Abstract
Objectives
To evaluate the efficiency of chromosomal microarray analysis (CMA) in the prenatal diagnosis of foetuses with isolated absent or hypoplastic nasal bone (NB) in the first and second trimester.
Methods
From January 2015 to April 2021, foetuses with isolated absent or hypoplastic NB who received invasive prenatal diagnosis were enrolled. The results of CMA were analysed
Results
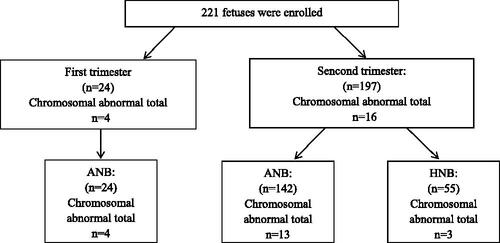
There were 221 foetuses, including 166 cases with isolated absent NB and 55 cases with isolated hypoplastic NB. Twenty-four foetuses (10.9%, 24/221) had an ultrasonic diagnosis in the first trimester and 197 (89.1%, 197/221) had a ultrasonic diagnosis in the second trimester. The overall diagnostic yield of CMA was 9.0% (20/221). Aneuploidies were detected in 13 (5.9%, 13/221) foetuses, including 10 Down syndrome, 2 Klinefelter's syndrome and 1 trisomy 18. Pathogenic copy number variations (CNVs) were detected in seven foetuses (3.2%, 7/221). In addition, variants of unknown significance (VOUS) were detected in four foetuses. The foetuses with isolated absent NB had a higher detection rate of chromosome abnormality than the isolated hypoplastic NB, but the difference was not significant in the statistical analysis (10.2% vs. 5.5%, χ2 =0.642, p = .423). No significant difference was observed in the detection rate between the first trimester and the second trimester (16.6% vs. 8.1%, χ2 = 1.002, p = .317, Chi-square test).
Conclusion
CMA can increase the diagnostic yield of chromosome abnormality, especially pathogenic CNVs for foetuses with isolated absent or hypoplastic NB. CMA should be recommended when isolated absent or hypoplastic NB is suspected antenatally.7
Introduction
Absent or hypoplastic foetal nasal bone (NB) found in both first and second trimester has been proven to be one of the strongest markers for Down syndrome [Citation1–3]. Absent or hypoplastic foetal NB is also associated with other common aneuploidies such as trisomy 18, trisomy 13 and Turner syndrome. Rare conditions such as Cri du chat (5p-) syndrome, Wolf-Hirshhorn syndrome (4p-) and Fryns Syndrome have also been reported [Citation4–6].
Chromosomal microarray analysis (CMA), which is capable of simultaneously detecting numerical chromosomal abnormalities and submicroscopic chromosomal imbalances at the whole-genome level, has been applied to identify chromosomal abnormalities in foetuses with structural abnormalities [Citation7–9]. It is well known that if additional anomalies are detected, CMA should be offered to the foetuses with non-isolated absent or hypoplastic foetal NB since the risk of microdeletion/microduplication syndromes is increased [Citation10–13]. However, data on the yield of CMA for isolated absent or hypoplastic foetal NB is controversial. Lostchuck et al. studied 80 cases of isolated hypoplastic foetal NB and showed no cases of pathogenic copy number variations (CNVs) detected [Citation14]. However, recent study with small sample size reported that the rate of pathogenic CNVs was 5.45% (3/55) in the foetuses with isolated absence or hypoplasia NB [Citation15]. As a result, should CMA be offered to isolated absent or hypoplastic foetal NB remain unclear. Therefore, the aim of this study was to evaluate the efficiency of CMA in the prenatal diagnosis of foetuses with isolated absent or hypoplastic NB, to provide further practical evidence for pre-testing consultation.
Materials and methods
From January 2015 to April 2021, cases with isolated absent or hypoplastic foetal NB detected by ultrasound, whose parents opted to have an invasive prenatal diagnosis by CMA were included in this study. The study was approved by our institutional review board and clinical research ethics committee.
Definitions were the following: (1) Diagnosis of absent NB in the first trimester is considered when NB is not visualized on a mid-sagittal view of the profile. (2) Diagnosis of absent NB in the second trimester was considered when NB is not visualized on any appropriate view (including sagittal, transverse and coronal sections). (3) Diagnosis of hypoplastic NB in the second trimester was defined as one or both sides of the nasal bone length below the 2.5th percentile of Chinese population [Citation16], including those with unilateral absence of nasal bone.
Following prenatal detection of an absent NB in the first trimester, detailed anomaly scans were scheduled in the second trimester to confirm persistence of it and assess for associated anomalies. Foetus was considered isolated if no other associated anomalies including soft markers (such as increased nuchal translucency (NT)) or structural abnormality were noted.
Following prenatal detection of an absent or hypoplastic foetal NB in the second trimester, a systematic sonographic assessment for associated anomalies was performed. Foetus was considered isolated if no other associated anomalies including soft markers (such as single umbilical artery) or structural abnormality were noted. All sonographic findings were confirmed by experienced sonologists dedicated to obstetric sonography. Therefore, the classifications in this study are based specifically upon the phenotype at initial presentation before invasive prenatal diagnosis ().
Figure 1. Prenatal diagnosis for foetuses with prenatally diagnosed with isolated absent or hypoplastic nasal bone.
Prenatal genetic testing was recommended for the parents, and the potential benefit and risk of invasive prenatal diagnosis and CMA were explained. Written informed consent was obtained before invasive test.
Microarray analyses were performed using a high-resolution genotyping single nucleotide polymorphism microarray, Affymetrix CytoScan 750 K Array (Affymetrix, Santa Clara, CA, USA). CNVs were identified based on associated records of the human reference genome 37(NCBI37hg19) of the National Centre for Biotechnology Information. Data were analyzed in accordance with American College of Medical Genetics guidelines.
Statistical analysis
Quantitative variables are expressed as the mean ± standard deviation, and categorical variables are expressed as the frequency and percentage. Fisher’s Exact test or χ2 were used to test the differences between CMA yield in relation to different parameters and compared to the background risk. p < .05 was considered statistically significant.
Results
There were 1267 foetuses with prenatally diagnosed with isolated absent or hypoplastic NB during the study period. Totally, 221 cases chose invasive prenatal diagnosis by CMA, including 166 cases with isolated absent NB and 55 cases with isolated hypoplastic NB. Twenty-four foetuses (10.9%, 24/221) had an ultrasonic diagnosis in the first trimester and 197 (89.1%, 197/221) had an ultrasonic diagnosis in the second trimester. The mean maternal age was 24.0 ± 4.0 years. Mean gestational age at diagnosis was 29.4 ± 5.3 weeks. Overall, 3.6% (8/221) of prenatal samples were performed following chorionic villus sampling, 67.4% (149/221) from amniocentesis and 29.0% (64/221) from cordocentesis sampling.
The overall diagnostic yield of CMA testing for foetuses with isolated absent or hypoplastic NB was 9.0% (20/221). Aneuploidies were detected in 13 (5.9%, 13/221) foetuses, including 10 Down syndrome (4.5%, 10/221), 2 Klinefelter's syndrome (0.9%, 2/221) and 1 trisomy 18 (0.5%, 1/221) (). Pathogenic CNVs were detected in seven foetuses (3.2%, 7/221). In addition, VOUS were detected in 4 foetuses (1.8%, 4/221). The details of the identified pathogenic CNVs and VOUS are presented in and .
Table 1. Aneuploidies in foetuses with isolated absent or hypoplastic nasal bone.
Table 2. Pathogenic CNVs in foetuses with isolated absent or hypoplastic nasal bone.
Table 3. VOUS in foetuses with isolated absent or hypoplastic nasal bone.
Seven pathogenic CNVs () sized from 713 kb to 4.9 Mb. The pathogenic CNVs were included Xp22.33 or Yp11.32 microdeletion, 14q22.1q22.3 microdeletion, 6p21.1p12.3 microdeletion, 1q21.1 microduplication, Xp22.31 microdeletion, 1p36 microdeletion and 16p11.2 microdeletion ().
In 166 foetuses with isolated absent NB, there were 13 cases of aneuploidies and 4 cases of pathogenic CNVs, and the chromosome abnormality rate was 10.2% (17/166). There were three cases of pathogenic CNVs in isolated hypoplastic NB. The foetuses with isolated absent NB had a higher detection rate of chromosome abnormality than the isolated hypoplastic NB group, but the difference was not significant in the statistical analysis (10.2% vs. 5.5% %, χ2 =0.642, p = .423).
Four foetuses (4/24, 16.7%) with chromosomal abnormalities were identified in the first trimester, all were aneuploidies. Sixteen foetuses with chromosomal abnormalities (16/197, 8.1%) were detected in the second trimester, including nine aneuploidies and seven pathogenic CNVs. No significant difference was observed in the detection rate between the first trimester and the second trimester (16.7% vs. 8.1%, χ2 =1.002, p = .317).
Discussion
Previous studies have demonstrated that the absence or hypoplasia of NB may be one of the strongest ultrasound markers for Down syndrome and other chromosomal abnormalities in both the first and second trimesters. However, studies of foetuses with isolated absent or hypoplastic NB by CMA were limited. We specifically focussed on CMA as first-tier testing in foetuses with isolated absent or hypoplastic NB and attempted to elaborate the relationship between pathogenic CNVs and isolated absent or hypoplastic NB. We identified chromosomal abnormalities in 9.0% foetuses, including 13 aneuploidies and 7 pathogenic CNVs. Down syndrome was the most common chromosomal abnormalities in our study and detected in 10 (4.5%) foetuses. This result further supports the idea that absent or hypoplastic foetal NB is a strong marker for Down syndrome.
Despite an increased diagnostic yield, the use of CMA in foetuses with isolated absent or hypoplastic NB is still controversial. Lostchuck et al. reported 80 cases for isolated hypoplastic NB. There were no cases of pathogenic CNVs in the 47/80 cases that were analyzed by CMA [Citation14]. In contrast, Gu et al. reported their tertiary-centre experience with isolated hypoplastic NB and found a non-trisomy-21 abnormality in 2 of the 39 foetuses, resulting in a frequency of pathogenic CNVs in isolated hypoplastic NB of 5.1% [Citation15]. Du et al. reported 42 cases with isolated absence NB during the second trimester and found 1 had a microdeletion [Citation17]. Wu et al. studied 111 cases with abnormal foetal NB (59 cases with absence NB and 52 cases with hypoplastic NB) and found 4 foetuses with pathogenic CNVs (3.6%, 4/111)[Citation18]. Huang et al. reported 32 cases with isolated hypoplastic NB, and found three foetuses with pathogenic CNVs and four with VOUS [Citation12]. To our best knowledge, this study is the first report describing the rate of chromosome abnormality in foetuses with isolated absent or hypoplastic NB. Aside from aneuploidy, CMA showed seven pathogenic CNVs, for a detection rate of 3.2%. It has been reported by Callaway et al. that pathogenic/likely pathogenic CNVs were detected in approximately 1.0% (94/9272) of the foetuses with normal ultrasound examination results and normal karyotyping [Citation19]. The diagnostic yield of CMA for pathogenic CNVs in foetuses with isolated absent or hypoplastic NB in our study was 3.2%, which is much higher than the control population (χ2 =10.673, p = .001, Chi-square test). Thus, CMA can increase the diagnostic yield of chromosomal abnormalities for foetuses with isolated absent or hypoplastic NB.
Cell-free DNA or so-called non-invasive prenatal testing (NIPT) is now widely used in clinical practice as a prenatal screening method for common aneuploidies. It also has the potential to detect foetal CNVs, but with false-positive and false-negative results. Recent studies have showed that the accuracy of NIPT for CNVs is still unsatisfactory and needs to be improved [Citation20,Citation21]. CMA is still the most effective method for CNVs detection. On the other hand, conventional karyotyping can identify the majority of foetal chromosomal abnormalities, at a resolution of greater than 10 Mb. In this study, the deletion or duplication sizes of the seven cases were ranged from 713 kb to 4.9 Mb, and all of them may not be diagnosed by the G-banding karyotyping. Finally, from a patient's point of view, they want to exclude as many abnormalities as possible to ensure that the foetus is healthy. Therefore, for prenatal genetic counselling, counsellors should discuss the possibility of pathogenic CNVs to parents when foetuses with isolated absent or hypoplastic NB. CMA should be recommended when isolated absent or hypoplastic NB is suspected antenatally.
Fantasia et al. analyzed the association of first trimester absent NB and genetic abnormalities at G-banding karyotype and CMA according to the NT thickness. The results showed pathogenic CNVs were found only in the group with NT > 99th centile [Citation22]. In this study, we also found no pathogenic CNVs in the first trimester isolated absent NB. These results demonstrated no increase in the risk for pathogenic CNVs in cases with isolated absent NB in the first trimester. However, due to the small sample size, prospective well-adjusted studies are needed to guide the optimal management of these foetuses. Zhang et al. compared 35 cases with isolated absent NB and 20 cases with isolated hypoplastic NB, and found the chromosomal abnormalities rate was increased in the foetuses with isolated hypoplastic NB [Citation15]. However, in our study, the foetuses with isolated absent NB had a higher detection rate of chromosomal abnormalities than the isolated hypoplastic NB, although the difference was not significant in the statistical analysis. The possible reasons for the discrepancy between two studies were the considerably little sample size in the previous study (55 vs. 229 foetuses) and the difference of inclusion criteria (just second trimester vs. first and second trimester).
Seven cases were reported as pathogenic CNVs and the fragment sizes ranged from 713 kb to 4.9 Mb. In case 4, a duplication of 1.3 Mb at distal 1q21.1 region was found. The phenotype of duplication of 1q21.1 region is variable, ranging from macrocephaly, autism spectrum disorder, congenital anomalies, to a normal phenotype. In this study, after comprehensive genetic counselling, the couples ultimately chose to continue the pregnancy. Ji et al. reported three foetuses with 1q21.1 duplication and found two with NB loss, indicating absent foetal NB may be related to 1q21.1 duplication [Citation23]. Zhang et al. also reported 55 foetuses with isolated absent or hypoplastic NB and found one foetus with 1q21.1 duplication [Citation15]. Combined with the three cases of the previous studies and our research, these results identify a further expansion of the prenatal presentation of 1q21.1 duplication. Accordingly, there may be a connection between this duplication and absent or hypoplastic NB, although this remains to be proven by an enlarged study.
Several additional limitations of the current study should be acknowledged. The clinical data are based on a single-centre, and were collected retrospectively. Follow-up data of long-term after childbirth were unavailable.
Conclusion
In summary, the results of the present study demonstrate that CMA can increase the diagnostic yield of chromosome abnormality for foetuses with isolated absent or hypoplastic NB. Counsellors should discuss the possibility of pathogenic CNVs to parents when foetuses with isolated absent or hypoplastic NB. CMA should be recommended when isolated absent or hypoplastic NB is suspected antenatally.
Ethical approval
The study was approved by the Medical Ethics Committee of Guangdong Women and Children Hosiptal. All pregnant women received genetic counselling and signed a written consent before the test. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author contributions
Shi Xiaomei: analyzed and interpreted the data, and wrote the final version of article. Lu Jian: CMA analysis. Li Ling and Wei Ran: data collection and manuscript editing. Wu Jing: project development and manuscript writing.
Acknowledgements
We thank all the project participants for their contributions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data included in this study are available upon request by contact with the corresponding author.
Additional information
Funding
References
- Moreno-Cid M, Rubio-Lorente A, Rodríguez MJ, et al. Systematic review and meta-analysis of performance of second-trimester nasal bone assessment in detection of fetuses with down syndrome. Ultrasound Obstet Gynecol. 2014;43(3):247–253.
- Agathokleous M, Chaveeva P, Poon LC, et al. Meta-analysis of second-trimester markers for trisomy 21. Ultrasound Obstet Gynecol. 2013;41(3):247–261.
- Cicero S, Rembouskos G, Vandecruys H, et al. Likelihood ratio for trisomy 21 in fetuses with absent nasal bone at the 11–14 week scan. Ultrasound Obstet Gynecol. 2004;23(3):218–223.
- Dukhovny S, Wilkins-Haug L, Shipp T, et al. Absent fetal nasal bone: what does it mean for the euploid fetus?. J Ultrasound Med. 2013;32(12):2131–2134.
- Sherer DM, Eugene P, Dalloul M, et al. Second-trimester diagnosis of cri du chat (5p-) syndrome following sonographic depiction of an absent fetal nasal bone. J Ultrasound Med. 2006;25(3):387–388.
- Xing Y, Holder J, Liu Y, et al. Prenatal diagnosis of Wolf-Hirschhorn syndrome: from ultrasound findings, diagnostic technology to genetic counseling. Arch Gynecol Obstet. 2018;298(2):289–295.
- Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–2184.
- Hillman SC, McMullan DJ, Hall G, et al. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2013;41(6):610–620.
- Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764.
- Ting YH, Lao TT, Lau T, et al. Isolated absent or hypoplastic nasal bone in the second trimester fetus: is amniocentesis necessary? J Matern Fetal Neonatal Med. 2011;24(4):555–558.
- Du Y, Ren Y, Yan Y, et al. Absent fetal nasal bone in the second trimester and risk of abnormal karyotype in a prescreened population of Chinese women. Acta Obstet Gynecol Scand. 2018;97(2):180–186.
- Huang H, Cai M, Ma W, et al. Chromosomal microarray analysis for the prenatal diagnosis in fetuses with nasal bone hypoplasia: a retrospective cohort study. Risk Manag Healthc Policy. 2021;14:1533–1540.
- Hou l, Wang x, Jiang h, et al. Application of chromosomal analysis for 29 cases of fetuses with nasal bone absence or hypoplasia. Zhonghua Yi Xue Za Zhi. 2018;98(43):3532–3535.
- Lostchuck E, Hui L. Should second trimester hypoplastic nasal bone be sole indication for diagnostic testing with chromosomal microarray analysis? Ultrasound Obstet Gynecol. 2019;53(6):848–850.
- Zhang F, Long W, Zhou Q, et al. Is prenatal diagnosis necessary for fetal isolated nasal bone absence or hypoplasia? Int J Gen Med. 2021;14:4435–4441.
- Hung JH, Fu CY, Chen CY, et al. Fetal nasal bone length and down syndrome during the second trimester in a Chinese population. J Obstet Gynaecol Res. 2008;34(4):518–523.
- Gu YZ, Nisbet DL, Reidy KL, et al. Hypoplastic nasal bone: a potential marker for facial dysmorphism associated with pathogenic copy number variant on microarray. Prenat Diagn. 2019;39(2):116–123.
- Wu LJ, Cao L, Hu P, et al. Research on the ultrasonographic diagnosis of fetal nasal bone dysplasia and choromsome microarray analysis results. Chin J Ultrasound Med. 2021;5(37):567-570.
- Callaway JL, Shaffer LG, Chitty LS, et al. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenat Diagn. 2013;33(12):1119–1123.
- Duan HL, Li J, Wang WJ, et al. Cell-free DNA test for pathogenic copy number variations: a retrospective study. Taiwan J Obstet Gynecol. 2021;60(6):1066–1071.
- Pei Y, Hu L, Liu J, et al. Efficiency of noninvasive prenatal testing for the detection of fetal microdeletions and microduplications in autosomal chromosomes. Mol Genet Genomic Med. 2020;8(8):e1339.
- Stampalija T, Sirchia F, Della Pietà I, et al. First-trimester absent nasal bone: is it a predictive factor for pathogenic CNVs in the low-risk population? Prenat Diagn. 2020;40(12):1563–1568.
- Ji X, Pan Q, Wang Y, et al. Prenatal diagnosis of recurrent distal 1q21.1 duplication in three fetuses with ultrasound anomalies. Front Genet. 2018;9(9):275.
