Abstract
Background
Circular RNA microarray analysis showed hsa_circ_0010235 (circ_0010235) was highly upregulated in non-small-cell lung cancer (NSCLC) patients; however, its role in carcinogenesis and development of NSCLC cells was unrevealed. Here, we intended to investigate role and mechanism of circ_0010235 in NSCLC proliferation, migration and invasion.
Methods and results
Expression of circ_0010235, microRNA (miR)-338-3p and kinesin family member 2A (KIF2A) was detected by quantitative real-time PCR, western blotting and immunohistochemistry (IHC). Cell progression was measured by cell-counting kit-8 assay, 5-ethynyl-2-deoxyuridine (EdU) assay, flow cytometry, transwell assay, western blotting, IHC and xenograft experiment. The relationship among circ_0010235, miR-338-3p and KIF2A was determined by dual-luciferase reporter assay, RNA immunoprecipitation and Pearson’s correlation analysis. Expression of circ_0010235 was increased in human NSCLC tissues and cells, accompanied with miR-338-3p downregulation and KIF2A upregulation. Essentially, circ_0010235 could sponge miR-338-3p via target binding, and miR-338-3p downstream targeted KIF2A. Functionally, exhaustion of circ_0010235 induced apoptosis rate of NSCLC cells and curbed cell viability, EdU incorporation, migration rate and invasion rate, accompanied with higher E-cadherin and lower N-cadherin expression. Additionally, re-expression of miR-338-3p prompted above similar effects in NSCLC cells in vitro. Contrarily, miR-338-3p blockage partially counteract the effects of circ_0010235 exhaustion; plus, restoration of KIF2A could attenuate miR-338-3p role, as well. Notably, interfering circ_0010235 delayed tumour growth of NSCLC cells by promoting miR-338-3p and E-cadherin expression, and depressing KIF2A, ki-67 and N-cadherin expression.
Conclusions
circ_0010235 could be a novel identified oncogenic circRNA in NSCLC, and targeting miR-338-3p/KIF2A axis was one regulatory mechanism underlying circ_0010235.
Circ_0010235 was an upregulated circRNA in NSCLC patients and cells.
Interfering circ_0010235 restrained NSCLC cell proliferation and metastasis in vitro and in vivo.
miR-338-3p per se suppressed NSCLC in vitro and its downregulation diminished the tumour-suppressive role of circ_0010235 blockage in NSCLC cells.
miR-338-3p could downstream target KIF2A and be sponged by circ_0010235.
KEY MESSAGE
Keywords:
Introduction
Non-small-cell lung cancer (NSCLC) is the most widespread type of lung cancers and is disposed with lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and lung large cell carcinoma (LULC) [Citation1]. The overall 5-year survival rate of patients with lung cancer remains less than 17% [Citation1,Citation2], partially due to the scarcity of valuable molecular biomarker. It is indispensable to further probe the profound mechanism regulating NSCLC development to improve outcomes in NSCLC.
Circular RNAs (circRNAs) are a comparatively new type of noncoding RNAs with loop structures resulting from pre-mRNA back splicing, and researches on circRNAs in lung cancers is gathering momentum [Citation3]. Dysregulation of circRNAs is closely related to proliferation, migration and invasion of NSCLC cells [Citation4]. Moreover, circRNAs in liquid biopsies are suggested to be candidate biomarkers for the prognosis and therapeutic response of NSCLC [Citation5]. Human circRNAs microarray analysis screens multiple tumour-specific candidate circRNAs in NSCLC patients [Citation6–8]. Hsa_circ_0010235 (circ_0010235) is an exon-derived circRNA from aldehyde dehydrogenase 4 family member A1 (ALDH4A1) and it was highly increased in human NSCLC tissues compared to para-cancer lung tissues [Citation6]. However, its contribution to NSCLC tumour progression remains undisclosed to date.
microRNAs (miRNAs), a classic endogenous noncoding RNAs, are versatile in NSCLC tumour initiation, progression, metastasis and as well as drug resistance. miRNAs show great potential for clinical applications [Citation9,Citation10]. miRNA (miR)-338-3p (miR-338-3p) functions anti-cancer efficacy in diverse human cancers including lung cancer. In NSCLC, miR-338-3p could restrain cell growth and metastasis by targeting different oncogenes [Citation11–13].
Furthermore, NSCLC is an oncogene-addicted malignancy, and actionable oncogenes such as epidermal growth factor receptor (EGFR) have become the standard of treatment in NSCLC [Citation14]. Alteration in KRAS is another oncogenic driver for NSCLC [Citation15], and mutant KRAS repurposes kinesin family members (KIFs) KIF2A and KIF2C to support the transformed phenotypes, including enhanced migration and invasion [Citation16,Citation17]. KIF2A is an M-type nonmotile microtubule depolymerase and an independent prognostic biomarker in NSCLC [Citation18,Citation19].
Thus, we attempted to explore the activities of circ_0010235 in proliferation, migration and invasion of NSCLC cells, and to explore whether there is a competing endogenous RNA (ceRNA) regulatory mechanism among circ_0010235, miR-338-3p and KIF2A.
Materials and methods
NSCLC biopsies
Fifty-three NSCLC patients were recruited from Shandong Provincial Chest Hospital in this study, and they received no preoperative chemo-/radio-therapy. The tumour-node-metastasis (TNM) stage was classified according to the 8th edition of the American Joint Committee on Cancer (AJCC) lung cancer staging system. Tumour specimens and corresponding adjacent normal tissues were collected and stored in liquid nitrogen immediately after resection. All patients provided informed consents before the surgery, and this study was approved by the Medical Ethics Committee of Shandong Provincial Chest Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
Cells
LUAD cell line A-549 (CRM-CCL-185) and LULC cell lines NCI-H2342 (CRL-5941) and NCI-H1299 (CRL-5803) were originally from ATCC (Manassas, VA); LUAD cell line HCC827 (CRL-2868) and human bronchial epithelioid cells 16HBE (CL-0249) were from Procell (Wuhan, China). HCC827 and NCI-H1299 cells were cultured in the commendatory RPMI-1640 medium (72400120; Gibco, Grand Island, NY), NCI-H2342 cells were in HITES medium (ATCC), and A-549 cells were in F-12K medium (21127030; Gibco, Grand Island, NY). NCI-H2342 cells were maintained in foetal bovine serum (FBS; 10100147; Gibco, Grand Island, NY) to a final concentration of 5%, and other cells were incubated in 10% FBS. All cells were in a cell culture incubator at 37 °C with 5% CO2.
Quantitative real-time PCR (qPCR)
Total RNA isolation from tissues and cells was performed using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s manual. Maxima First Strand cDNA Synthesis Kit (K1642; Thermo Scientific, Shanghai, China) and Maxima SYBR Green/Fluorescein qPCR Master Mix (K0241; Thermo Scientific, Shanghai, China) were used to synthesize cDNA and amplify special RNAs. The qPCR primers for circ_0010235, ALDH4A1, miR-338-3p, KIF2A, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and RNU6B (U6) are shown in . GAPDH and U6 were the internal references for the detection of RNA levels. Data of qPCR were analysed using comparative cycle threshold method (2–ΔΔCT).
Table 1. The sequence of primers, siRNAs, shRNAs and miRNAs.
CircRNA characterization
The circular construction of circ_0010235 was detected using ribonuclease R (RNase R) digestion compared with the linear host gene. An aliquot of total RNA (2.5 μg) isolated from NCI-H1299 and A-549 cells was treated with 10 U RNase R (GENESEED, Guangzhou, China) for 30 min at 37 °C. With enzyme inactivation (10 min at 70 °C), the RNase R-treated RNAs were for qPCR. The equal volume of the 1× reaction buffer was used as negative control. Next, the half-life of circ_0010235 and ALDH4A1 was measured under actinomycin D (ActD) treatment. The culture media of NCI-H1299 and A-549 cells were added with 2 mg/mL ActD (Sigma-Aldrich, St. Louis, MO) for 0 h, 6 h, 12 h and 16 h at 37 °C, and total RNA was isolated at indicated time-points for qPCR.
Cell transfection
Two specific small interfering RNAs (siRNAs) targeting circ_0010235 (si-circ_0010235#1 and si-circ_0010235#2) and one scrambled control siRNA were purchased from RIBOBIO (Guangzhou, China), as well as the small hairpin RNA (shRNA) targeting circ_0010235 (sh-circ_0010235) and control sh-NC. The pSilencer2.1-U6 hygro (EK-Bioscience, Shanghai, China) was employed to express shRNAs. The miRNA mimic (miR-338-3p) and miR-338-3p inhibitor (in-miR-338-3p) were obtained from RIBOBIO, as well as negative controls miR-NC and in-miR-NC. The coding domain sequence of KIF2A mRNA (NM_001243952) was inserted into pcDNA3.1(+) plasmid (Genemeditech, Shanghai, China) to overexpress KIF2A. Similarly, fragment (NM_001243952; 2591-4080) of 3′untranslated region (3′UTR) of KIF2A mRNA was cloned into pcDNA3.1(+) plasmid (Genemeditech, Shanghai, China), and circ_0010235 was cloned into pCD5-ciR plasmid (GENESEED, Guangzhou, China). Above mentioned nucleotides were transfected into NCI-H1299 and A-549 cells at cell density at 60%, and the transfection reagent was Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Post-transfection for 48 h, cells were collected for further analysis.
Cell-counting kit (CCK)-8 assay and 5-ethynyl-2-deoxyuridine (EdU) assay
Enhanced CCK-8 (Beyotime, Shanghai, China) and BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime, Shanghai, China) were utilized to measure NSCLC cell proliferation. Briefly, 3000 cells were transferred in 96-well plate, and five paralleled wells were set in each group. These adherent cells were further cultivated for 72 h, and 10 μL CCK-8 solution was added in each well at different time-points 0 h, 24 h, 48 h and 72 h. With 2 h-incubation of CCK-8, optical density (OD) at 450 nm was measured on a microplate reader. Cell proliferation curve was drawn according to OD values. For EdU assay, 5000 cells were transferred in 96-well plate in triplicate. After cell adherence, cells were dyed with 10 μM EdU reagent for 2 h and re-dyed with DAPI Staining Solution (Beyotime, Shanghai, China). Fluorescence detections of Alexa Fluor 488-labelled EdU and DAPI were performed on an inverted fluorescence microscope (Nikon Microsystems, Shanghai, China). EdU positive cell rate was calculated according to three random fields.
Flow cytometry (FCM)
Dead Cell Apoptosis Kit with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (V13242; Invitrogen, Carlsbad, CA) was utilized to evaluate apoptosis using FCM method. In brief, 106 cells were precooled with phosphate buffer saline, re-suspended in the binding buffer, and then stained with Annexin V-FITC and PI. Last, stained cells were subjected to a flow cytometer (BD Biosciences, San Jose, CA), and analysed on Annexin V-FITC/PI quadrant using CytExpert software (BD Biosciences, San Jose, CA). Bio-repeats were run in triplicate.
Transwell migration and invasion assay
Transwell insert (Thermo Fisher Scientific, Waltham, MA) was used to measure cell migration ability, and Matrigel transwell chamber was for invasion capacity examination. The chamber for invasion assay was pre-coated with Matrigel by incubating with 50 µL diluted Matrigel (BD Biosciences, San Jose, CA) for 2 h at 37 °C. Cells were starved for 12 h and 104 cells (for migration) or 105 cells (for invasion) were placed in the upper chamber with serum-free medium; the lower chambers were filled with culture medium with 15% FBS (Gibco, Grand Island, NY). Bio-repeats were run in triplicate. After 48 h, cells on transwell member’s lower surface were visualized using crystal violet staining, and counted under a microscope (Nikon Microsystems, Shanghai, China). Migration/invasion rate was calculated from five fields under random selection.
Western blotting
Protein extraction from tissues and cells was executed utilizing RIPA (Beyotime, Shanghai, China). An aliquot of proteins (20 μg) was separated by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis, supported on polyvinylidene difluoride membranes, and then incubated with antibodies; eventually, protein bands were visualized by the ECL kit (Beyotime, Shanghai, China). Antibody to KIF2A (anti-KIF2A; sc-398010, 1:500), anti-E-cadherin (sc-8426, 1:500), anti-N-cadherin (sc-59987, 1:500) and anti-GAPDH (sc-365062; 1:1000) were purchased from Santa Cruz Biotechnology (Shanghai, China), as well as m-IgGκ BP-HRP (sc-516102; 1:10,000). Relative protein expression was normalized to GAPDH, and grey density of all protein bands was analysed by ImageJ (NIH, Bethesda, MD).
Prediction of circRNA-miRNA and miRNA-mRNA connection
CircInteractome (https://circinteractome.nia.nih.gov/mirna_target_sites.html), circBank (http://www.circbank.cn/index.html) and StarBase (http://starbase.sysu.edu.cn/index.php) software were used to predict miRNA binding sites in circ_0010235 and to predict miR-338-3p response elements in KIF2A mRNAs. The Venn diagram was drawn to determine the overlapping computational miRNAs for circ_0010235.
Dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay
The wide-type (WT) sequences of circ_0010235 and fragment sequence of KIF2A 3′UTR containing the intact predicted binding sequences of miR-338-3p were sub-cloned into the psiCHECK2 vectors (Promega, Madison, WI). The circ_0010235 WT vector and KIF2A 3′UTR WT vector were subjected to QuickChange Lightning multisite-directed mutagenesis kit (Agilent Technologies, Palo Alto, CA) to produce the corresponding mutants (MUT) vectors. Above WT or MUT vectors were co-transfected into NCI-H1299 and A-549 cells with commercial synthetic miR-338-3p or miR-NC. After transfection for 48 h, the luciferase activities of hluc and hRluc were measured by dual-luciferase reporter assay system (Promega, Madison, WI).
Argonaute 2 (Ago2) RIP assay was performed to measure the co-enrichment of circ_0010235, miR-338-3p and KIF2A, and Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) was used. NCI-H1299 and A-549 cells at 80% confluence were harvested and lysed in RIP lysis buffer and subjected to sonicate. Then, cell extracts were incubated with magnetic beads which were pre-immersed in anti-Ago2 (ab186733, 1:50, Abcam, Cambridge, UK). Ago2-mediated precipitation was lysed in TRIzol reagent (Invitrogen, Carlsbad, CA), and RNA levels in that were analysed by qPCR. Control immunoglobulin G (IgG) RIP was performed using Anti-Ago2 (ab186733, 1:50, Abcam, Cambridge, UK).
Establishment of xenograft mice models
A-549 cells expressing sh-NC or sh-circ_0010235 vectors were re-suspended in serum-free medium and then inoculated into BALB/c nude mice (sex, male; age, 5-week-old); these mice were purchased from Beijing Vital River Laboratory Animal Technology Company (Beijing, China). 5 × 106 cells were for one mice and five mice were for each group. Following the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006), these mice were injected subcutaneously into the flanks. Tumour volume was measured after seven day-xenograft, and then measured every four days to provide tumour growth curves. Volume (mm3)=0.5 × length × width2. At the 27th after cell inoculation, mice were sacrificed with 2% methoxyflurane anaesthesia and cervical dislocation. Xenograft tumours were completely dissected and photographed, and tumour weight were measured. This animal experiment was approved by the Animal Ethics Committee of the Shandong Provincial Chest Hospital.
Immunohistochemistry (IHC)
In situ expression of KIF2A and proliferation marker ki-67 in human tumours and xenograft mice tumours was measured using IHC method. Tissues were fixed with 4% paraformaldehyde and embedded in paraffin. Paraffin-embedded tissues were sliced into 4 µm slides. Antigen retrieval was performed using 0.01 M citrate buffer for 20 min at 95 °C, and slides were incubated in 3% H2O2 solution for 15 min at 25 °C. Blocked with 5% goat serum for 30 min incubation at 25 °C, the slides were incubated with primary antibodies overnight at 4 °C and secondary antibody for 30 min at 25 °C, and re-stained with diaminobenzidine and haematoxylin; eventually, tissue protein expression of KIF2A and ki-67 was observed under microscope (Nikon Microsystems, Shanghai, China) at an appropriate magnification (100×). Anti-KIF2A (sc-398010, 1:100) and anti-ki-67 (ARG53222; 1:200) were from Santa Cruz Biotechnology and arigo Biolaboratories (Taiwan, China), respectively. The m-IgGκ BP-HRP (sc-516102; 1:100) and mouse anti-rabbit IgG-HRP (sc-2357; 1:100) were from Santa Cruz Biotechnology (Shanghai, China).
Data collection and analysis
Data analysis was performed on GraphPad Prism version 6.0 (GraphPad Software, La Jolla, CA), as well as the drawing of scatter diagram, Venn diagram and histograms. Differences were determined by Student’s t-test and one-way analysis of variance. Correlation between two RNA levels was confirmed by Pearson’s correlation analysis. The p values were *p< .05, **p< .01, ***p< .001 or ****p< .0001.
Results
Expression of circ_0010235 was stably elevated in NSCLC tissues and cells
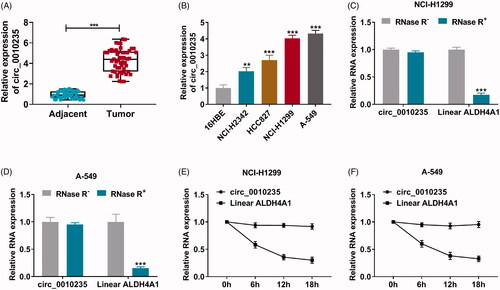
Data of qPCR showed that circ_0010235 expression was 4.2-fold high in NSCLC patients’ tumour tissues than in para-cancer tissues (); besides, its level was overall elevated in human NSCLC cell lines than normal 16HBE cell line (), and NCI-H1299 and A-549 cells exhibited the most upregulation of circ_0010235. With RNase R treatment in total RNAs from NCI-H1299 and A-549 cells, the linear gene ALDH4A1 was extremely downregulated, whereas circ_0010235 expression was unaltered (). Furthermore, half-life of ALDH4A1 was less than 12 h in NCI-H1299 and A-549 cells under ActD treatment, and meanwhile circ_0010235 expression was stable within 18 h of ActD treatment (). These data manifested that circ_0010235 was stably expressed and upregulated in NSCLC patients and cells.
Figure 1. Expression of circ_0010235 was stably elevated in NSCLC tissues and cells. (A, B) qPCR detected circ_0010235 expression (A) in 53 paired NSCLC tumour tissues (tumour) and adjacent normal tissues (adjacent) and (B) in NSCLC cell lines (NCI-H2342, HCC827, NCI-H1299 and A-549) and normal lung cell line (16HBE). (C–F) qPCR detected RNA expression of circ_0010235 and linear ALDH4A1 (C, D) in NCI-H1299 and A-549 cells-derived total RNAs treated with RNase R (RNase R+) and un-treated with RNase R (RNase R–) and (E, F) in NCI-H1299 and A-549 cells treated with ActD during consecutive 18 h. **p< .01 and ***p< .001.
Exhaustion of circ_0010235 curbed proliferation, migration and invasion of NSCLC cells
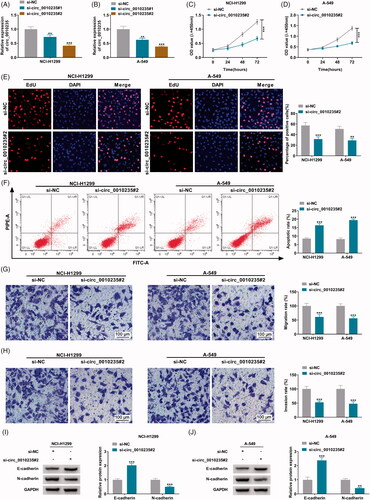
Two siRNAs targeting circ_0010235 were designed to silence its expression, and si-circ_0010235#2 showed a better silencing efficiency in both NCI-H1299 and A-549 cells (). CCK-8 assay depicted that OD values of si-circ_0010235#2-transfected cells were lower than si-NC-transfected cells during consecutive 72 h (). EdU assay demonstrated less EdU positive cells happened in circ_0010235-silenced NCI-H1299 and A-549 cells via si-circ_0010235#2 transfection comparing to control cells (). The two assays indicated that exhausting circ_0010235 suppressed cell proliferation of NSCLC cells, and this anti-proliferation effect was accompanied with apoptosis rate enhancement (). Cell migration rate and invasion rate were consistently lowered according to transwell assays (), which was paralleled with increased E-cadherin expression and decreased N-cadherin expression (). These results demonstrated a suppressive effect of circ_0010235 exhaustion on migration and invasion of NSCLC cells.
Figure 2. Exhaustion of circ_0010235 curbed proliferation, migration and invasion of NSCLC cells. (A, B) qPCR detected circ_0010235 expression in NCI-H1299 and A-549 cells transfected with si-NC, si-circ_0010235#1 or si-circ_0010235#2. (C–J) In si-NC- and si-circ_0010235#1-transfected NCI-H1299 and A-549 cells, (C, D) CCK-8 assay measured OD value at 450 nm during consecutive 72 h, (E) EdU assay determined EdU positive rate, (F) FCM evaluated apoptotic rate, (G, H) transwell assay confirmed migration rate and invasion rate, and the scale bar was 100 μm in transwell images, (I, J) western blotting detected protein expression of E-cadherin and N-cadherin, normalized to GAPDH. **p< .01 and ***p< .001.
circ_0010235 was responsive to miR-338-3p via target binding
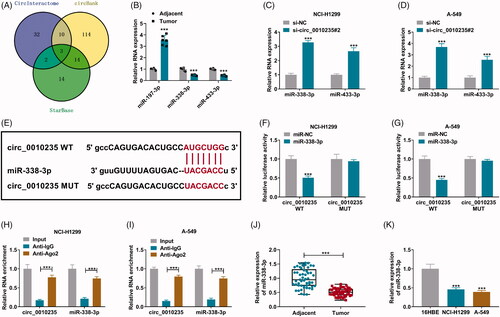
CircInteractome, circBank and StarBase software were used to predict miRNA binding sites in circ_0010235, and three overlapping miRNAs were obtained according to the Venn diagram (). Among these, three miRNAs, miR-338-3p and miR-433-3p were downregulated in NSCLC patients’ tumours (), and their expression was highly induced in circ_0010235-blocked NCI-H1299 and A-549 cells (). Here, we selected miR-338-3p as candidate target for circ_0010235. Due to mutations of miR-338-3p response elements, circ_0010235 MUT report vectors were unresponsive to miR-338-3p ectopic expression (). Meanwhile, luciferase activity of circ_0010235 WT report vectors was reduced in miR-338-3p-transfected NCI-H1299 and A-549 cells. Moreover, RIP assay and qPCR detected that circ_0010235 and miR-338-3p were concurrently enriched in anti-Ago2-immunoprecipitated RNAs in NCI-H1299 and A-549 cells (). These results indicated that there was a target relationship between circ_0010235 and miR-338-3p in NSCLC cells. Additionally, miR-338-3p was a downregulated miRNA in human NSCLC tumours and cells ().
Figure 3. The target relationship between circ_0010235 and miR-338-3p. (A) Venn diagram showed the overlapping miRNAs that were predicted to be responsive to circ_0010235 by CircInteractome, circBank and StarBase software. (B) qPCR detected RNA expression of miR-197-3p, miR-338-3p and miR-433-3p in human tumour (n = 6) and adjacent (n = 6) samples. (C, D) qPCR detected RNA expression of miR-338-3p and miR-433-3p in si-NC- and si-circ_0010235#2-transfected NCI-H1299 and A-549 cells. (E) The miR-338-3p response elements are shown in circ_0010235 WT and MUT according to StarBase. (F, G) Dual-luciferase reporter assay measured luciferase activity of circ_0010235 WT and MUT report vectors in NCI-H1299 and A-549 cells transfected with miR-338-3p or miR-NC. (H, I) RIP assay and qPCR detected RNA enrichment of circ_0010235 and miR-338-3p in anti-IgG- and anti-Ago2-precipitated RNAs in cell extract of NCI-H1299 and A-549 cells. (J, K) qPCR detected miR-338-3p expression in human tumour (n = 53) and adjacent (n = 53) tissues, and in cells (16HBE, NCI-H1299 and A-549). ***p< .001.
Blockage of miR-338-3p attenuated the effects of circ_0010235 exhaustion in NSCLC cells
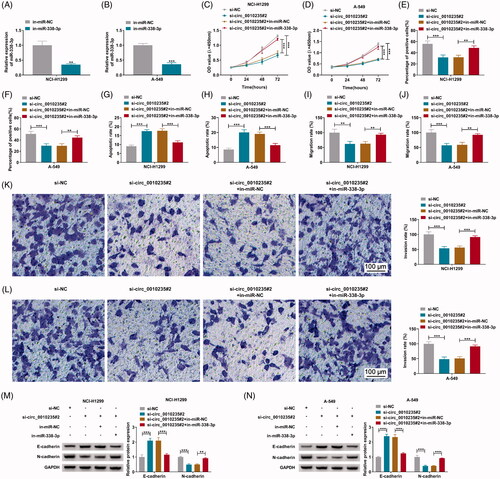
Commercial in-miR-338-3p was used to inhibit miR-338-3p expression in NCI-H1299 and A-549 cells via transfection (). Anti-proliferation effect of circ_0010235 blockage in NCI-H1299 and A-549 cells was weakened by in-miR-338-3p transfection than in-miR-NC transfection, as evidenced by the increase of OD values during 72 h () and the promotion of EdU positive cells (), as well as the decrease of apoptotic rate (). Similarly, silencing circ_0010235-mediated anti-migration and anti-invasion roles in NCI-H1299 and A-549 cells were partially counteracted by silencing miR-338-3p via transfection, as described by the augmented transwell migration rate and invasion rate (), paralleling with diminished E-cadherin and restored N-cadherin (). These results demonstrated that blocking miR-338-3p could attenuate suppressive effects of circ_0010235 exhaustion on NSCLC proliferation, migration and invasion.
Figure 4. Blockage of miR-338-3p attenuated the effects of circ_0010235 exhaustion in NSCLC cells. (A, B) qPCR detected miR-338-3p expression in in-miR-NC- and in-miR-338-3p-transfected NCI-H1299 and A-549 cells. (C–N) In NCI-H1299 and A-549 cells transfected with si-NC, si-circ_0010235#2 or si-circ_0010235#2 along with in-miR-NC or in-miR-338-3p, (C, D) CCK-8 assay measured OD value at 450 nm during consecutive 72 h, (E, F) EdU assay determined EdU positive rate, (G, H) FCM evaluated apoptotic rate, (I–L) transwell assay confirmed migration rate and invasion rate, and the scale bar was 100 μm in transwell images and (M, N) western blotting detected protein expression of E-cadherin and N-cadherin, normalized to GAPDH. **p< .01 and ***p< .001.
miR-338-3p could target on KIF2A
StarBase software showed computational miR-338-3p binding sites in KIF2A 3′UTR (), and their mutations led to the outcome that luciferase activity of KIF2A 3′UTR MUT report vectors was not like its WT vectors which were responsive to miR-338-3p overexpression and their luciferase activity was damaged with commercial miR-338-3p transfection (). Expression of KIF2A mRNA was promoted in human NSCLC tumour tissues and cells (); consistently, in situ expression of KIF2A in NSCLC patients’ tumours and KIF2A protein expression in extract of NSCLC tumour tissues and cells were overall upregulated than normal tissues and cells (). These data illuminated that KIF2A was highly expressed in NSCLC patients and cells and could be a target for miR-338-3p.
Figure 5. MiR-338-3p could target KIF2A. (A) miR-338-3p response elements are shown in KIF2A 3′UTR WT and MUT according to StarBase software. (B, C) Dual-luciferase reporter assay measured luciferase activity of KIF2A 3′UTR WT and MUT report vectors in NCI-H1299 and A-549 cells transfected with miR-338-3p or miR-NC. (D, G) qPCR detected KIF2A mRNA expression, (E) IHC measured in situ expression of KIF2A, and the scale bar was 50 μm in IHC images and (F, H) western blotting examined KIF2A protein expression in human tumour and adjacent tissues and cells (16HBE, NCI-H1299 and A-549). **p< .01 and ***p< .001.
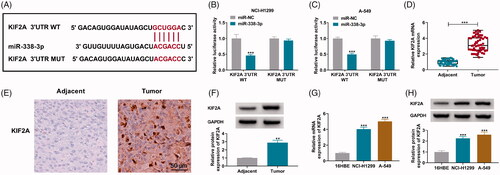
Restoration of KIF2A partially abated the inhibiting effect of miR-338-3p re-expression on NSCLC cell proliferation, migration and invasion
Transfecting miR-338-3p and KIF2A vectors resulted in the overexpression of miR-338-3p and KIF2A in NCI-H1299 and A-549 cells, respectively (). Re-expressing miR-338-3p restrained OD values of NCI-H1299 and A-549 cells during consecutive 72 h (), and lessened EdU positive cells (), and as well as facilitated apoptotic rate (). Furthermore, co-transfecting miR-338-3p and KIF2A vectors could diminish above effects and rescued cell proliferation of NCI-H1299 and A-549 cells with miR-338-3p overexpression (). Transwell migration rate and invasion rate of NCI-H1299 and A-549 cells were synchronously descended by overexpressing miR-338-3p via transfection, and this descent was salvaged by additionally restoring KIF2A via transfection as well (), accompanying with downregulated E-cadherin and upregulated N-cadherin (). These outcomes illustrated that miR-338-3p re-expression suppressed NSCLC proliferation, migration and invasion, and this suppressive role could be attenuated by restoring KIF2A.
Figure 6. Restoration of KIF2A partially abated the inhibiting effect of miR-338-3p re-expression on NSCLC cell proliferation, migration and invasion. (A) qPCR detected miR-338-3p expression in miR-NC- and miR-338-3p-transfected NCI-H1299 and A-549 cells. (B) Western blotting examined KIF2A protein expression in pcDNA- and pcDNA-KIF2A (KIF2A)-transfected NCI-H1299 and A-549 cells. (C–N) In NCI-H1299 and A-549 cells transfected with miR-NC, miR-338-3p or miR-338-3p combined with pcDNA or KIF2A, (C, D) CCK-8 assay measured OD value at 450 nm during consecutive 72 h, (E, F) EdU assay determined EdU positive rate, (G, H) FCM evaluated apoptotic rate, (I–L) transwell assay confirmed migration rate and invasion rate, and the scale bar was 100 μm in transwell images and (M, N) western blotting detected protein expression of E-cadherin and N-cadherin. *p< .05, **p< .01 and ***p< .001.
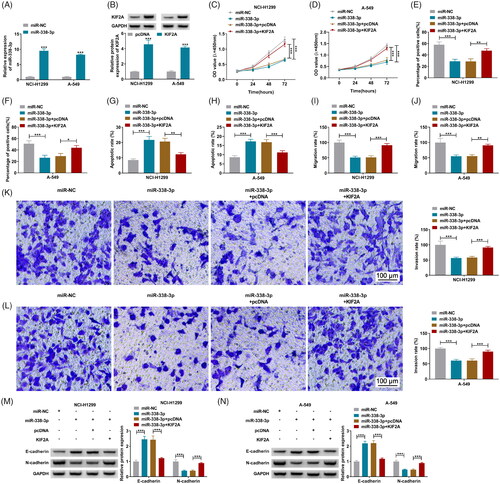
miR-338-3p modulated the regulatory effect of circ_0010235 on KIF2A expression
Pearson’s correlation analysis determined that miR-338-3p expression was inversely correlated with circ_0010235 or KIF2A mRNA in NSCLC patients’ tumours (), and that there was a positive correlation between circ_0010235 and KIF2A mRNA expression in these samples (). In terms of protein expression, KIF2A could be depressed by circ_0010235 exhaustion in NCI-H1299 and A-549 cells, and this depression was partially abated by simultaneously exhausting miR-338-3p ().
Figure 7. The relationship among circ_0010235, miR-338-3p and KIF2A. (A–C) Pearson’s correlation analysis determined the correlation among circ_0010235, miR-338-3p and KIF2A. (D, E) Western blotting examined KIF2A protein expression in NCI-H1299 and A-549 cells transfected with si-NC, si-circ_0010235#2 or si-circ_0010235#2 along with in-miR-NC or in-miR-338-3p. ***p< .001.
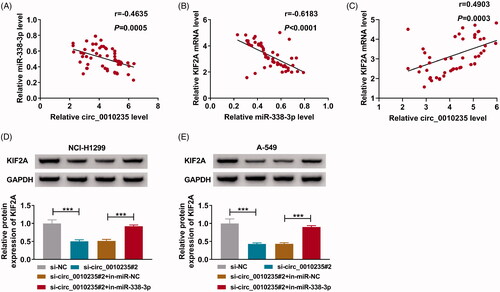
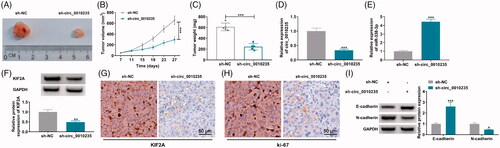
Interference of circ_0010235 might delay tumour growth and suppress metastasis of NSCLC cells by altering miR-338-3p and KIF2A
Stably transfecting sh-circ_0010235 rather than sh-NC could control tumorigenicity of A-549 cells in vivo, as evidenced by the delayed tumour growth in nude mice (n = 5) (). More importantly, circ_0010235 expression was blocked in extracts of xenograft tumour tissues in sh-circ_0010235 group compared to sh-NC group (), paralleled with promoted miR-338-3p and inhibited KIF2A (). In addition, according to in situ analysis of the gene expression, KIF2A labelled cells and ki-67 labelled cells were lesser in xenograft tumours induced by circ_0010235-interfered A-549 cells (). Regarding tumour metastasis, E-cadherin protein was enhanced and N-cadherin protein was inhibited in above delayed tumours (). These results indicated that interfering circ_0010235 might delay tumour growth and suppress metastasis of NSCLC cells in vivo by altering miR-338-3p and KIF2A.
Figure 8. Interference of circ_0010235 delayed tumour growth and metastasis of NSCLC cells by altering miR-338-3p and KIF2A. (A–I) A-549 cells expressing sh-NC or sh-circ_0010235 vectors were inoculated into nude mice (n = 5). (A–C) Tumour volume and weight were examined. (D, E) qPCR detected expression of circ_0010235 and miR-338-3p, (F, I) western blotting examined protein expression of KIF2A, E-cadherin and N-cadherin and (G, H) IHC measured in situ expression of KIF2A and ki-67, and the scale bar was 50 μm in IHC images in xenograft tumour tissues. *p< .05, **p< .01 and ***p< .001.
Discussion
CircRNAs were critical regulators of NSCLC cells, representing potent diagnostic, prognostic and therapeutic markers in NSCLC [Citation4,Citation20]. Here, we confirmed the abnormal upregulation of circ_0010235 in NSCLC patients and the tumour-suppressive functions of blocking circ_0010235 on NSCLC cell proliferation, migration and invasion both in vitro and in vivo. Moreover, harbouring multiple miRNA binding sites seemed to be one typical feature of circRNAs [Citation21]. We further validated circ_0010235 as a sponge for miR-338-3p, and miR-338-3p could downstream target KIF2A. This research might conclude that circ_0010235 was oncogenic in NSCLC progression partially through circ_0010235/miR-338-3p/KIF2A ceRNA axis.
First of all, expression of circ_0010235 was detected to be highly expressed in this cohort of NSCLC biopsies, which was in consistent with data from Jiang et al. [Citation6]. Then, role of circ_0010235 in NSCLC cells was investigated. Its level was increased in human NSCLC cell lines, and silencing circ_0010235 via siRNA attenuated cell viability, EdU incorporation, transwell migration and invasion rates of NSCLC cells, but facilitated apoptosis rate. These results demonstrated an inhibiting effect of circ_0010235 deficiency on NSCLC proliferation, migration and invasion phenotypes. In addition, interfering circ_0010235 suppressed NSCLC progression both in vitro and in vivo by upregulating E-cadherin and downregulating endothelial–mesenchymal transition (EMT)-related protein N-cadherin. Above outcomes indicated an oncogenic role of circ_0010235 in NSCLC, and suggested that circ_0010235 might be a therapeutic target in treating NSCLC. However, whether the expression of circ_0010235 was tissue- and developmental-stage specific was unrevealed in this study, as well as its linkage to clinical diagnosis and prognosis.
In terms of circRNAs inherent features, we observed that circ_0010235 expression was unresponsive to RNase R and ActD treatments, suggesting a circular construction and a long half-life of circ_0010235. This validation prompted that circ_0010235 might exist in circulating body fluids such as blood, and thus it could be a potential but promising non-invasive biomarker in NSCLC. This suspect should be further verified by analysing the correlation between circulating circ_0010235 expression and deleterious clinicopathological characteristics. Mechanically, circ_0010235 possessed several miRNA response elements in the primary sequence. Among these candidate miRNAs, miR-197-3p was enormously raised in plasma of lung cancer patients and tissues of NSCLC patients [Citation22,Citation23], and plasma miR-197-3p was involved with NSCLC tumour stage and metastasis [Citation22]; miR-433-3p was downregulated in different NSCLC cell lines [Citation24]; miR-338-3p was a relatively better-studied miRNA in lung cancers through serving as tumour suppressor. Therefore, miR-338-3p was chose to be candidate target for circ_0010235, and this connection was further identified using dual-luciferase reporter assay and Ago2 RIP assay. Thus, we considered circ_0010235 acted as a sponge to absorb miR-338-3p and further modulated KIF2A expression. Of note, there were diverse noncoding RNAs sponging this miRNA, such as circRNA circ_0000003 [Citation25], and lncRNAs SBF2-AS1 and LINC00525 [Citation26,Citation27]. Here, expression of miR-338-3p was downregulated in NSCLC patients’ tissues and cells, and its restoration could counteract NSCLC cell progression by suppressing cell viability, proliferation, migration and invasion. This tumour-suppressive role of miR-338-3p in NSCLC cells had been declared by many researchers [Citation11–13].
Except for KIF2A, there were various target genes for miR-338-3p in regulation of NSCLC behaviours, including EMT regulator SOX4 [Citation11], adaptor protein IRS2 [Citation12,Citation25,Citation27] and rate-limiting enzyme SphK2 [Citation13], as well as metalloprotease ADAM17 [Citation26]. KIF2A together with KIF2B and KIF2C constituted KIFs family in human, and these three membranes were highly conserved in their primary sequences but displayed distinct localization and nonoverlapping functions [Citation28]. KIF2A was located in centrosomes [Citation28], and had been discovered to be oncogenic in multiple cancer progressions [Citation29–31]. Here, we showed that KIF2A downregulation was the common result between blocking circ_0010235 and re-expressing miR-338-3p in curbing NSCLC proliferation, migration and invasion phenotypes. Additionally, in NSCLC biopsies, KIF2A mRNA expression was linear and negatively correlated with miR-338-3p but positively correlated with circ_0010235. Functionally, restoring KIF2A was conductive to NSCLC cell malignancies, and oncogenic role of KIF2A had been demonstrated in LUSC and LUAD [Citation18,Citation19,Citation32]. Furthermore, hyperexpression of KIF2A was previously uncovered to be closely correlated with higher stage and lymph node metastasis in NSCLC patients, as well as worse overall survival and disease-free survival [Citation18,Citation19,Citation32]. Chemoresistance to cisplatin in NSCLC seemed also acquired with KIF2A overexpression [Citation32]. By the way, similar to KIF2A, KIF2C was also a computational target for miR-338-3p according to StarBase software. Herein, we left the interaction between miR-338-3p and KIF2C to be unsubstantiated, and this assumption was still very worthy due to the big significance of KIF2C in NSCLC [Citation17,Citation33,Citation34].
Conclusions
Collectively, we demonstrated that circ_0010235 was highly expressed in NSCLC patients’ tumours and its silencing could suppress NSCLC malignant transformation by inhibiting proliferation, migration and invasion partially via circ_0010235/miR-338-3p/KIF2A ceRNA pathway.
Acknowledgements
Ethical approval: All patients provided informed consents before the surgery, and this study was approved by the Medical Ethics Committee of Shandong Provincial Chest Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
Disclosure statement
The authors declare that they have no financial conflicts of interest.
Data availability statement
All data generated or analysed during this study are included in this article.
References
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454.
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311.
- Ma Y, Zhang X, Wang YZ, et al. Research progress of circular RNAs in lung cancer. Cancer Biol Ther. 2019;20(2):123–129.
- Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889.
- de Fraipont F, Gazzeri S, Cho WC, et al. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet. 2019;10:390.
- Jiang MM, Mai ZT, Wan SZ, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–674.
- Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):2878.
- Ma D, Qin Y, Huang C, et al. Circular RNA ABCB10 promotes non-small cell lung cancer progression by increasing E2F5 expression through sponging miR-584-5p. Cell Cycle. 2020;19(13):1611–1620.
- Ahn YH, Ko YH. Diagnostic and therapeutic implications of microRNAs in non-small cell lung cancer. Int J Mol Sci. 2020;21(22):8782.
- Leonetti A, Assaraf YG, Veltsista PD, et al. MicroRNAs as a drug resistance mechanism to targeted therapies in EGFR-mutated NSCLC: current implications and future directions. Drug Resist Updates. 2019;42:1–11.
- Li Y, Chen P, Zu L, et al. MicroRNA-338-3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am J Cancer Res. 2016;6(2):127–140.
- Zhang P, Shao G, Lin X, et al. MiR-338-3p inhibits the growth and invasion of non-small cell lung cancer cells by targeting IRS2. Am J Cancer Res. 2017;7(1):53–63.
- Zhang G, Zheng H, Zhang G, et al. MicroRNA-338-3p suppresses cell proliferation and induces apoptosis of non-small-cell lung cancer by targeting sphingosine kinase 2. Cancer Cell Int. 2017;17:46.
- Tsakonas G, Ekman S. Oncogene-addicted non-small cell lung cancer and immunotherapy. J Thorac Dis. 2018;10(Suppl. 13):S1547–S1555.
- Schulze AB, Evers G, Kerkhoff A, et al. Future options of molecular-targeted therapy in small cell lung cancer. Cancers (Basel). 2019;11(5):690.
- Zaganjor E, Weil LM, Gonzales JX, et al. Ras transformation uncouples the kinesin-coordinated cellular nutrient response. Proc Natl Acad Sci U S A. 2014;111(29):10568–10573.
- Zaganjor E, Osborne JK, Weil LM, et al. Ras regulates kinesin 13 family members to control cell migration pathways in transformed human bronchial epithelial cells. Oncogene. 2014;33(47):5457–5466.
- Uchida A, Seki N, Mizuno K, et al. Regulation of KIF2A by antitumor miR-451a inhibits cancer cell aggressiveness features in lung squamous cell carcinoma. Cancers (Basel). 2019;11(2):258.
- Xie T, Li X, Ye F, et al. High KIF2A expression promotes proliferation, migration and predicts poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2018;497(1):65–72.
- Pei X, Chen SW, Long X, et al. circMET promotes NSCLC cell proliferation, metastasis, and immune evasion by regulating the miR-145-5p/CXCL3 axis. Aging (Albany NY). 2020;12(13):13038–13058.
- Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial–mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17(1):140.
- Zheng D, Haddadin S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4(6):575–586.
- Yang T, Li H, Chen T, et al. LncRNA MALAT1 depressed chemo-sensitivity of NSCLC cells through directly functioning on miR-197-3p/p120 catenin axis. Mol Cells. 2019;42(3):270–283.
- Weng L, Qiu K, Gao W, et al. LncRNA PCGEM1 accelerates non-small cell lung cancer progression via sponging miR-433-3p to upregulate WTAP. BMC Pulm Med. 2020;20(1):213.
- Li S, Niu X, Li H, et al. Circ_0000003 promotes the proliferation and metastasis of non-small cell lung cancer cells via miR-338-3p/insulin receptor substrate 2. Cell Cycle. 2019;18(24):3525–3539.
- Chen Q, Guo SM, Huang HQ, et al. Long noncoding RNA SBF2-AS1 contributes to the growth and metastatic phenotypes of NSCLC via regulating miR-338-3p/ADAM17 axis. Aging (Albany NY). 2020;12(18):17902–17920.
- Yang Z, Lin X, Zhang P, et al. Long non-coding RNA LINC00525 promotes the non-small cell lung cancer progression by targeting miR-338-3p/IRS2 axis. Biomed Pharmacother. 2020;124:109858.
- Welburn JP, Cheeseman IM. The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol Cell. 2012;23(24):4786–4795.
- Zhang X, Wang Y, Liu X, et al. KIF2A promotes the progression via AKT signaling pathway and is upregulated by transcription factor ETV4 in human gastric cancer. Biomed Pharmacother. 2020;125:109840.
- Wang J, Ma S, Ma R, et al. KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer. 2014;14(1):461.
- Wang CQ, Qu X, Zhang XY, et al. Overexpression of Kif2a promotes the progression and metastasis of squamous cell carcinoma of the oral tongue. Oral Oncol. 2010;46(1):65–69.
- Wang G, Wang Z, Yu H. Kinesin family member 2A high expression correlates with advanced tumor stages and worse prognosis in non-small cell lung cancer patients. J Clin Lab Anal. 2020;34(4):e23135.
- Gan H, Lin L, Hu N, et al. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem Funct. 2019;37(6):424–431.
- Bai Y, Xiong L, Zhu M, et al. Co-expression network analysis identified KIF2C in association with progression and prognosis in lung adenocarcinoma. Cancer Biomark. 2019;24(3):371–382.
