Abstract
Background
Long non-coding RNAs (lncRNAs) are involved in the progression of various cancers, including clear cell renal cell carcinoma (ccRCC). This study aimed to investigate the expression and prognostic value of long intergenic non-protein coding RNA (LINC) 01232 in ccRCC and preliminary explore the molecular mechanism underlying the role of LINC01232 in ccRCC progression.
Methods
Tumour tissues and adjacent normal tissues of 122 patients with ccRCC were collected in this study. The levels of LINC01232, microRNA (miR)-204-5p and RAB22A were measured by quantitative real-time PCR. The proliferation, migration and invasion of ccRCC cells were detected by cell counting kit-8 assay and Transwell assay, respectively. The interaction among LINC01232, miR-204-5p and RAB22A was confirmed by bioinformatics analysis, dual-luciferase reporter assay and Pearson correlation analysis. The association of LINC01232 and miR-204-5p with ccRCC patient survival was verified by the Kaplan–Meier method and log-rank test. The prognostic value of LINC01232 in ccRCC was confirmed by Cox regression analysis.
Results
LINC01232 expression was increased in ccRCC tumour tissues and ccRCC cells and independently predicted the prognosis of ccRCC patients. In addition, LINC01232 silencing inhibited ccRCC cell proliferation, migration and invasion. Moreover, LINC01232 served as a sponge for miR-204-5p, and miR-204-5p reduction reversed the inhibitory effect of LINC01232 silencing on ccRCC cell function. Furthermore, LINC01232 could sponge miR-204-5p, causing the elevation of RAB22A in ccRCC, thereby promoting ccRCC cell function.
Conclusion
LINC01232 may be an independent prognostic biomarker in ccRCC and plays an oncogenic role in ccRCC progression by sponging miR-204-5p and upregulating RAB22A.
Introduction
Renal cell carcinoma (RCC), a cancer that occurs in renal epithelial cells, is one of the most common and lethal malignancies [Citation1]. The most common subtype of RCC is clear cell RCC (ccRCC), which accounts for approximately 80% of adult clinical RCC cases [Citation2]. Localized ccRCC can be treated by partial or total resection, but most patients are initially diagnosed at an advanced stage, and advanced ccRCC remains a clinical challenge with a 5-year overall survival rate of less than 20% and a poor prognosis [Citation3,Citation4]. Thus, there is an urgent need to explore reliable prognostic biomarkers and novel approaches to improve the prognosis and treatment of patients with ccRCC.
Long non-coding RNAs (lncRNAs) are RNAs greater than 200 nucleotides in length that cannot encode proteins [Citation5]. At present, the roles of lncRNAs in malignant tumours have been gradually unravelled, providing some new potential targets for the treatment of diseases [Citation6–8]. However, there have been fewer functional lncRNA analyses in ccRCC. Our previous studies have demonstrated that long intergenic non-protein coding RNA (LINC) 01232 has important biological functions and clinical significance in pancreatic adenocarcinoma (PAAD) [Citation9,Citation10]. In addition, Zhao et al. have reported that LINC01232 is highly expressed in oesophageal squamous cell carcinoma (ESCC) tissues and ESCC cell lines and can regulate the biological functions of ESCC cells [Citation11]. Moreover, similar results were reported regarding LINC01232 in oral carcinoma (OC) [Citation12]. In this study, by using a bioinformatics analysis platform, we found that the expression levels of LINC01232 in ccRCC tumour tissues were significantly upregulated and significantly correlated with disease prognosis. However, until now, there has been no report on LINC01232 in ccRCC.
Therefore, the purpose of this study was to analyse the expression level of LINC01232 in ccRCC tumour tissues and ccRCC cell lines, explore the role of LINC01232 in ccRCC tumour progression using in vitro experiments, and provide a preliminary exploration of its molecular mechanisms. This study may provide a new and effective biomarker for prognosis prediction of ccRCC patients and a potential target for ccRCC therapy.
Material and methods
Patients and tissue collection
Tissue samples, including tumour tissues and adjacent normal tissues (at least 5 cm away from the tumour), were collected from 122 ccRCC patients who underwent surgical resection at Zibo Maternal and Child Health Hospital from 2010 to 2016. None of the patients had received radiotherapy or chemotherapy before surgery, and all patients were diagnosed with ccRCC by pathological examination. All tissues were collected during surgery and immediately stored in liquid nitrogen until use. Each patient has signed informed consent and the procedures for all experiments were approved by the Ethics Committee of Zibo Maternal and Child Health Hospital. All patients were followed up for five years, and survival information was recorded.
Cell culture and transfection
A human normal renal tubular epithelial cell line HK-2 and five ccRCC cell lines (Caki-1, A498, KN-41, 786-O and ClearCa-1) were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Thermo Fisher Scientific, Waltham, MA) containing 10% foetal bovine serum (FBS, Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin.
The short hairpin RNA (shRNA) against LINC01232 (sh-LINC01232), the corresponding negative control (sh-NC), microRNA (miR)-204-5p mimic, mimic NC, miR-204-5p inhibitor and inhibitor NC were synthesized by GenePharma. The sequences were as follows: sh-LINC01232, CCGCGACACGTCATCTAGAATAACTCHAGTTATTCTAGATGACGTGTCTTTTTG, and sh-NC, CCGCGGACTTGCCTCCTACACTACTCHAGTAGTGTAGGAGGCAAGTCCTTTTTG; miR-204-5p mimic, 5′-UUCCCUUUGUCAUCCUAUGCCU-3′, and mimic NC, 5′-UUCUCCGAACGUGUCACGU-3′; miR-204-5p inhibitor, 5′-AGGCAUAGGAUGACAAAGGGAA-3′, and inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′. The above fragments were respectively transfected into Caki-1 and 786-O cells using Lipofectamine 3000 (Invitrogen, USA). The pcDNA3.1 and pcDNA3.1-LINC01232 expression vectors were synthesized by GenePharma and were transfected into Caki-1 cells by Lipofectamine 3000 (Invitrogen, USA).
Bioinformatics analysis
In this study, the expression level of LINC01232 and its relationship with ccRCC patient survival in TCGA database were analysed using starBase v3.0 [Citation13] platform (http://starbase.sysu.edu.cn/index.php), and the combination of miR-204-5p with LINC01232 was predicted by this platform. In addition, by using the starBase v3.0 platform, we analysed the expression level of miR-204-5p and its correlation with survival of ccRCC patients in TCGA database, and further analysed the correlation among LINC01232, miR-204-5p and RAB22A in ccRCC patients. Moreover, starBase v3.0 platform was used to analyse the relationship of RAB22A with ccRCC patient survival in TCGA database.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA). A NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA) was utilized to verify RNA purity and concentration. Then, a Reverse Transcription kit (Thermo Fisher Scientific, Waltham, MA) was used for reverse transcription to synthesize the cDNA. The qRT-PCR was carried out using a 7500 Real-Time PCR System (Applied Biosystems, USA) with SYBR green I Master Mix kit (Invitrogen, Carlsbad, CA) for the detection of LINC01232, RAB22A and miR-204-5p levels. The levels of LINC01232 and RAB22A were normalized to GAPDH, and miR-204-5p levels were normalized to U6. Their levels were calculated using the 2−ΔΔCt method [Citation14].
Cell proliferation analysis
Proliferation of Caki-1 and 786-O cells was assessed by cell counting kit-8 (CCK-8) assay. The ccRCC cells were seeded in 96-well cell culture plates at a cell density of 3 × 103 cells/well. After incubation at 37 °C for 24, 48 and 72 h, 10 µl CCK-8 reagent was added to each well. Cells were then placed in an incubator at 37 °C for another 2 h. Then, the optical density (OD) value at 450 nm was detected using a microplate analyser (Bio-Rad Laboratories, Inc.).
Cell migration and invasion analysis
The migration and invasion abilities of Caki-1 and 786-O cells were measured using Transwell assays. For invasion assays, Transwell chambers were precoated with Matrigel (Corning, USA), and Transwell chambers without Matrigel precoating were used for migration assays. For each analysis, cells (5 × 104) were added to the upper chamber with serum-free medium; meanwhile, the lower chamber was filled with serum medium. After 24-h incubation at 37 °C, cells on the upper chamber membrane were wiped away. Cells on the bottom side were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 20 min at room temperature. The number of cells was counted under an inverted light microscope (Olympus Corporation, Tokyo, Japan).
Dual-luciferase reporter assay
The wild-type (WT)-LINC01232, mutant-type (MUT)-LINC01232, WT-RAB22A and MUT-RAB22A were inserted into the pmirGLO dual luciferase reporter vector (Promega, WI). The WT or MUT vector was then co transfected with miR-204-5p mimic or mimic NC into Caki-1 cells using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA). After 48 h, the luciferase activity was measured using the dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. All procedures followed the manufacturer’s instructions.
Statistical analysis
Data analysis results were expressed as mean ± SD. All analyses were performed by SPSS 22.0 (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad Software, Inc.), and repeated independently at least three times. Differences in measurement data between two groups and among multiple groups were analysed by using Student’s t-test and one-way ANOVA followed by Tukey’s post hoc test, respectively. Comparison between categorical variables was conducted by Chi-square test. Pearson correlation analysis was performed to analyse the correlation between two variables (including LINC01232, RAB22A and miR-204-5p). Kaplan–Meier survival curves and log-rank tests were utilised to investigate the relationship of LINC01232 and miR-204-5p expression with overall survival of ccRCC patients. Cox regression analysis was used to identify the prognostic value of LINC01232 in ccRCC patients. p < .05 indicated a statistically significant difference.
Results
Overexpression of LINC01232 in ccRCC tissues and cell lines
The expression data of 535 ccRCC tissues and 72 normal tissues from TCGA database were analysed in starBase v3.0 platform, and the results indicated that LINC01232 was significantly upregulated in ccRCC tissues compared with that in normal tissues (<.001). The expression of LINC01232 in other RCC subtypes from TCGA dataset was analysed in starBase v3.0 platform and presented in . LINC01232 expression in kidney chromophobe (KICH) was decreased (, p < .001), and LINC01232 expression in kidney renal papillary cell carcinoma (KIRP) was increased (, p < .001). In addition, the expression of LINC01232 in other types of cancer was also assessed using the TCGA dataset (). In addition to ccRCC and KIRP, the significantly increased LINC01232 expression was also found in colon adenocarcinoma (COAD; , p < .001), oesophageal carcinoma (ESCA; , p < .01), lung squamous cell carcinoma (LUSC; , p < .001) and prostate adenocarcinoma (PRAD; , p < .001). Consistent with this result, tumour tissues from 122 ccRCC patients in this study had significantly higher LINC01232 levels than adjacent normal tissues (<.001), and LINC01232 expression was similarly increased in ccRCC cell lines compared with that in normal cells (, all p < .001).
Figure 1. Overexpression of LINC01232 in ccRCC tissues and cell lines. (A) LINC01232 expression in 535 ccRCC tissues and 72 normal tissues from TCGA database by starBase v3.0 analysis. (B) LINC01232 expression in tumour tissues and normal tissues from 122 ccRCC patients from our study cohort (fold change is 0.89). (C) LINC01232 expression in ccRCC cell lines and normal renal tubular epithelial cell line (fold changes are 2.64, 2.25, 1.98, 2.54 and 1.75, respectively). ***p < .001 vs. ccRCC tissues from TCGA database or normal tissues from 122 ccRCC patients or HK-2. LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
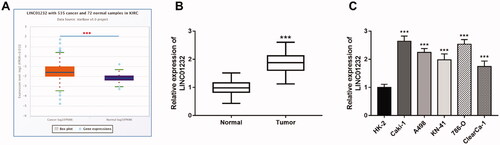
Figure 2. LINC01232 is associated with overall survival of ccRCC patients. (A) According to the analysis by starBase, high LINC01232 was associated with poor overall survival of ccRCC patients (log-rank p = .002). (B) The results of our study cohort revealed that patients with high LINC01232 level had short survival time than the patients with low LINC01232 level (log-rank p = .006). LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
Association of LINC01232 with the clinicopathological characteristics of ccRCC
In this study, the median value of LINC01232 levels (1.85) was used to divide high and low levels of LINC01232. The analysis results of showed that LINC01232 was significantly correlated with tumour size (p = .020), lymph node metastasis (p = .006) and TNM stage (p = .001). In addition, no association was found between LINC01232 expression and age or gender (all p > .05).
Table 1. Association of LINC01232 with the clinicopathological characteristics of ccRCC.
High LINC01232 is associated with poor overall survival in ccRCC patients
According to the results of starBase analysis, the overall survival of ccRCC patients with high LINC01232 levels was significantly worse than that of patients with low LINC01232 levels (, log-rank p = .002). In our study cohort, we found that high level of LINC01232 was significantly associated with short survival time, which was consistent with the results of starBase analysis (, log-rank p = .006). Cox regression analysis was performed and the results are shown in . The results of univariate Cox analysis revealed that lymph node metastasis, TNM stage and LINC01232 were associated with overall survival in ccRCC patients. The significant variables from the univariate analysis were then included in the multivariate analysis. Multivariate Cox analysis results demonstrated that TNM stage [hazard ratio (HR)=2.084, 95% confidence interval (CI)=1.280–2.841, p = .007] and LINC01232 (HR = 2.207, 95% CI = 1.433–3.086, p < .001) were independently correlated with the overall survival of ccRCC patients.
Table 2. Cox regression analysis results for patients with ccRCC.
LINC01232 reduction inhibits ccRCC cell proliferation, migration and invasion
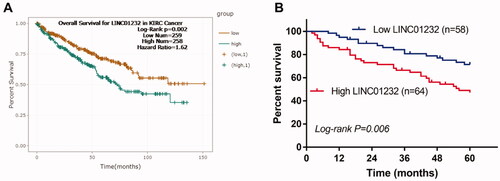
As presented in , sh-LINC01232 significantly suppressed the levels of LINC01232 in the Caki-1 and 786-O cells (all p < .001). In addition, the proliferation of Caki-1 and 786-O cells was suppressed by LINC01232 reduction (, all p < .01). In Caki-1 and 786-O cells, the migration (, all p < .001) and invasion (, all p < .001) were all inhibited by LINC01232 reduction.
Figure 3. Effects of LINC01232 on the proliferation, migration and invasion of ccRCC cells. (A) LINC01232 expression was inhibited by sh-LINC01232 in Caki-1 (fold change is 0.22) and 786-O cells (fold change is 0.25). (B–D) LINC01232 silencing inhibited the proliferation, migration and invasion of Caki-1 and 786-O cells. **p < .01, ***p < .001 vs. Control. sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
LINC01232 sponges miR-204-5p in ccRCC
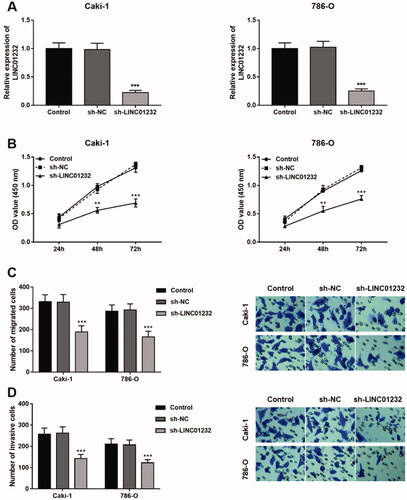
Through the starBase platform, the binding site of LINC01232 to miR-204-5p was predicted (), and the expression levels of LINC01232 and miR-204-5p were also found to be significantly negatively correlated in ccRCC (, r= −0.220, p < .001). As shown in , the luciferase reporter assay results showed that the luciferase activity of the WT-LINC01232 group was significantly inhibited by miR-204-5p overexpression (p < .05), and no change was found in the luciferase activity of the MUT-LINC01232 group. In addition, sh-LINC01232 inhibited and pcDNA3.1-LINC01232 promoted relative expression of LINC01232 (, all p < .001). Moreover, the expression of miR-204-5p was promoted by LINC01232 silencing and was inhibited by LINC01232 overexpression in Caki-1 cells (, all p < .001). Analysis of the miR-204-5p expression quantity data in starBase showed that miR-204-5p was significantly lower in ccRCC tissues than that in normal tissues (<.001), and the overall survival of ccRCC patients with low miR-204-5p levels was significantly poor (, log-rank p < .001). Analysis of a study of 122 patients with ccRCC similarly demonstrated that miR-204-5p levels were downregulated in ccRCC tumour tissues compared with that in normal tissues (<.001) and significantly negatively correlated with LINC01232 levels in tumour tissues (, r= −0.600, p < .001), and that patients with low miR-204-5p levels had significantly lower five-year survival rates (, log-rank p = .002).
Figure 4. LINC01232 sponges miR-204-5p in ccRCC. (A) The binding site of LINC01232 to miR-204-5p was predicted. (B) LINC01232 was found to be negatively correlated with miR-204-5p in ccRCC by starBase platform (r= −0.220, p < .001). (C) In Caki-1 cells, relative luciferase activity was significantly inhibited by miR-204-5p upregulation in WT-LINC01232 group. (D) LINC01232 expression was downregulated by sh-LINC01232 and was upregulated by pcDNA3.1-LINC01232 (fold changes are 0.21 and 2.91). (E) miR-204-5p levels were promoted by LINC01232 silencing and were suppressed by the overexpression of LINC01232 in Caki-1 cells (fold changes are 1.97 and 0.4). (F) Through starBase platform, miR-204-5p was significantly downregulated in ccRCC. (G) Through the starBase platform, ccRCC patients with low miR-204-5p levels had a poor overall survival (log-rank p < .001). (H) miR-204-5p levels in ccRCC tumour tissues and normal tissues (fold change is 0.39). (I) miR-204-5p levels were negatively correlated with LINC01232 levels in tumour tissues (r= −0.600, p < .001). (J) Patients with low miR-204-5p levels had a poor five-year survival (log-rank p = .002). *p < .05, ***p < .001 vs. Control or ccRCC tissues from TCGA database or normal tissues from 122 ccRCC patients. WT: wide-type; MUT: mutant-type; sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
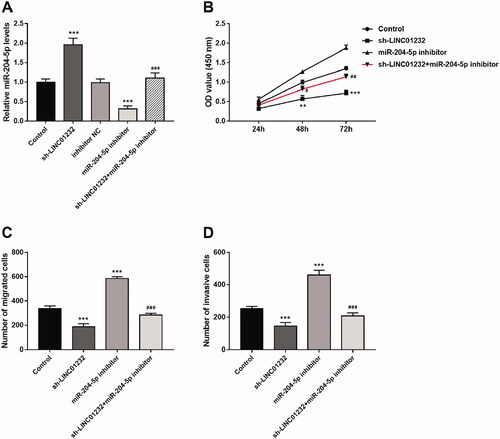
miR-204-5p mediates the regulatory effects of LINC01232 on ccRCC proliferation, migration and invasion
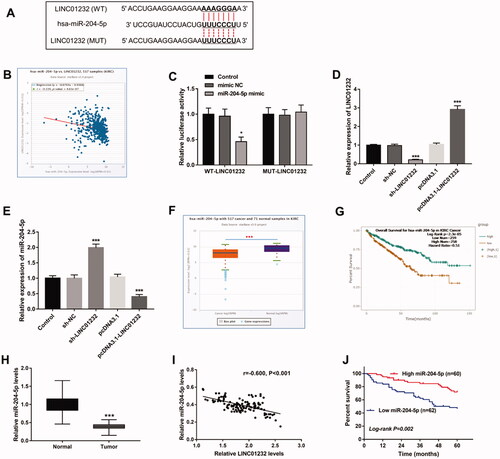
As shown in , miR-204-5p levels were significantly promoted by sh-LINC01232 and repressed by miR-204-5p inhibitor in Caki-1 cells, and miR-204-5p inhibitor reversed the promoting effect of sh-LINC01232 on miR-204-5p expression (all p < .001). In addition, downregulation of miR-204-5p was found to promote the proliferation, migration and invasion of Caki-1 cells, and miR-204-5p downregulation reversed the inhibitory effects of LINC01232 silencing on Caki-1 cell proliferation, migration and invasion (), all p < .05).
Figure 5. miR-204-5p mediates the effects of LINC01232 on ccRCC cell proliferation, migration and invasion. (A) The upregulation of miR-204-5p caused by LINC01232 silencing was reversed by miR-204-5p downregulation in Caki-1 cells (fold changes are 1.96, 0.31 and 1.10, respectively). (B–D) miR-204-5p downregulation reversed the inhibitory effects of LINC01232 silencing on Caki-1 cell proliferation, migration and invasion. **p < .01, ***p < .001 vs. Control; #p < .05, ##p < .01, ###p < .001 vs. sh-LINC01232. sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
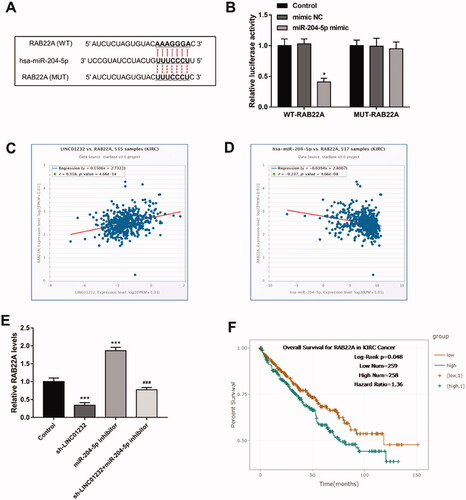
LINC01232 positively regulates RAB22A through sponging miR-204-3p in ccRCC
As shown in , the 3′-UTR region of RAB22A had a binding site for miR-204-5p. Luciferase reporter assay results illustrated that miR-204-5p overexpression markedly inhibited the luciferase activity of the WT-RAB22A group (<.05). Analysis of the molecular correlations of the starBase platform demonstrated that LINC01232 was positively correlated with RAB22A in ccRCC (, r = 0.318, p < .001) and miR-204-5p was negatively correlated with RAB22A in ccRCC (, r= −0.237, p < .001). As presented in , silencing of LINC01232 significantly suppressed, whereas downregulation of miR-204-5p significantly promoted the level of RAB22A in Caki-1 cells (all p < .001). Besides, we observed that RAB22A inhibition originally caused by LINC01232 silencing was relieved by miR-204-5p downregulation in Caki-1 cells (p < .001). Through the results of the analysis of TCGA database by starBase platform, we found that high RAB22A level was associated with poor overall survival in ccRCC patients (, log-rank p = .048).
Figure 6. LINC01232 positively regulates RAB22A through sponging miR-204-3p in ccRCC. (A) The binding sites of miR-204-5p and RAB22A were predicted. (B) miR-204-5p upregulation significantly inhibited relative luciferase activity in the WT-RAB22A group. (C–D) Through the starBase platform, in ccRCC, LINC01232 was positively correlated with RAB22A (r = 0.318, p < .001) and miR-204-5p was negatively correlated with RAB22A (r= −0.237, p < .001). (E) miR-204-5p downregulation reversed the suppressive effects of LINC01232 silencing on RAB22A levels in Caki-1 cells (fold changes are 0.34, 1.87 and 0.77, respectively). (F) Through the starBase platform, high RAB22A levels were associated with poor overall survival in ccRCC patients (log-rank p = .048). *p < .05, ***p < .001 vs. Control; ###p < .001 vs. sh-LINC01232. WT: wide-type; MUT: mutant-type; sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
Discussion
This study is the first to investigate the role of LINC01232 in ccRCC. Our study demonstrated that LINC01232 was upregulated in ccRCC tumour tissues and cells and could independently predict the prognosis of ccRCC patients. In addition, it was further found that knockdown of LINC01232 inhibited the proliferation, migration and invasion of ccRCC cells in vitro. Moreover, mechanistic studies indicated that LINC01232 positively regulated the levels of RAB22A via sponging miR-204-3p to affect ccRCC tumour progression.
Various types of cancer have been found to be affected by lncRNAs [Citation6,Citation15,Citation16]. In addition, some lncRNAs have been revealed to play crucial roles in ccRCC. For example, it has been reported that lncRNA small nucleolar RNA host gene 16 (SNHG16) expression is increased in RCC tissues and ccRCC cells, and the function of ccRCC cells is suppressed by SNHG16 knockdown [Citation17]. Yang et al. have shown that lncRNA homo sapiens HLA complex group (HCG) 18 promotes the development and progression of ccRCC [Citation18]. By using the starBase platform, LINC01232 showed a significant increase in ccRCC. In our study cohort, we also found a markedly higher level of LINC01232 in ccRCC tumour tissues and cell lines, and LINC01232 was markedly correlated with tumour size, lymph node metastasis and TNM stage. Besides, LINC01232 has been found to be involved in other cancers, such as PAAD [Citation9], pancreatic cancer (PC) [Citation19] and ESCC [Citation11]. PAAD is a type of PC that begins in the ducts of the pancreas. As the most common form of PC, PAAD accounts for approximately 95% of all pancreatic malignancies. Therefore, LINC01232 may be involved in the progression of ccRCC. Further cellular experimental results demonstrated that LINC01232 knockdown suppressed the proliferation, migration and invasion of ccRCC cells. In addition, inhibition of LINC01232 can inhibit ESCC cell proliferation, migration and invasion in vitro [Citation11]. Li et al. have shown that LINC01232 regulates cell proliferation and migration in PAAD, suggesting an oncogenic role of LINC01232 for PAAD [Citation9]. Thus, LINC01232 may function as a tumour promotor in ccRCC progression and pathogenesis.
Considering the important role of LINC01232 in ccRCC progression, the clinical significance of LINC01232 in ccRCC was explored. lncRNAs have been widely reported as prognostic predictors in different types of cancer [Citation20–22]. In addition, some lncRNAs have been reported to be of value for predicting the prognosis of ccRCC, such as lncRNA AGAP2 antisense RNA 1 (AGAP2-AS1) [Citation23] and lncRNA colorectal neoplasia differentially expressed (CRNDE) [Citation24]. The results of the analysis of the TCGA database and our cohort indicated that LINC01232 was independently associated with survival in ccRCC patients. In addition, LINC01232 has been demonstrated to serve as an independent prognostic predictor for PAAD patients [Citation10] and PC patients [Citation19]. Moreover, Chen et al. have shown that LINC01232 levels are associated with shorter overall survival of OC patients [Citation12]. Thus, LINC01232 may be an independent prognostic biomarker for patients with ccRCC.
Mechanically, an increasing number of studies have shown that lncRNAs can serve as competing endogenous RNAs (ceRNAs) through sponging miRNAs in cancer progression, thereby releasing these miRNAs bound mRNAs from degradation [Citation25]. Some lncRNAs have been found to sponge miRNAs to affect the progression of cancers, including ccRCC [Citation17,Citation18,Citation26]. In addition, LINC01232 can promote disease progression through sponging miR-654-3p in ESCC [Citation11]. Du et al. have reported that LINC01232 may sponge miR-370-5p, miR-654-3p and miR-204-5p in PAAD [Citation10]. In this study, we found that LINC01232 could directly bind to miR-204-5p. LINC01232 knockdown promoted and LINC01232 overexpression repressed the level of miR-204-5p in ccRCC cells. Several studies have shown that miR-204-5p is aberrant in ccRCC [Citation27,Citation28] and is involved in ccRCC progression [Citation29]. Consistently, the results of TCGA dataset analysis and our cohort analysis demonstrated that miR-204-5p was decreased in ccRCC tissues and associated with the survival of ccRCC patients, indicating that miR-204-5p may be involved in ccRCC progression. More importantly, miR-204-5p knockdown was found to promote ccRCC cell function and reverse the effect of LINC01232 on ccRCC cell function. Notably, Wu et al. have reported that lncRNA SNHG4 contributes to RCC progression by sponging miR-204-5p [Citation27]. Therefore, LINC01232 may play an oncogenic role in ccRCC by sponging miR-204-5p.
RAB22A has been shown to act as a direct functional target of miR-204-5p, the levels of which are regulated by miR-204-5p in several cancers, such as gastric cancer [Citation30] and glioma [Citation31]. In addition, it is noteworthy that a study by Xiong et al. has revealed that miR-204-5p can suppress RCC cell proliferation and invasion by suppressing RAB22A [Citation32]. By starBase platform analysis and our cohort analysis, we found that miR-204-5p could directly bind to RAB22A and negatively regulate the level of RAB22A. In addition, LINC01232 was found to be positively correlated with RAB22A by starBase platform analysis. Moreover, the repression of RAB22A, caused by LINC01232 knockdown, was abolished by miR-204-5p downregulation. Furthermore, ccRCC patients with high RAB22A were found to have a poor survival prognosis by the analysis of TCGA database in starBase platform. Thus, RAB22A expression may be associated with the prognosis of ccRCC patients, which will be verified in our future study. The above data indicated that LINC01232 may regulate the progression of ccRCC by regulating the miR-204-5p/RAB22A axis. Notably, RAB22A was shown to have pro-immunogenic functions in cancer cells [Citation33]. In line with these findings, overexpression of immune-related genes such as PD-L1 was shown to mark high-risk subgroups of ccRCC patients in several cohorts [Citation34], including TCGA ccRCC patients [Citation35]. Given the fact that immune therapy is a crucial part of systemic treatment for metastatic RCC, this information appears relevant and underlines the potential role of the LINC01232/miR-204-5p/RAB22A axis in ccRCC.
However, this study has some limitations. First, the study sample size was small, and a large sample should be used for further studies. Second, in vivo experiments were not performed in our study, and further investigation of the role of LINC01232 in ccRCC in vivo is warranted. Third, when performing knockdown with shRNA, at least two separate shRNA designs should be used in parallel to study the gene knockdown effects. However, this study only used a shRNA, which is a limitation. Thus, we will use at least two separate shRNA designs for knockdown in future studies. Fourth, miR-204-5p has been found to target other downstream target genes, such as Sirtuin-1 (SIRT1) [Citation36] and runt-related transcription factor 2 (RUNX2) [Citation28], which will be studied in future studies.
In conclusion, this study indicates that LINC01232, which is upregulated in ccRCC tissues and cells, may act as an independent prognostic biomarker for ccRCC patients. In addition, LINC01232 facilitates ccRCC tumour progression via sponging miR-204-5p and increasing RAB22A, suggesting the critical role of the LINC01232/miR-204-5p/RAB22A axis in ccRCC progression. Thus, our study may provide potential prognostic biomarker and therapeutic targets for ccRCC.
Ethics approval and consent to participate
The experimental procedures were all in accordance with the guideline of the Ethics Committee of Zibo Maternal and Child Health Hospital and has approved by the Ethics Committee of Zibo Maternal and Child Health Hospital, and this study follows the Declaration of Helsinki.
A signed written informed consent was obtained from each patient.
Consent for publication
Written informed consent for publication was obtained from each participant.
Author contributions
QL carried out the research design and conception; QL and CL analysed and interpreted the data regarding, performed the examination of cell; QL and CL wrote and revised the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download ()Acknowledgements
The TCGA data (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga/citing-tcga) were used in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Escudier B, Porta C, Schmidinger M, et al. Renal cell carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up†. Ann Oncol. 2019;30(5):706–720.
- Wolf MM, Kimryn Rathmell W, Beckermann KE. Modeling clear cell renal cell carcinoma and therapeutic implications. Oncogene. 2020;39(17):3413–3426.
- Xue J, Zhu S, Qi F, et al. RUNX1/miR-582-5p pathway regulates the tumor progression in clear cell renal cell carcinoma by targeting COL5A1. Front Oncol. 2021;11:610992.
- Petitprez F, Ayadi M, de Reynies A, et al. Review of prognostic expression markers for clear cell renal cell carcinoma. Front Oncol. 2021;11:643065.
- Li Y, Huo FF, Wen YY, et al. Screening and identification of an immune-associated lncRNA prognostic signature in ovarian carcinoma: evidence from bioinformatic analysis. Biomed Res Int. 2021;2021:6680036.
- Wang B, Tang D, Liu Z, et al. LINC00958 promotes proliferation, migration, invasion, and epithelial-mesenchymal transition of oesophageal squamous cell carcinoma cells. PLoS One. 2021;16(5):e0251797.
- Song K, Yu P, Zhang C, et al. The LncRNA FGD5-AS1/miR-497-5p axis regulates septin 2 (SEPT2) to accelerate cancer progression and increase cisplatin-resistance in laryngeal squamous cell carcinoma. Mol Carcinog. 2021; 60:469-480
- Xie FW, Liu JC. LncRNA SNHG12 regulates the miR-101-3p/CUL4B axis to mediate the proliferation, migration and invasion of non-small cell lung cancer. Kaohsiung J Med Sci. 2021; 37:664-674.
- Li Q, Lei C, Lu C, et al. LINC01232 exerts oncogenic activities in pancreatic adenocarcinoma via regulation of TM9SF2. Cell Death Dis. 2019;10(10):698.
- Du W, Lei C, Wang Y, et al. LINC01232 sponges multiple miRNAs and its clinical significance in pancreatic adenocarcinoma diagnosis and prognosis. Technol Cancer Res Treat. 2021;20:1533033820988525.
- Zhao M, Cui H, Zhao B, et al. Long intergenic non‑coding RNA LINC01232 contributes to esophageal squamous cell carcinoma progression by sequestering microRNA‑654‑3p and consequently promoting hepatoma‑derived growth factor expression. Int J Mol Med. 2020;46(6):2007–2018.
- Chen M, Xu X, Ma H. Identification of oncogenic long noncoding RNAs CASC9 and LINC00152 in oral carcinoma through genome-wide comprehensive analysis. Anticancer Drugs. 2019;30(4):356–362.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- He L, Wang J, Zhou L, et al. LncRNA PCAT18 promotes non-small cell lung cancer progression by sponging miR-4319. Cancer Manag Res. 2021;13:3761–3774.
- Ma X, Li Y, Song Y, et al. Long noncoding RNA CCDC26 promotes thyroid cancer malignant progression via miR-422a/EZH2/Sirt6 axis. Onco Targets Ther. 2021;14:3083–3094.
- Cheng T, Shuang W, Ye D, et al. SNHG16 promotes cell proliferation and inhibits cell apoptosis via regulation of the miR-1303-p/STARD9 axis in clear cell renal cell carcinoma. Cell Signal. 2021;84:110013.
- Yang Y, Gong P, Yao D, et al. LncRNA HCG18 promotes clear cell renal cell carcinoma progression by targeting miR-152-3p to upregulate RAB14. Cancer Manag Res. 2021;13:2287–2294.
- Meng LD, Shi GD, Ge WL, et al. Linc01232 promotes the metastasis of pancreatic cancer by suppressing the ubiquitin-mediated degradation of HNRNPA2B1 and activating the A-Raf-induced MAPK/ERK signaling pathway. Cancer Lett. 2020;494:107–120.
- Geers R, Ingram DL, Dauncey MJ. Time course of the change in nuclear 3,5,3'-triiodothyronine receptors of skeletal muscle in relation to energy intake. Q J Exp Physiol. 1988;73(3):447–449.
- Zhang Y, Sun Y, Ding L, et al. Long non-coding RNA LINC00467 correlates to poor prognosis and aggressiveness of breast cancer. Front Oncol. 2021;11:643394.
- Jia X, Wei L, Zhang Z. NEAT1 overexpression indicates a poor prognosis and induces chemotherapy resistance via the miR-491-5p/SOX3 signaling pathway in ovarian cancer. Front Genet. 2021;12:616220.
- Gao L, Zhao A, Wang X. Upregulation of lncRNA AGAP2-AS1 is an independent predictor of poor survival in patients with clear cell renal carcinoma. Oncol Lett. 2020;19(6):3993–4001.
- Ding C, Han F, Xiang H, et al. LncRNA CRNDE is a biomarker for clinical progression and poor prognosis in clear cell renal cell carcinoma. J Cell Biochem. 2018;119(12):10406–10414.
- Jiang Y, Li W, Yan Y, et al. LINC01094 triggers radio-resistance in clear cell renal cell carcinoma via miR-577/CHEK2/FOXM1 axis. Cancer Cell Int. 2020;20:274.
- Huang Y, Zheng Y, Shao X, et al. Long non-coding RNA TPT1-AS1 sensitizes breast cancer cell to paclitaxel and inhibits cell proliferation by miR-3156-5p/caspase 2 axis. Hum Cell. 2021; 34:1244-1254.
- Wu J, Liu T, Sun L, et al. Long noncoding RNA SNHG4 promotes renal cell carcinoma tumorigenesis and invasion by acting as ceRNA to sponge miR-204-5p and upregulate RUNX2. Cancer Cell Int. 2020;20:514.
- Shu X, Hildebrandt MA, Gu J, et al. MicroRNA profiling in clear cell renal cell carcinoma tissues potentially links tumorigenesis and recurrence with obesity. Br J Cancer. 2017;116(1):77–84.
- Gowrishankar B, Ibragimova I, Zhou Y, et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15(3):329–341.
- Zhang B, Yin Y, Hu Y, et al. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol. 2015;32(1):331.
- Xia Z, Liu F, Zhang J, et al. Decreased expression of MiRNA-204-5p contributes to glioma progression and promotes glioma cell growth, migration and invasion. PLoS One. 2015;10(7):e0132399.
- Xiong F, Liu K, Zhang F, et al. MiR-204 inhibits the proliferation and invasion of renal cell carcinoma by inhibiting RAB22A expression. Oncol Rep. 2016;35(5):3000–3008.
- Mayorga LS, Cebrian I. Rab22a: a novel regulator of immune functions. Mol Immunol. 2019;113:87–92.
- Flaifel A, Xie W, Braun DA, et al. PD-L1 expression and clinical outcomes to cabozantinib, everolimus, and sunitinib in patients with metastatic renal cell carcinoma: analysis of the randomized clinical trials METEOR and CABOSUN. Clin Cancer Res. 2019;25(20):6080–6088.
- Marquardt A, Solimando AG, Kerscher A, et al. Subgroup-independent mapping of renal cell carcinoma-machine learning reveals prognostic mitochondrial gene signature beyond histopathologic boundaries. Front Oncol. 2021;11:621278.
- Zhang M, Cao M, Kong L, et al. MiR-204-5p promotes lipid synthesis in mammary epithelial cells by targeting SIRT1. Biochem Biophys Res Commun. 2020;533(4):1490–1496.
