina HiSeq2500 systems for single read 50 base pairs according to manufacturer’s instructions. Dual indexed samples were demultiplexed using bcl2fastq software (Illumina).
MeD-seq data processing
Data processing was carried out using specifically created scripts in Python. Raw fastq files were subjected to illumina adaptor trimming and reads were filtered based on LpnPI restriction site occurrence between 13 and 17 bp from either 5’ or 3’ end of the read. Reads that passed the filter were mapped to mm10 using bowtie2. Mapped reads were used to assign read count scores to each individual LpnPI site in the mm10 genome. Sam and bam files were generated using samtools for visualization. Gene and CpG island annotations were downloaded from ENSEMBL (www.ensembl.org). Genome-wide individual LpnPI site scores were used to generate read count scores for the following annotated regions: TSS (1 kb before and 1 kb after), CpG islands and gene body (1 kb after TSS till TES).
Data analysis was carried out in Python. Correlations between datasets were calculated using the Pearson coefficient after log10 transformation of read counts, outliers (Z-score above 5.0) were excluded and graphs were created using matplotlib. DMR detection was performed between two datasets containing the regions of interest (TSS, gene body or CpG islands) using the Chi-Squared test on read counts. Significance was called by either Bonferroni or FDR. Differently methylated regions were used for unsupervised hierarchical clustering, the Z-score of the read counts per sample was used in heatmaps. Genome-wide detection of DMRs was done using a sliding window. This sliding window was fixed at one specific LpnPI site and added the read counts of 50 LpnPI sites up – and downstream of the initially selected LpnPI site. Using the read counts in this window, significant differences were called using the Chi-squared test with a Bonferroni correction. By sliding this window across the genome, all neighboring LpnPI sites that were called differentially methylated were summarized and reported as a DMR, overlap with TSS, gene bodies and CpG Islands were also reported.
Quantification of heart structural parameters
Heart structural parameters were measured using the Nanozoomer Digital Pathology Imaging Software, as described here. For each heart we selected equivalent sections, to allow comparison. Area of the LV and RV was calculated as the mean of three independent measurements per heart. Thickness of LV wall and RV wall were calculated as the mean of at least four measurements per heart, spread along the whole wall. Cardiomyocyte area was calculated as the mean of 50 cardiomyocytes per heart, in total 200 cardiomyocytes per group.
Statistical analysis
Statistical tests were performed using a two-tailed T-test when comparing to experimental groups. Statistical analysis regarding the DNA methylation data is described above.
Results
A jet lag protocol for circadian rhythm disturbance during gestation
We investigated the impact of CRD during development on later life health of the offspring, using repeated gestational jet lag as a model. Pregnant female mice were subjected to a chronic jet lag protocol, composed of an 8 hr delayed (DEL) or advanced (ADV) light-dark cycle, repeated every 3 days (). As a control, pregnant mice were kept under a regular light-dark cycle (CTRL). Offspring was born and raised under normal light-dark conditions (). Although it was previously shown that jet lag impinges on pregnancy outcome in the mouse (Summa et al. Citation2012), we did not observe significant changes in the percentage of successful pregnancies, litter size, or male/female sex ratio in offspring (Figure S1).
Gestational jet lag impairs growth and affects circadian behavior
To minimize phenotypic differences resulting from hormonal variation, as occurs in female mice as a consequence of the estrus cycle, this study focused on male offspring. At weaning (26 days afterbirth), body weight of DEL and ADV males was similar to that of CTRL males (). Interestingly, subsequent weight measurements revealed that DEL and ADV males gained weight slower than CTRL males, resulting in a lifelong weight reduction in the ADV group ().
Repeated jet lag causes the circadian clock of the pregnant mouse to continuously adjust, affecting food intake and metabolism. Maternal control of the fetal clock occurs via metabolic cues and is therefore likely affected by the repeated jet lag (Canaple et al. Citation2018; Ohta et al. Citation2008), which accordingly could impinge on the development of the circadian system. Starting at the age of 12 weeks, the circadian behavior of the male offspring was assessed by monitoring voluntary wheel running activity under various light conditions (representative actograms are shown in Figure S1D). As shown in , ADV males had a significantly longer (p = 0.03) free-running period (τ) in constant dark conditions (τ = 23.75 ± 0.06 h) than CTRL males (τ = 23.64 ± 0.09 h), while the free-running period of DEL males (τ = 23.68 ± 0.08 h) was not significantly altered (p = 0.3). In order to determine the capacity to recover from a jet lag, male offspring were sequentially subjected to a 6-h phase advance and a 6-h phase delay and allowed to re-entrain. Adjustment to a phase advance is known to require more days than adjustment to a phase delay (Davidson et al. Citation2006; Gundel and Wegmann Citation1987). In line with the literature, CTRL offspring adjusted slower to a phased advance (5.9 ± 0.3 days; mean ± SD) than to a phase delay (3.4 ± 0.7 days) (,). Surprisingly, the speed at which jet lag offspring readjusted to the new light-dark cycle was related to the direction of the gestational jet lag. As shown in , ADV males adjusted to a phase advance in 4.3 ± 0.5 days, which is significantly faster (p = 0.004) than observed for CTRL males, while adjustment to a phase delay was not affected. Oppositely, DEL males took 2.3 ± 0.8 days to adjust to a phase delay, significantly shorter (p = 0.009) than the time needed by the CTRL males (). These results indicate that the gestational jet lag protocol primed the offspring to more rapidly adjust to a phase shift in the same direction as the jet lag experienced in utero. Taken together, these data show that developmental CRD impairs postnatal growth and affects circadian behavior.
Chronic gestational jet lag, bone structure, and strength
Next, we analyzed the impact of gestational jet lag on bone structure of the offspring, using micro-computed tomography (µCT). Regarding the trabecular bone, no significant differences were observed between the jet lag offspring the CTRL group ( and S2). Interestingly, cortical bone size in ADV males was reduced compared to the CRTL offspring, since both perimeter and endocortical volume were significantly lower, whilst cortical volume and thickness were unaltered (, , and S2). The moment of inertia was also significantly reduced in the ADV males, indicative for compromised bone strength (). Although not significant, energy to fracture appeared to be affected in the ADV males compared to the CTRL males (). We did not observe differences in cortical bone measures between the DEL and CTRL males (, , and S2). Our data suggest that chronic gestational exposure to an advance jet lag protocol (ADV) reduces cortical bone size.
Figure 2. Gestational jet lag affects bone structure and strength.
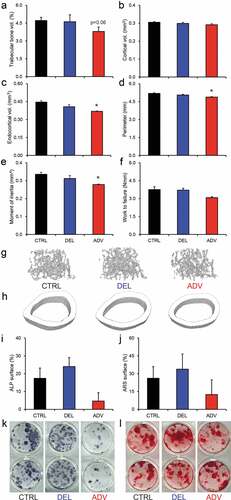
Defects in bone mineralization and structure can be due to an altered osteoblast and/or osteoclast function. To investigate this, bone marrow-derived osteoclast and osteoblast cultures were made from CTRL, DEL and ADV mice. We did not observe any notable differences in osteoclast number or osteoclast-mediated bone resorption, which was assessed by quantifying the resorbed areas in von Kossa-stained Osteoassay plates (Figure S2). On the other hand, osteoblast differentiation from ADV mouse bone marrow appeared to be hampered, as demonstrated by reduced surface percentage of alkaline phosphatase positive colonies (,) and to a lesser extent that of alizarin red positive colonies (,). These data suggest that reduced osteoblast function underlies the reduced cortical bone size and trabecular bone mass seen in ADV males.
Chronic gestational jet lag affects heart morphology and function
Gross analysis of the hearts of DEL and ADV males did not reveal major differences in mass (Figure S3), the weight of jet lag offspring hearts was similar to that of CTRL hearts. However, histological analysis did reveal striking differences in morphology. As shown in and S3B, the left ventricle (LV) cavity of male offspring hearts was clearly enlarged in ADV males, and the LV wall was thinner. Concerning the right ventricle (RV), the wall was thinner in both DEL and ADV males. We quantified structural parameters of both LV and RV ().
Figure 3. Impaired cardiac structure and function in the jet lag offspring.
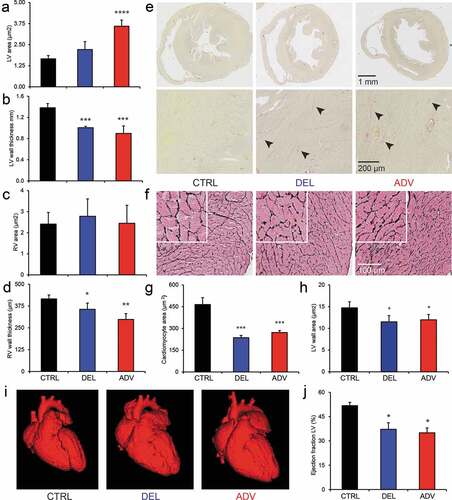
To further analyze the structure of the heart tissue, heart sections were stained with Picrosirius red and Gomorri silver. The Picrosirius red staining procedure allows detection of collagen deposition, indicative of fibrosis, which appears as a pink/red color. In the hearts of 20-week old CTRL males, we did not detect signs of fibrosis, as indicated by the lack of staining (, left panel). However, both DEL and ADV males showed a clear Picrosirius red staining (, middle and right panels). Collagen deposits are indicated by arrowheads in . In DEL and ADV groups, all hearts showed positive Picrosirius red staining, while this was not detected in the CTRL group.
In order to determine cardiomyocyte area, we stained heart slices with Gomorri silver. With this staining method, the cell membrane is clearly visible, making it possible to accurately measure cell area. As shown in , the myocardial tissue structure is altered in both DEL and ADV offspring, and the cardiomyocytes are smaller (). Because the cross-sectional area of the LV wall is smaller in DEL and ADV males than in the CTRL males (), we conclude that there is no myocardial hyperplasia (more cells) in the jet lag offspring.
The altered heart morphology observed in DEL and ADV males is quite severe and suggestive of functional cardiac impairment. Heart function can be studied in sedated mice using micro-computed tomography (μCT). Although male offspring had been sacrificed for above-mentioned studies, female offspring was still available, allowing us to study heart function at 40 weeks of age. The LV was enlarged in both DEL and ADV females (), and the ejection fraction was reduced by 30% in both DEL and ADV offspring (). The functional data obtained indicated reduced cardiac output. Structural analysis of the female offspring hearts (Figure S3) indicate a weaker and distended heart muscle, leading to an enlargement of the heart cavities (LV and RV), as observed in dilated cardiomyopathy. The effect observed was stronger in the ADV group, which is in line with the knowledge that a phase-advance jet lag is more disruptive than a phase-delay jet lag.
The DNA methylation epigenetic landscape is disturbed in gestational jet lag offspring
According to the DOHaD theory, adverse conditions during early development induce epigenetic alterations which are at the basis of an increased risk of non-communicable disease (Ladd-Acosta and Fallin Citation2016). In order to determine whether epigenetic alterations could be at the basis of the circadian and developmental defects observed in the jet lag offspring, we performed genome-wide DNA methylation analysis using the MeD-seq method (Boers et al. Citation2018). This method allows the identification of alterations in the epigenetic landscape, which can be related to altered gene expression. Considering that the jet lag protocol affects maternal feeding and timing of maternal metabolic cues (Canaple et al. Citation2018; Ohta et al. Citation2008), we chose to analyze liver tissue from 4-week old pups. The heat map in shows DNA methylation levels of differentially methylated gene promotors between CTRL, DEL and ADV groups, and was generated using hierarchical clustering on read counts from a region of 2 Kb around the transcription start site (TSS) after z-score normalization. Interestingly, we observed major differences in DNA (promotor) methylation between the control and jet lag offspring. As highlighted on the right side of , there is a cluster of approximately 500 gene promoters which are hypermethylated in the jet lag offspring. We performed gene ontology analysis using this group of differentially methylated genes and identified biological processes which are affected in the jet lag offspring (). A feature that all these processes have in common is that they are regulated by micro-RNA cluster miR17-92 (Gu et al. Citation2017; Mendell Citation2008; Mohan et al. Citation2015), which is hypermethylated in the DEL and ADV groups (). A graphic representation of the methylation levels in miR17 is shown in Figure S4A.
Figure 4. Differential DNA methylation between the control and gestational jet lag offspring.
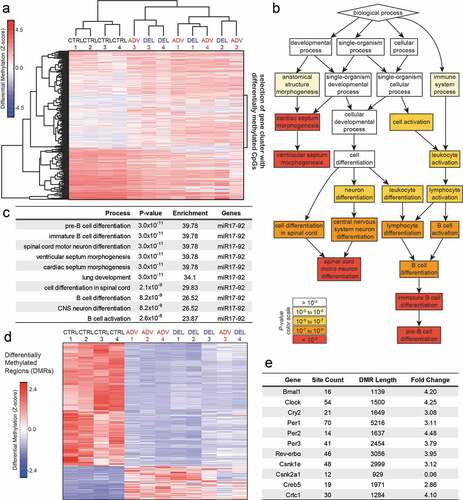
Next step was to further analyze possible differential DNA methylation outside of gene promoters. To this end, we applied a genome-wide scanning window to detect Differentially Methylated Regions (DMRs), which were then used to perform hierarchical clustering (). In order to determine whether the molecular circadian oscillator could be affected by DNA methylation changes, we next searched for overlap between DMRs and circadian clock gene loci. Interestingly, although circadian clock gene promoters were not identified as differentially methylated, most core clock genes overlap with DMRs in their gene bodies (). Furthermore, Creb5 and Crtc1, genes encoding proteins which are involved in circadian clock synchronization (Koyanagi et al. Citation2011; Sakamoto et al. Citation2013), are also located in DMRs (). Most interestingly, Casein kinase 2 (CK2α) is among the genes with the highest level of differential methylation (methylation is about 20 times lower in the control) (). CK2 was recently shown to regulate CLOCK/BMAL1 activity in a CRY dependent manner (Tamaru et al. Citation2015). Importantly, mouse studies have shown that altered expression of CK2 causes developmental abnormalities, among which cardiac malformation and skeletal defects (Bragdon et al. Citation2011; Hauck et al. Citation2008; Landesman-Bollag et al. Citation2011; Lou et al. Citation2008). As an example, graphic representations of the methylation levels in Creb5, Crtc1 and CK2α are shown in Figure S4 (B-D). The differential methylation observed suggests that expression of the affected genes could be altered. To verify this possibility, we have performed gene expression analysis by quantitative RT-PCR. As shown in , the differential gene expression observed between the control and jet lag offspring supports the changes in DNA methylation. Importantly, there is a significant change in the expression levels of the clock genes Bmal1, Clock and Cry2, and of Creb5.
Figure 5. Differential gene expression between the control and gestational jet lag offspring.
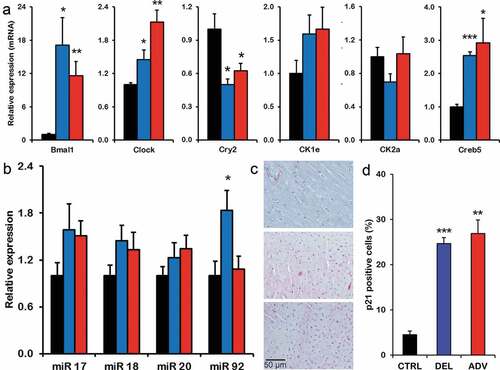
The expression levels of miR17-92 cluster in the liver are only marginally affected (). It has been reported that deletion of the miR17-92 cluster causes a ventricular septum defect (Ventura et al. Citation2008). To investigate whether expression levels of miR17-92 were altered in heart tissue during development and could contribute to the pathology observed, we quantified expression of one of the downstream targets, P21, in the hearts of the offspring. Importantly, we observe that the percentage of P21 positive cells in the hearts of the jet lag offspring is increased in comparison to the control group ().
Our data show that gestational jet lag induces alterations in DNA methylation and gene expression, which underlie the skeletal and cardiac defects observed in the jet lag offspring, indicating that gestational CRD is a DOHaD factor.
Discussion
In the present study, we analyzed the short- and long-term effects of gestational CRD, using chronic gestational jet lag as an experimental model. Recent animal studies have shown that a repeated jet lag during pregnancy is a good model for development of metabolic disorders and neuropsychiatric disorders in the offspring (Mendez et al. Citation2016; Varcoe et al. Citation2011; Zhang et al. Citation2017). However, to our knowledge, this is the first study showing a pathological phenotype as a result of developmental jet lag (e.g. altered heart structure and function, altered bone thickness and strength). (Mendez et al. Citation2016) show altered hormone levels and gene expression, but do not show disease pathology. The current study identifies heart and bone pathological features as a consequence of gestational jet lag and provides mechanistic evidence for DOHaD. Our data show that offspring of mice exposed to repeated jet lag during pregnancy have structural alterations of bone and heart tissue, and have reduced cardiac function. We identify DNA methylation changes which correlate with altered gene expression, and which underlie the pathology observed. In particular, the differential gene expression observed for the clock genes Bmal1, Clock and Cry2 indicates a disturbance in the circadian oscillator.
We undoubtedly show that repeated jet lag during pregnancy is a valid model for developmental CRD, inducing cardiac and skeletal structural alterations. Notably, both DEL and ADV offspring showed reduced weight gain, which resulted in a life-long lower body weight in the ADV group. Adult ADV male offspring displayed thinner and weaker bones, along with altered cardiac structure and function. Interestingly, similar pathological features are observed in BMAL1 deficient mice (Lefta et al. Citation2012). In addition, ADV animals showed an increased circadian period, indicative for a more evening-like chronotype, which in humans has been associated with a higher incidence of non-communicable disease, such as obesity, metabolic syndrome and diabetes type II (Merikanto et al. Citation2013; Yu et al. Citation2015). Phase advances are known to be more deleterious than phase delays (Davidson et al. Citation2006), and this could explain the differences observed between DEL and ADV offspring.
A recent study has shown that chronic exposure of adult mice to continuous light negatively affects bone structure and muscle performance and induces a pro-inflammatory state (Lucassen et al. Citation2016). Yet, the latter effect was reversible as health conditions rapidly improve after re-exposing animals to a robust light-dark regime. In our study, we have identified life-long effects of gestational CRD, which is in line with the DOHaD theory. Importantly, we identified epigenetic alterations as a consequence of gestational CRD which support the phenotypes observed. The promoter region of miR17-92, required for heart and bone development (Gu et al. Citation2017; Mohan et al. Citation2015), was hypermethylated in the jet lag offspring, and increased expression of one of the targets of this miR cluster, P21, was increased in the hearts of the jet lag offspring. Additionally, DMRs were identified within circadian clock genes and genes involved in clock resetting, resulting in altered gene expression. Such alterations were likely induced by the gestational jet lag and are at the basis of the observed phenotypes. We have provided evidence that gestational CRD is a DOHaD-related risk factor that affects the epigenetic landscape and predisposes to later-life adverse health effects, such as cardiovascular disease and skeletal defects.
Author contributions
I.C. carried out all animal experiments and ex vivo cardiac measurements.
M.S.K. and B.v.d.E carried out the bone-related analyses.
Y.R., I.v.d.P and J.E performed the in vivo cardiac analysis.
R.B, J.B, and J.G performed the epigenetic analysis.
I.C. and A.A.S. conducted the qPCR analysis.
I.C., I.R., and G.T.J.v.d.H. conceived the work.
M.V. and I.R. provided clinical background knowledge.
I.C. and G.T.J.v.d.H. coordinated the working team, prepared the figures, and wrote the manuscript with contributions from all authors.
All authors discussed the results and commented on the manuscript.
Supplemental Material
Download PDF (997.2 KB)Disclosure Statement
The authors declare they have no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM, Lehnert H, Oster H. 2012. Circadian desynchrony promotes metabolic disruption in a mouse model of shift work. PLoS One. 7:e37150.
- Barker DJ. 2007. The origins of the developmental origins theory. J Intern Med. 261:412–417.
- Boers R, Boers J, de Hoon B, Kockx C, Ozgur Z, Molijn A, van IJcken W, Laven J, Gribnau J. 2018. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 28:88–99.
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 25:1468–1486.
- Bragdon B, Thinakaran S, Moseychuk O, Gurski L, Bonor J, Price C, Wang L, Beamer WG, Nohe A. 2011. Casein kinase 2 regulates in vivo bone formation through its interaction with bone morphogenetic protein receptor type Ia. Bone. 49:944–954.
- Canaple L, Gréchez-Cassiau A, Delaunay F, Dkhissi-Benyahya O, Samarut J. 2018. Maternal eating behavior is a major synchronizer of fetal and postnatal peripheral clocks in mice. Cell Mol Life Sci. 75:3991–4005.
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. 2006. Chronic jet lag increases mortality in aged mice. Curr Biol. 16:R914–R916.
- Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, Haraguchi S, Emoto N, Okuno Y, Tsujimoto G, Kanematsu A, et al. 2010. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med. 16:67–74.
- Fontanetti PA, Nervegna MT, Vermouth NT, Mandalunis PM. 2014. Prenatal exposure to continuous constant light alters endochondral ossification of the tibiae of rat pups. Cells Tissues Organs. 200:278–286.
- Foster RG, Wulff K. 2005. The rhythm of rest and excess. Nat Rev Neurosci. 6:407–414.
- Fu L, Pelicano H, Liu J, Huang P, Lee C. 2002. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 111:41–50.
- Galdames HA, Torres-Farfan C, Spichiger C, Mendez N, Abarzua-Catalan L, Alonso-Vazquez P, Richter HG. 2014. Impact of gestational chronodisruption on fetal cardiac genomics. J Mol Cell Cardiol. 66:1–11.
- Godfrey KM, Costello PM, Lillycrop KA. 2015. The developmental environment, epigenetic biomarkers and long-term health. J Dev Orig Health Dis. 6:399–406.
- Gu H, Liu Z, Zhou L. 2017. Roles of miR-17-92 cluster in cardiovascular development and common diseases. Biomed Res Int. 2017:9102909.
- Gundel A, Wegmann HM. 1987. Resynchronization of the circadian system following a 9-hr advance or a delay zeitgeber shift: real flights and simulations by a van-der-Pol oscillator. Prog Clin Biol Res. 227B:391–401.
- Hauck L, Harms C, An J, Rohne J, Gertz K, Dietz R, Endres M, von Harsdorf R. 2008. Protein kinase CK2 links extracellular growth factor signaling with the control of p27(Kip1) stability in the heart. Nat Med. 14:315–324.
- Haus EL, Smolensky MH. 2013. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med Rev. 17:273–284.
- Knutsson A. 2003. Health disorders of shift workers. Occup Med (Lond). 53:103–108.
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. 2006. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 20:1868–1873.
- Koyanagi S, Hamdan AM, Horiguchi M, Kusunose N, Okamoto A, Matsunaga N, Ohdo S. 2011. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J Biol Chem. 286:32416–32423.
- Ladd-Acosta C, Fallin MD. 2016. The role of epigenetics in genetic and environmental epidemiology. Epigenomics. 8:271–283.
- Landesman-Bollag E, Belkina A, Hovey B, Connors E, Cox C, Seldin DC. 2011. Developmental and growth defects in mice with combined deficiency of CK2 catalytic genes. Mol Cell Biochem. 356:227–231.
- Lefta M, Campbell KS, Feng HZ, Jin JP, Esser KA. 2012. Development of dilated cardiomyopathy in Bmal1-deficient mice. Am J Physiol Heart Circ Physiol. 303:H475–H485.
- Lou DY, Dominguez I, Toselli P, Landesman-Bollag E, O’Brien C, Seldin DC. 2008. The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol Cell Biol. 28:131–139.
- Lucassen EA, Coomans CP, van Putten M, de Kreij SR, van Genugten JH, Sutorius RP, de Rooij KE, van der Velde M, Verhoeve SL, Smit JW, et al. 2016. Environmental 24-hr cycles are essential for health. Curr Biol. 26:1843–1853.
- Mahoney MM. 2010. Shift work, jet lag, and female reproduction. Int J Endocrinol. 2010:813764.
- Mendell JT. 2008. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 133:217–222.
- Mendez N, Halabi D, Spichiger C, Salazar ER, Vergara K, Alonso-Vasquez P, Carmona P, Sarmiento JM, Richter HG, Seron-Ferre M, et al. 2016. Gestational chronodisruption impairs circadian physiology in rat male offspring, increasing the risk of chronic disease. Endocrinology. 157:4654–4668.
- Merikanto I, Lahti T, Puolijoki H, Vanhala M, Peltonen M, Laatikainen T, Vartiainen E, Salomaa V, Kronholm E, Partonen T. 2013. Associations of chronotype and sleep with cardiovascular diseases and type 2 diabetes. Chronobiol Int. 30:470–477.
- Mohan S, Wergedal JE, Das S, Kesavan C. 2015. Conditional disruption of miR17-92 cluster in collagen type I-producing osteoblasts results in reduced periosteal bone formation and bone anabolic response to exercise. Physiol Genomics. 47:33–43.
- Ohta H, Xu S, Moriya T, Iigo M, Watanabe T, Nakahata N, Chisaka H, Hanita T, Matsuda T, Ohura T, et al. 2008. Maternal feeding controls fetal biological clock. PLoS One. 3:e2601.
- Roenneberg T, Merrow M. 2016. The circadian clock and human health. Curr Biol. 26:R432–R443.
- Sakamoto K, Norona FE, Alzate-Correa D, Scarberry D, Hoyt KR, Obrietan K. 2013. Clock and light regulation of the CREB coactivator CRTC1 in the suprachiasmatic circadian clock. J Neurosci. 33:9021–9907.
- Summa KC, Vitaterna MH, Turek FW. 2012. Environmental perturbation of the circadian clock disrupts pregnancy in the mouse. PLoS One. 7:e37668.
- Tamaru T, Hattori M, Honda K, Nakahata Y, Sassone-Corsi P, van der Horst GT, Ozawa T, Takamatsu K. 2015. CRY drives cyclic CK2-mediated BMAL1 phosphorylation to control the mammalian circadian clock. PLoS Biol. 13:1002293.
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. 1995. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 308:1043–1045.
- van Deel E, Ridwan Y, van Vliet JN, Belenkov S, Essers J. 2016. In vivo quantitative assessment of myocardial structure, function, perfusion and viability using cardiac micro-computed tomography. J Vis Exp. 16:53603.
- van der Eerden BC, Oei L, Roschger P, Fratzl-Zelman N, Hoenderop JG, van Schoor NM, Pettersson-Kymmer U, Schreuders-Koedam M, Uitterlinden AG, Hofman A, et al. 2013. TRPV4 deficiency causes sexual dimorphism in bone metabolism and osteoporotic fracture risk. Bone. 57:443–454.
- van Dycke KC, Rodenburg W, van Oostrom CT, van Kerkhof LW, Pennings JL, Roenneberg T, van Steeg H, van der Horst GTJ. 2015. Chronically alternating light cycles increase breast cancer risk in mice. Curr Biol. 25:1932–1937.
- Varcoe TJ, Gatford KL, Kennaway DJ. 2018. Maternal circadian rhythms and the programming of adult health and disease. Am J Physiol Regul Integr Comp Physiol. 314:R231–R241.
- Varcoe TJ, Wight N, Voultsios A, Salkeld MD, Kennaway DJ. 2011. Chronic phase shifts of the photoperiod throughout pregnancy programs glucose intolerance and insulin resistance in the rat. PLoS One. 6:e18504.
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 132:875–886.
- Voiculescu SE, Le Duc D, Roșca AE, Zeca V, Chiţimuș DM, Arsene AL, Drăgoi CM, Nicolae AC, Zăgrean L, Schöneberg T, et al. 2016. Behavioral and molecular effects of prenatal continuous light exposure in the adult rat. Brain Res. 1650:51–59.
- Yu JH, Yun CH, Ahn JH, Suh S, Cho HJ, Lee SK, Yoo HJ, Seo JA, Kim SG, Choi KM, et al. 2015. Evening chronotype is associated with metabolic disorders and body composition in middle-aged adults. J Clin Endocrinol Metab. 100:1494–1502.
- Zhang P, Li G, Li H, Tan X, Cheng HM. 2017. Environmental perturbation of the circadian clock during pregnancy leads to transgenerational mood disorder-like behaviors in mice. Sci Rep. 7:12641.
