Abstract
Background: Cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) genes are frequently altered in acute lymphoblastic leukaemia (ALL) patients. The aim of this meta-analysis was to comprehensively assess the prognostic value of CDKN2A/B deletions in ALL patients.
Methods: Systematic literature review was conducted in PubMed, Embase and Cochrane databases up to July 2018. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated with fixed-effects or random-effects models.
Results: A total of thirteen studies including 2857 patients were eligible for this meta-analysis. Combined HRs suggested that CDKN2A/B deletions were poor prognostic factors for both overall survival (OS) (HR = 2.15, 95% CI 1.82–2.54) and event-free survival (EFS)/disease-free survival (DFS)/relapse-free survival (RFS) (HR = 2.16, 95% CI 1.73–2.69). The adverse impact remained significant in both adult and paediatric ALL patients, and also in subgroups by ethnicity, ALL type, detection method of CDKN2A/B deletions, statistical method and endpoint.
Conclusions: Our findings suggested that CDKN2A/B deletions were associated with poor prognosis independently in both adult and childhood ALL patients. Inclusion of CDKN2A/B status may further improve the risk stratification of ALL patients.
Although numerous studies have explored the prognostic significance of cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) deletions in acute lymphoblastic leukaemia (ALL) patients, the results remain conflicting.
In this meta-analysis, we found that CDKN2A/B deletions were independent poor prognostic markers for both adult and paediatric ALL patients.
Our findings justify the inclusion of CDKN2A/B status in the risk stratification of ALL patients.
Key Messages
Introduction
Acute lymphoblastic leukaemia (ALL) is a heterogeneous hematologic malignancy in terms of genetic background and clinical manifestation and prognosis, featured by the overproduction of clonal immature lymphoblasts [Citation1]. It occurs in about 20% of adult leukaemias and 75–80% of childhood leukaemias [Citation2]. In the past decades, remarkable progress in treatment strategies has improved the management of ALL, but disease resistance and relapse is still a problem for a substantial proportion of patients [Citation3,Citation4]. Thus, accurate risk stratification is crucial for the optimization of treatment regimens for individual patients. Routinely used poor prognostic factors include hypodiploidy, MLL rearrangement, t(9;22)(q34;q11.2): BCR-ABL1, Ph-like ALL, complex karyotype, and intrachromosomal amplification of chromosome 21 (iAMP21) [Citation3]. But these factors cannot clarify all the high-risk patients. Hence, additional genetic insights are being required for more precise prediction of the disease outcome. In the past decades, genome-wide analyses have brought breakthroughs in the genetics of ALL and provided a plenty of potential prognostic markers [Citation5].
One genetic marker of potential prognostic value is the cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) genes at chromosome arm 9p21 [Citation6]. These are two tumour suppressor genes lying adjacent to each other, which encode three proteins: p16INK4A (inhibitor of CDK4) and p14ARF (alternative reading frame) by CDKN2A and p15INK4B by CDKN2B. p16INK4A and p15INK4B are CDK inhibitors critical for cell cycle regulation, whereas p14ARF functions as a stabilizer of the tumour suppressor protein p53 via antagonizing p53 ubiquitin ligase MDM2 [Citation7]. Inactivation of the genes can lead to the malignant proliferation of tumour cells, and plays an important role in the pathogenesis and drug resistance of ALL. In vitro and in vivo studies revealed that CDKN2A/B inactivation could endow pre-B cells with increased self-renewal capacity and guarantee the acquisition of full leukemic potential, as well as contribute to resistance to tyrosine kinase inhibitors (TKIs) therapy in BCR-ABL1 positive ALL mouse models [Citation8–Citation11]. Deletions of CDKN2A/B occur frequently in both childhood and adult ALL, with an incidence of 30–50%. The prognostic value of CDKN2A/B deletions has been widely investigated in numerous studies, but the results remain controversial [Citation12–18]. Hence, we performed a meta-analysis to comprehensively assess the prognostic significance of CDKN2A/B deletions in ALL patients. To the best of our knowledge, no meta-analysis has been published previously evaluating the association between CDKN2A/B deletions and ALL outcome.
Methods
The present meta-analysis was reported according to the PRISMA statement [Citation19].
Literature search and study selection
We performed a comprehensive literature search of PubMed, Embase, and Cochrane databases up to July 2018 with the following terms: “acute lymphoblastic leukaemia” or “Precursor Cell Lymphoblastic Leukaemia-Lymphoma [Mesh]”, “CDKN2A/B” or “CDKN2A” or “Genes, p16“[Mesh] or “CDKN2B” or “Cyclin-Dependent Kinase Inhibitor p15“[Mesh]. Reference lists were also screened to identify eligible studies.
The inclusion criteria were as follows: (1) prospective or retrospective cohort studies assessing the prognostic value of CDKN2A/B deletions in ALL patients; (2) CDKN2A/B status was classified into “CDKN2A/B-normal” and “CDKN2A/B-deletion”; (3) the outcome was time-dependent endpoints, disease-free survival (DFS)/event-free survival (EFS)/relapse-free survival (RFS) and overall survival (OS); (4) hazard ratios (HRs) with corresponding 95% confidence intervals (CIs) either had to be reported or able to be calculated from the data presented. Studies were excluded if they were: (1) case reports, reviews, meta-analysis, comments, meeting abstracts and non-human studies; (2) without sufficient data to calculate HRs and 95% CIs. If overlapping cohorts were reported in several studies, only the latest and most intact version was included. Two reviewers independently screened the titles and abstracts of all identified studies to assess their eligibility for inclusion.
Data extraction and quality assessment
Two reviewers independently extracted the data from each eligible study using a piloted form including the following information: first author, publication year, study region, recruitment time, sample size, sex distribution, age range, ethnicity, ALL type, treatment protocol, follow-up time, CDKN2A/B detection method, the number of cases with CDKN2A/B deletions, statistical method, outcomes and HRs with 95% CIs. A third reviewer was consulted if any discrepancy occurred.
The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analysis (available at http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp) was applied to assess the quality of included studies by two reviewers independently. The NOS is composed of nine items classified into three dimensions including selection (four items), comparability (two items), and outcome (three items). Based on the NOS, the quality of studies can be classified into high-quality (scores 7–9), intermediate-quality (scores 4–6) and low-quality (scores 1–3).
Statistical analysis
We pooled the HRs and 95% CIs for OS and DFS/EFS/RFS. If both univariate and multivariate results were reported, we utilized the latter. If HRs and 95% CIs were not explicitly provided, we estimated them from the Kaplan–Meier survival curves using the methods described by Tierney et al. [Citation20]. In addition, subgroup analyses were preformed according to age group, ethnicity, ALL type, detection method of CDKN2A/B deletions, statistical method and endpoint.
Heterogeneity among studies was assessed using χ2-based Q-tests and I2 tests. Random-effects model was used to calculate the pooled HRs and 95% CIs if significant heterogeneity existed (p < .10 or I2 > 50%); otherwise, fixed-effects model was applied.
Publication bias was evaluated by Begg's and Egger's tests. A single study’s influence on pooled HRs was analyzed with sensitivity analysis by sequentially omitting one study at a time. All statistical analyses were conducted using Stata version 12.0 software (Stata Corp., College Station, TX).
Results
Study selection and characteristics
The literature screening process is shown as a flow diagram in . Finally, 13 studies with a total number of 2857 patients were included in this meta-analysis [Citation14–18,Citation21–28]. These studies were published between 2005 and 2018 and the sample size ranged from 54 to 864 with a median of 81. The details of included studies are shown in . These studies were conducted in 10 countries (USA, UK, Germany, Italy, Netherlands, Finland, Spain, Lithuania, China and India). Among these studies, eight enrolled adult patients, four included paediatric patients, and one covered both adults and children. Three studies included unselected ALL patients, seven studies of B-ALL, one study of Ph-negative ALL and two studies of Ph-positive ALL.
Table 1. Characteristics of studies included in the Meta-analysis.
Table 1. Continued.
CDKN2A/B deletions was detected by different methods in these studies, including multiplex ligation-dependent probe amplification (MLPA), single nucleotide polymorphism (SNP) arrays, comparative genomic hybridization (CGH) arrays and interphase fluorescence in situ hybridization (I-FISH), among which MLPA was most commonly used. CDKN2A/B deletion rates in individual studies ranged from 22.1 to 48.6%.
The NOS score of included studies ranged from 6 to 9 with a median of 7, indicating acceptable quality of these studies.
Relationship between CDKN2A/B deletions and OS
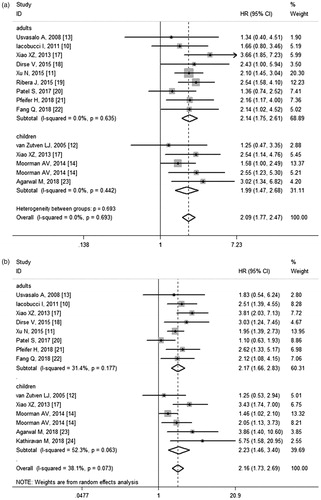
Twelve studies involving 2782 subjects assessed the influence of CDKN2A/B deletions on OS. The pooled HR for OS was 2.15 (95% CI 1.82–2.54) (), suggesting the adverse impact of CDKN2A/B deletions. The test for heterogeneity revealed no significant heterogeneity (p = .785, I2 = 0.0%), so fixed-effects model was employed. Further subgroup analyses were performed according to age group, ethnicity, ALL type, CDKN2A/B detection method, and uni/multivariate analysis (). The negative impact retained for all stratified analyses, indicating the robustness of the relevance.
Figure 2. (a) Meta-analysis of the association between CDKN2A/B deletions and OS in ALL patients. (b) Meta-analysis of the association between CDKN2A/B deletions and EFS/DFS/RFS in ALL patients.
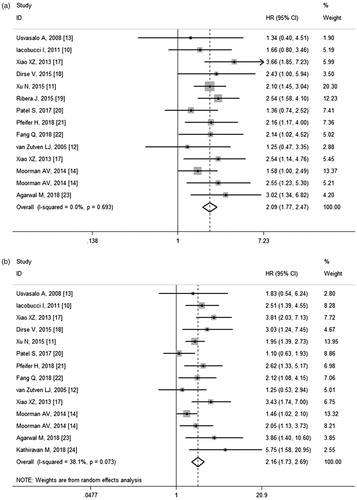
Table 2. Subgroup analyses of CDKN2A/B deletions for prognosis of ALL.
In subgroup analysis by age group, the pooled HR was 2.14 (95% CI 1.75–2.61) for adult patients and 1.99 (95% CI 1.47–2.68) for paediatric patients (). When stratified by ethnicity, the HR was slightly higher for Asians than for Caucasians, with HR values of 2.43 (95% CI 1.87–2.15) and 1.88 (95% CI 1.51–2.34), respectively. According to ALL type, significant relevance persisted among unselected ALL, B-ALL and Ph-positive ALL cohorts. When stratified by CDKN2A/B detection method, the combined HR for MLPA was 2.16 (95% CI 1.70–2.75), and for non-MLPA was 2.03 (95% CI 1.61–2.55). In subgroup analysis by statistical method, the pooled HR was 2.15 (95% CI 1.76–2.62) by univariate analysis and 1.95 (95% CI 1.44–2.65) by multivariate analysis.
Relationship between CDKN2A/B deletions and EFS/DFS/RFS
Overall, 12 studies with a total of 2715 patients reported results on EFS/DFS/RFS. Meta-analysis revealed that CDKN2A/B deletions were associated with worse EFS/DFS/RFS (HR = 2.16, 95% CI 1.73–2.69) (). Mild heterogeneity existed among studies (p = .073, I2 = 38.1%) and random-effects model was applied to combine the results. Subgroup analyses were also conducted according to age group, ethnicity, ALL type, CDKN2A/B detection method, uni/multivariate analysis and endpoint (). The heterogeneity was non-significant for subgroups of ethnicity, indicating that ethnicity might be the main source of heterogeneity.
In subgroup analyses, the deleterious impact of CDKN2A/B deletions on EFS/DFS/RFS was still significant in both adults (HR = 2.11, 95% CI 1.72–2.60) and childhood patients (HR = 2.23, 95% CI 1.46–3.40) (), in univariate (HR = 2.15, 95% CI 1.71–2.71) or multivariate (HR = 1.96, 95% CI 1.42–2.72) analysis, and in studies using MLPA (HR = 1.96, 95% CI 1.53–2.50) or non-MLPA (HR = 2.16, 95% CI 1.57–2.96) methods. As observed for OS, the HR was higher in Asians (HR = 2.51, 95% CI 1.98–3.20) than in Caucasians (HR = 1.73, 95% CI 1.40–2.14). Subgroup analysis was also conducted according to ALL type stratification. The pooled HRs indicated that CDKN2A/B deletions were poor prognostic factor for EFS/DFS/RFS in unselected ALL, B-ALL, Ph-positive ALL and Ph-negative B-ALL patients. In subgroup analysis by endpoint, the pooled HR for EFS was 2.34 (95% CI 1.63–3.36) and 2.05 (95% CI 1.59-2.65) for DFS, respectively ().
Publication bias and sensitivity analyses
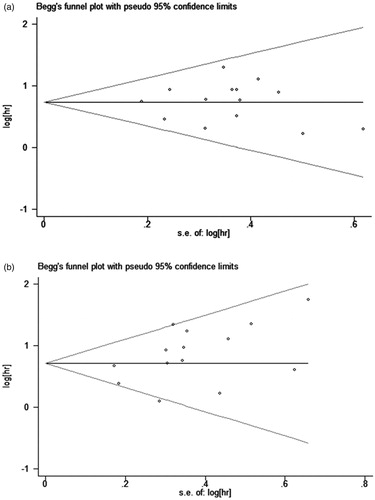
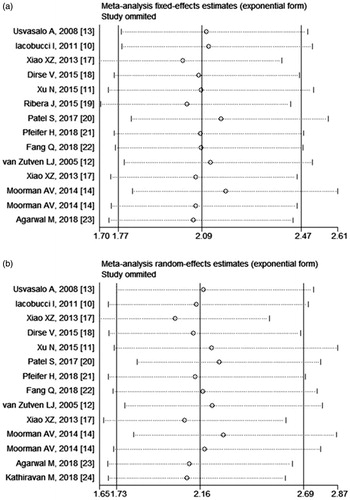
The Egger's and Begg's tests indicated no statistically significant publication bias for both OS (p = .988 for Egger's test and 0.913 for Begg's test) and EFS/DFS/RFS (p = .073 for Egger's test and 0.063 for Begg's test) (). In sensitivity analyses, after excluding one study at a time, the pooled effect sizes were not significantly affected by the exclusion of any specific study ().
Discussion
CDKN2A/B are frequently deleted in both adult and childhood ALL. Although numerous studies have been conducted thus far evaluating the prognostic significance of CDKN2A/B deletions in ALL patients, the correlation is still under debate [Citation12–18]. Thus, it is valuable to perform a meta-analysis to comprehensively assess the association between CDKN2A/B deletions and clinical outcome in ALL patients.
The present meta-analysis included thirteen studies containing a total of 2857 patients. CDKN2A/B deletions were found to be an adverse indicator for both OS and EFS/DFS/RFS. In subgroup analysis, the negative impact persisted in both adult and childhood patients. The prognosis of paediatric ALL is much superior to that of adults [Citation29,Citation30], partly due to more intensive treatment regimens and less occurrence of unfavourable cytogenetic subtypes [Citation5,Citation31]. Our results suggested that inclusion of CDKN2A/B status would further improve risk stratification in both adult and childhood ALL patients. In subgroup analysis by ethnicity, the HR for Asians was slightly higher than that for Caucasians, implying that the adverse impact of CDKN2A/B deletions may be more prominent in Asian patients. In respect to statistical method, the unfavourable impact was statistically significant in either univariate or multivariate model, suggesting the independent prognostic significance of CDKN2A/B deletions.
The past decade has seen remarkable advances in the dissection of genetic basis of ALL [Citation32–37], and what we should keep in mind is that the prognostic value of a single genetic factor can be limited. Genetic alterations in ALL can be classified into two major categories: initiating lesions including structural chromosome alterations or aneuploidy, and cooperative alterations in genes that regulate lymphoid development (IKZF1, PAX5, EBF1), cell-cycle progression and tumour suppression (CDKN2A/B, RB1), and epigenetic modification [Citation5,Citation38]. Moorman et al. [Citation18] once developed a novel risk classification strategy of paediatric B-ALL by integrating copy-number alterations (CNAs) of eight most commonly deleted genes with established cytogenetic risk groups. According to their new strategy, patients could be classified into two genetic risk groups. Good-risk genetic group (GEN-GR) included ETV6-RUNX1, high hyperdiploidy, normal copy-number status for all eight genes, isolated deletions of ETV6/PAX5/BTG1 and ETV6 deletions with a single additional deletion of BTG1/PAX5/CDKN2A/B. Patients with other genetic features were classified as poor-risk genetic group (GEN-PR). The genetic risk was independently associated with clinical outcomes in both original cohort and validation cohort. One limitation of this study was that other genetic heterogeneities such as sequence variants (e.g. RAS, JAK2 and TP53 mutations) and Ph-like signatures were not included in the risk classification. Thus, further studies are required to define how they interface with the novel classification and whether similar stratification could be generated in adult patients.
Aside from genetic alterations, minimal residual disease (MRD) is another crucial factor that has prognostic and therapeutic implications for both paediatric and adult ALL patients [Citation39]. And integration of MRD response and genetic alterations may further improve the risk stratification of ALL patients. Several studies have made the attempt to develop integrated models [Citation40–44]. Nevertheless, most of the studies were carried out in paediatric patients and consensus is still yet to be made around the best method for integrating genetic and MRD data to stratify patients.
The three proteins encoding by CDKN2A/B belong to the RB1 and TP53 pathways and act as tumour suppressors by regulating the G1/S checkpoint of cell cycle. Inactivation of the genes leads to uncontrolled proliferation of differentiated cells and tumour formation [Citation45]. The CDKN2A/B locus can be inactivated by deletion, methylation, or mutation, with genomic deletion being the most frequent form in ALL [Citation46,Citation47]. Promoter methylation causing transcriptional silencing is another mechanism of CDKN2A/B inactivation, occurred in 0–40% of ALL patients from published studies, but seems to lack prognostic significance [Citation13,Citation46,Citation48–53]. The rate of point mutations appeared to be very low, with their prognostic value not fully clarified due to lack of large cohort studies [Citation46,Citation54–56].
Williams et al. [Citation10] revealed that p14ARF inactivation shortened the latency and increased the resistance to imatinib of BCR-ABL1-induced ALL in mouse models. Disruption of p16INK4 and p15INK4B results in constitutive activation of CDK4/6. Inhibitors of CDK4/6 have been developed and are currently being tested in clinical trials in a diversity of tumours, but not yet in ALL [Citation57,Citation58]. The Food and Drug Administration (FDA) has approved three CDK4/6 inhibitors (palbociclib, ribociclib, and abemaciclib) in the treatment of breast cancer [Citation58]. Further pre-clinical and clinical studies are required to test the effectiveness and safety of CDK4/6 inhibitors in ALL patients.
Several limitations of the present meta-analysis should be mentioned. Firstly, all of the studies included were retrospective observational studies but not prospective controlled trials. Secondly, some of the HRs were extracted from Kaplan–Meier curves, which might be less reliable than the original data or multivariate analysis results. Thirdly, as revealed by some studies [Citation46,Citation47,Citation59], the deletion rate of CDKN2A/B might vary across different phenotypic and genetic subgroups. However, due to the limited data available, we could not perform further subgroup analysis by different genetic subsets. Finally, therapeutic regimen is an important factor that could affect ALL outcomes, yet we were unable to conduct subgroup analysis by treatment because of the diverse regimens used in each study.
In conclusion, our meta-analysis indicated that CDKN2A/B deletions were poor prognostic markers for both adult and paediatric ALL patients. Combined with established and emerging risk factors, CDKN2A/B alterations might further improve the risk stratification and risk-adapted treatment strategies for ALL patients. Further large-scale studies with homogeneity are needed to confirm our findings, as well as preclinical and clinical studies to test the probable therapeutic effects of CDK4/6 inhibitors in ALL patients.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 2017;7:e577.
- Shen Z, Gu X, Mao W, et al. Influence of pre-transplant minimal residual disease on prognosis after Allo-SCT for patients with acute lymphoblastic leukemia: systematic review and meta-analysis. BMC Cancer. 2018;18:755.
- Jabbour E, Pui CH, Kantarjian H. Progress and Innovations in the Management of Adult Acute Lymphoblastic Leukemia. JAMA Oncol. 2018;4:1413–1420.
- Smith L, Glaser AW, Kinsey SE, et al. Long-term survival after childhood acute lymphoblastic leukaemia: population-based trends in cure and relapse by clinical characteristics. Br J Haematol. 2018;182:851–858.
- Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35:975–983.
- Wang Y, Chen G, Cao R, et al. Allogeneic hematopoietic stem cell transplantation improves the prognosis of p16-deleted adult patients with acute lymphoblastic leukemia. Pharmacogenomics. 2017;18:77–84.
- Bernt KM, Hunger SP. Current concepts in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia. Front Oncol. 2014;4:54.
- Kim DH, Moldwin RL, Vignon C, et al. TEL-AML1 translocations with TEL and CDKN2 inactivation in acute lymphoblastic leukemia cell lines. Blood. 1996;88:785–794.
- Notta F, Mullighan CG, Wang JC, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367.
- Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2006;103:6688–6693.
- Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–2287.
- Faderl S, Kantarjian HM, Manshouri T, et al. The prognostic significance of p16INK4a/p14ARF and p15INK4b deletions in adult acute lymphoblastic leukemia. Clin Cancer Res. 1999;5:1855–1861.
- Kim M, Yim SH, Cho NS, et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet. 2009;195:59–65.
- Iacobucci I, Ferrari A, Lonetti A, et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin Cancer Res. 2011;17:7413–7423.
- Xu N, Li YL, Zhou X, et al. CDKN2 gene deletion as poor prognosis predictor involved in the progression of adult B-lineage acute lymphoblastic leukemia patients. J Cancer. 2015; 6:1114–1120.
- van Zutven LJ, van Drunen E, de Bont JM, et al. CDKN2 deletions have no prognostic value in childhood precursor-B acute lymphoblastic leukaemia. Leukemia. 2005;19:1281–1284.
- Usvasalo A, Savola S, Raty R, et al. CDKN2A deletions in acute lymphoblastic leukemia of adolescents and young adults: an array CGH study. Leuk Res. 2008;32:1228–1235.
- Moorman AV, Enshaei A, Schwab C, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–1444.
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
- Xiao XZ, Xu N, Zhang JF, et al. Comparison of clinical implications of p16 deletion in childhood and adult B-lineage acute lymphoblastic leukemia. Zhonghua Xue Ye Xue Za Zhi. 2013;34:389–394.
- Dirse V, Bertasiute A, Gineikiene E, et al. A population-based single nucleotide polymorphism array analysis of genomic aberrations in younger adult acute lymphoblastic leukemia patients. Genes Chromosomes Cancer. 2015;54:326–333.
- Ribera J, Morgades M, Zamora L, et al. Prognostic significance of copy number alterations in adolescent and adult patients with precursor B acute lymphoblastic leukemia enrolled in PETHEMA protocols. Cancer. 2015;121:3809–3817.
- Patel S, Mason CC, Glenn MJ, et al. Genomic analysis of adult B-ALL identifies potential markers of shorter survival. Leuk Res. 2017;56:44–51.
- Pfeifer H, Raum K, Markovic S, et al. Genomic CDKN2A/2B deletions in adult Ph + ALL are adverse despite allogeneic stem cell transplantation. Blood. 2018;131:1464–1475.
- Fang Q, Yuan T, Li Y, et al. Prognostic significance of copy number alterations detected by multi-link probe amplification of multiple genes in adult acute lymphoblastic leukemia. Oncol Lett. 2018;15:5359–5367.
- Agarwal M, Bakhshi S, Dwivedi SN, et al. Cyclin dependent kinase inhibitor 2A/B gene deletions are markers of poor prognosis in Indian children with acute lymphoblastic leukemia. Pediatr Blood Cancer. 2018;65:e27001.
- Kathiravan M, Singh M, Bhatia P, et al. Deletion of CDKN2A/B is associated with inferior relapse free survival in pediatric B cell acute lymphoblastic leukemia. Leuk Lymphoma. 2018;1–9.
- Ma H, Sun H, Sun X. Survival improvement by decade of patients aged 0-14 years with acute lymphoblastic leukemia: a SEER analysis. Sci Rep. 2014;4:4227.
- Pulte D, Jansen L, Gondos A, et al. Survival of adults with acute lymphoblastic leukemia in Germany and the United States. PLoS One. 2014;9:e85554.
- Pui CH, Yang JJ, Hunger SP, et al. Childhood acute lymphoblastic leukemia: progress through collaboration. JCO. 2015;33:2938–2948.
- Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764.
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266.
- Kawamata N, Ogawa S, Zimmermann M, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784.
- Paulsson K, Cazier JB, Macdougall F, et al. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: unexpected similarities with pediatric disease. Proc Natl Acad Sci USA. 2008;105:6708–6713.
- Okamoto R, Ogawa S, Nowak D, et al. Genomic profiling of adult acute lymphoblastic leukemia by single nucleotide polymorphism oligonucleotide microarray and comparison to pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1481–1488.
- Safavi S, Hansson M, Karlsson K, et al. Novel gene targets detected by genomic profiling in a consecutive series of 126 adults with acute lymphoblastic leukemia. Haematologica. 2015;100:55–61.
- Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood. 2017;130:2064–2072.
- Berry DA, Zhou S, Higley H, et al. Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: a meta-analysis. JAMA Oncol. 2017;3:e170580.
- Kang H, Chen IM, Wilson CS, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405.
- Chen IM, Harvey RC, Mullighan CG, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2012;119:3512–3522.
- Beldjord K, Chevret S, Asnafi V, et al. Oncogenetics and minimal residual disease are independent outcome predictors in adult patients with acute lymphoblastic leukemia. Blood. 2014;123:3739–3749.
- O'Connor D, Enshaei A, Bartram J, et al. Genotype-specific minimal residual disease interpretation improves stratification in pediatric acute lymphoblastic leukemia. JCO. 2018;36:34–43.
- Stanulla M, Dagdan E, Zaliova M, et al. IKZF1(plus) defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. JCO. 2018;36:1240–1249.
- Carrasco Salas P, Fernandez L, Vela M, et al. The role of CDKN2A/B deletions in pediatric acute lymphoblastic leukemia. Pediatr Hematol Oncol. 2016;33:415–422.
- Sulong S, Moorman AV, Irving JA, et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100–107.
- Schwab CJ, Chilton L, Morrison H, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98:1081–1088.
- Wong IH, Ng MH, Huang DP, et al. Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood. 2000;95:1942–1949.
- Garcia-Manero G, Daniel J, Smith TL, et al. DNA methylation of multiple promoter-associated CpG islands in adult acute lymphocytic leukemia. Clin Cancer Res. 2002;8:2217–2224.
- Gutierrez MI, Siraj AK, Bhargava M, et al. Concurrent methylation of multiple genes in childhood ALL: correlation with phenotype and molecular subgroup. Leukemia. 2003;17:1845–1850.
- Tsellou E, Troungos C, Moschovi M, et al. Hypermethylation of CpG islands in the promoter region of the p15INK4B gene in childhood acute leukaemia. Eur J Cancer. 2005;41:584–589.
- Mirebeau D, Acquaviva C, Suciu S, et al. The prognostic significance of CDKN2A, CDKN2B and MTAP inactivation in B-lineage acute lymphoblastic leukemia of childhood. Results of the EORTC studies 58881 and 58951. Haematologica. 2006;91:881–885.
- Braun M, Pastorczak A, Fendler W, et al. Biallelic loss of CDKN2A is associated with poor response to treatment in pediatric acute lymphoblastic leukemia. Leuk Lymphoma. 2017;58:1162–1171.
- Rasool O, Heyman M, Brandter LB, et al. p15ink4B and p16ink4 gene inactivation in acute lymphocytic leukemia. Blood. 1995;85:3431–3436.
- Calero Moreno TM, Gustafsson G, Garwicz S, et al. Deletion of the Ink4-locus (the p16ink4a, p14ARF and p15ink4b genes) predicts relapse in children with ALL treated according to the Nordic protocols NOPHO-86 and NOPHO-92. Leukemia. 2002;16:2037–2045.
- Lemos JA, Defavery R, Scrideli CA, et al. Analysis of p16 gene mutations and deletions in childhood acute lymphoblastic leukemias. Sao Paulo Med J. 2003;121:58–62.
- Pan Q, Sathe A, Black PC, et al. CDK4/6 inhibitors in cancer therapy: a novel treatement strategy for bladder cancer. Bladder Cancer. 2017;3:79–88.
- Thu KL, Soria-Bretones I, Mak TW, et al. Targeting the cell cycle in breast cancer: towards the next phase. Cell Cycle. 2018;17:1871–1885.
- Barbosa TC, Terra-Granado E, Quezado Magalhaes IM, et al. Frequency of copy number abnormalities in common genes associated with B-cell precursor acute lymphoblastic leukemia cytogenetic subtypes in Brazilian children. Cancer Genet. 2015;208:492–501.