Abstract
Background
Epilepsy is a heterogeneous complex condition that involve the human brain. Genetic predisposition to epilepsy is a fundamental factor of the disorder aetiology. The sodium voltage-gated channel (SCN) genes variants are critical biomarker for the epilepsy development and progression. In this study, we aimed to investigate the association of several SNCs genetic polymorphisms with epilepsy risk and their intrudance of the disease prognosis.
Methods
Blood samples were withdrawn from 296 Epilepsy patients in addition to 293 healthy matched participants prior to DNA extraction. PCR-sequencing was used for genotyping analysis. Genotyping outputs were then statistically analysed for genotype/phenotype evaluation.
Results
Within SCN1A gene we found that the rs6432861 (p = 0.014) was in correlation with the risk of epilepsy. In addition, both rs4667485 and rs1469649 of SCN2A gene were significantly correlated to epilepsy risk for both allelic (4e-4 and 1e-3) and genotypic (1e-3 and 5e-3). Moreover, the haplotype analysis showed that the GATGCTCGGTTTCGCTACGCA haplotype of SCN2A gene was significantly related to epilepsy increased risk, p = 6e-3, OR (CI) = 2.02 (1.23–3.31). In relevant to our finding, many of the investigated SCNs variants in the current study were related to several clinical features of epilepsy.
Conclusion
In light of our results, we infer that SCN genes polymorphisms are strong candidates for epilepsy development and progression. Furthermore, these variant are essential for the disorder prognosis and medications outcomes.
Genetic polymorphisms of sodium channels SCN1A, SCN2A and SCN3A were found to be associated with the risk of epilepsy.
SCN1B polymorphisms were found to be correlated to epilepsy reduced risk.
SCNs variants are involved in the epilepsy prognosis and response to treatment.
Key Messages
Keywords:
Introduction
Epilepsy is a major neurological disorder in which heterogeneous conditions impact the human’s health [Citation1]. The main characterization of epilepsy is recurrent seizures that happen due to a sudden abnormal electrical activity in the central nervous system [Citation2]. There are several factors that contribute to the development of epilepsy including environmental and genetic factors . Precisely, epilepsy onset is attributed to the interaction between both the environment and the genetic factors. Besides, it considered as a complex disorder. Two major types of epilepsy were identified according to seizures; generalized seizures that produced through the entire brain and focal seizures that occur in a local part of the brain [Citation3].
Molecular studies have reported that genetic markers of crucial genes may influence epilepsy risk. These genes are involved in different neurological pathways such as voltage-gated sodium [Citation4], potassium, calcium, chloride channels, acetylcholine and γ-amino butyric acid [Citation5,Citation6]. The sodium voltage-gated channel (SCN) proteins play a fundamental roles in the generation of action potential and found in neuronal, endocrine cells and consisted of either alpha or beta subunit [Citation7]. SCN mediates the influx of sodium ions that are important for neurons to initiate the action potential. Therefore, mutations within SCN genes may impact epilepsy development and progression [Citation8].
SCN1A has been intensively studied regarding the risk of epilepsy. These genes are located on chromosome 2 and consist of 26 exons that spans over 100 kb of genomic DNA [Citation9]. This gene was implicated in Genetic Epilepsy with Febrile Seizures Plus (GEFS+) and Familial Febrile Seizures (FFS). GEFS + is an epilepsy syndrome that is detected within families, while FFS common in children from six months to five years [Citation1]. In addition, SCN1A was involved in Dravet syndrome (DS) which is severe genetic epileptic encephalopathy with an increased risk of sudden and unexpected death of someone with epilepsy [Citation10]. Regarding genetic vulnerability to epilepsy, different types of mutation have been reported in SCN1A including frameshift, nonsense and splice site mutations. Moreover, about 30 missense mutants that alter the SCN1A activity have been exposed. These missense mutations may also obliterate channel function by manipulating the properties of the channel, trafficking or subcellular localization [Citation11]. The association between several single variants with epilepsy risk and drug response has been elaborated. In Italy, three variants of SCN1A: rs6730344, rs6732655 and rs10167228 were reported to impact the drug resistance to epilepsy [Citation12]. On the other hand, a study in Hong Kong and Malaysia, Patino et al. [Citation13] and Ramadan et al. [Citation14] investigated the influence of common polymorphisms within SCN1AA (rs3812718 and rs2298771), SCN2A (rs17183814) on drug response to epilepsy and found no correlation between these variants and anti-epileptic drug (AEDs) [Citation15]. Recently, SCN2A mutations have been implicated with epilepsy of infancy with migrating focal seizures (EIMFS) syndrome [Citation16,Citation17], benign familial neonatal infantile seizures (BFNIS) and early infantile epileptic encephalo-pathy (EIEE) [Citation18]. Several variants of SCN2A gene (rs10197716, rs2119068, rs2119067, rs353116, rs353112 and rs6740895) have been proposed to be associated with Valproic Acid (VPA) responses in Chinese Han epileptic patients [Citation18].
The available data that describe the association between SCN3A variants and epilepsy are limited and few were correlated to focal epilepsy [Citation19]. rs1057518801 and rs1057520753 of SCN3A manifest a gain of function in sodium channels that results in more hyperpolarized potentials and had been linked to early infantile epileptic encephalopathy [Citation20]. SCN1B and SCN2B gene have been associated with GEFS [Citation21,Citation22] and dravet syndrome [Citation13,Citation23]. rs786205830 is a loss of function mutation in the SCN1B gene had been linked to early infantile developmental and epileptic encephalopathy (Aeby et al. 2019). SCN8A has been reported to influence epilepsy at four months of early life and can affect the development. This gene may also enhance several epilepsy syndromes such as Lennox-Gastaut syndrome, West syndrome and epileptic encephalopathies (e.g. Dravet syndrome) [Citation16]. Moreover, SCN8A also related to EIEE and BFIS (Makoff et al. 2009).
Hence, the aforementioned genes of sodium voltage-gated channels and their polymorphisms were represented as risk factor for epilepsy; this study aims to investigate the association between several genetic variants of SCN genes and epilepsy in Saudi population. In addition, in this study, genotype–phenotype analyses were conducted to reveal the relationship between the genetic variants of SCNs and several clinical and prognostic factors of epilepsy patients.
Materials and methods
Study subjects and design
This study involved 296 individuals who were diagnosed with Epilepsy in addition to 293 healthy matched participants. The study cohorts were Arabs from Saudi Arabia. Epileptic patients were recruited from neurology clinics at Asir Central Hospital, Abha Maternity and Childern Hospital, Khamis Mushait General Hospital and Primary Health Care Centre in Almansak in addition to their medical records. Written informed consent was obtained from all study subjects, and ethical approval to carry out this study was obtained from the Research Ethics Committee at King Khalid University (Reference No. ECM#2019-38).
DNA extraction and genotyping
Genomic DNA was purified from peripheral blood sample using the Wizard Genomic DNA Purification kit (Promega Corporation, Madison, WI, USA). The quality and quantity of the purified DNA were obtained using agarose gel electrophoresis and the Nano-Drop ND-1000 UV-Vis Spectrophotometer (BioDrop, UK), respectively. DNA samples were then diluted with nuclease-free water to achieve a final concentration of 20 ng/μl and a final volume ranging between 50 and 500 μl. Afterwards, samples were shipped on ice to Gehrmann Laboratories, The University of Queensland, QLD 4072 for custom genotyping on the Sequenom MassARRAY® system (iPLEX GOLD) (Sequenom, USA).
Statistical analyses
Both the Hardy–Weinberg equilibrium (p2 + 2pq + q2 = 1) (http://www.oege.org/software/hwe-mr-calc.html) was applied to test the SNPs. snpSTAT, © 2006 Institut Català d'Oncologia software was used to assess the genotypic and allelic frequencies in addition to the genetic association. Data description and phenotype-genotype analyses were conducted using the JASP 0.16 software. For the present study, statistical significance for single testing was set at p value <.05. Bonferroni correction was performed to the multiple testing using the sets of the significance cut-off at (α/n) where α = 0.05 and n number of tests, and to sustain the overall p-value at significance level of 0.025 or less.
Results
Patient’s characteristics
displays several clinical features of epilepsy in addition to the general parameters. The study cohort consisted of males and females at similar ratio. The mean age of the participants was 9.7 ± 7.986 and ranged from few months to 35 years old. Within patients, the average of the body mass index, BMI indicated a healthy weight (23.2 ± 7.789), as the table shows. The majority of the participants were diagnosed with generalized epilepsy. About 55% of the patients had relapse, while 79% had periodic epilepsy. The response to the first medication for the epileptic patients in the current study was good that accounted for 64%, while most of the cohort did not take any ADE medications. In this study, we estimated that 33.9% of patients recorded with family history of epilepsy; meanwhile 11.9% had a history of febrile seizure. Patients were classified also according to seizure type, and the table depicts that 94.6% of them had motor movement seizure; 52% of patients were observed with seizure free immediately after medication compared with 18% who needed six months to reach seizure-free status.
Table 1. General and clinical characteristic of the epilepsy cohort.
Candidate SNPs
describes the information regarding the genetic variants of SCN superfamily among epilepsy cases and healthy controls. Minor alleles and their frequencies, and p value of HWE in addition to the chromosomal position of the SNPs are listed in the table. The non-polymorphic and SNPs that did not fulfil the HWE were excluded from this study.
Table 2. Description of SCN genes variants among epilepsy cases and healthy controls.
Genetic association between SCN genetic variants and epilepsy
The correlation between the investigated SNPs and the risk of epilepsy is illustrated in . Both allelic and genotypic frequencies among patients and controls were calculated. The significant of the association was estimated depending on the p value. As the table shows, there was no statistically significant difference in the genotypic distribution among cases and controls of rs3812718 (SCN1A) (p = .039). However, the distribution of the variant allele among patients (13%) was lower than it among controls (21%). This indicates that the TT genotype may be a protective factor against epilepsy and may decrease the risk of the disease. Our results also illustrated that the variant genotypes of rs13383628 (TT) may influence the risk of epilepsy (p = .014). Both rs4667485 and rs1469649 of SCN2A gene were significantly correlated to epilepsy risk for both allelic (4e-4 and 1e-3) and genotypic (1e-3 and 5e-3) association according to . We propose that the C/CC of rs4667485 as well as the G/GG of rs1469649 may act as increased risk factor for epilepsy development and progression. This study findings also revealed that the minor allele A of the rs16850186 in SCN3A was higher within controls than within patients but no compelling association was detected (p = 0.03). Finally, the haplotype analysis revealed that the GATGCTCGGTTTCGCTACGCA haplotype of SCN2A polymorphisms (see ) was significantly related to epilepsy increased risk, p = 6e-3, OR (CI) = 2.02 (1.23–3.31). Linkage disequilibrium analysis was performed as shows, and seven SNPs were found as casual variants within the significant SCN2A haplotype.
Figure 1. Linkage disequilibrium (LD) plots of variants within SCN2A. Values in the boxes from top to bottom are D, D′, r and p value. Boxes without numbers have D′= 1. Red boxes indicate a significant, whereas yellow boxes indicate insignificant variants.
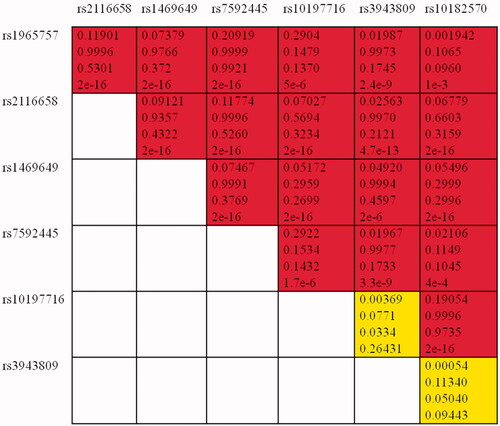
Table 3. Genetic association between the SCN polymorphisms and epilepsy.
Association of the clinical factors of epilepsy with SCN genes variants
and describe the relationship between different features of epilepsy and the investigated variants. Within SCN2A gene, we found that the rs10194956 and rs4667869 were related to the duration of first seizure (p = .04 and .01, respectively). The rs303778 of SCN8A was correlated to vitamin B12 concentration (p = 2e-3) ().
Table 4. Regression analysis of SCN variants and epilepsy features.
Table 5. Regression analysis of SCN variants and epilepsy features.
According to the results displayed in , rs2169312, rs13405797, rs2162600 of SCN1A and rs353139 of SCN2A were in association with history of febrile seizure of epilepsy (p = 6e-3, .023, .026 and .040, respectively), whereas rs1965757, rs7581811, rs7592445 rs3943809 and rs12614399 of SCN2A and rs7598098 of SCN3A were correlated to gender of epileptic patients (p = .022, .035,.021,.044,.049 and .024, respectively). rs4667485, rs1469649, rs353112 and rs10182570 of SCN2A reported to be connected with periodic epilepsy in this study, p values = 0.011, 0.040, 6e-3 and 1e-3 respectively. rs353139, rs16850331, rs12614399 of SCN2A found to influence the drug responsiveness (p = .047, .042 and .034, respectively). Drug level was significantly associated with rs10194956, rs13383628, rs11690959 of SCN1A, rs7596422 of SCN3A and rs11169883 of SCN8A (p values = 0.018, 0.040, 0.039, 0.011 and 0.019 respectively).
The rs13405797 of SCN1A and rs2119067 of SCN3A were related to family history of epilepsy (p = 0.035 and 0.023). The rs10182570 of SCN2A was correlated to Epilepsy classification (p = 0.039), while rs935403 was of the same gene found in association with seizure classification (p = 0.014) ().
Discussion
Epilepsy is a complex neurological condition that impacts the brain and cause seizure [Citation24]. Genetic predisposition to epilepsy has been a fundamental part of the disorder aetiology [Citation25]. Voltage-gated sodium channels are critical for genetics epilepsy, and these channels play a key role in mediating the electrical excitability. Thus, it is lucid that any genetic mutations in these gene coding channels can interfere the epilepsy development or progression. When the channels are activated by membrane depolarization, it will cause conformational change that increases the sodium ion influx in addition to cell depolarization and later the channels will be deactivated ending in resting of membrane potential [Citation11]. This study investigated several genetic variants of SCN genes (SCN1A, SCN2A, SCN3A, SCN1B, SCN2B, SCN3B and SCN8A) and their association with epilepsy risk; these genes have been studied in this regard and conflicted results were reported [Citation7]. rs3812718 is a common intronic variant that located in splice donor site, and it modifies alternative splicing of exon 5. We suggest that TT genotype of rs3812718 in SCN1A may be a protective factor against epilepsy and may decrease the risk of the disease in Saudi population. In contrast to our finding, rs3812718 was reported as a risk factor for GEFS + in Chinese population [Citation26,Citation27]. In one meta-analysis they revealed that the rs3812718 TT genotype was involved in high risk of developing drug resistance in epilepsy children [Citation28].
We also discovered that in SCN1A gene, the variant genotypes of rs6432861 (CC) may influence the risk of epilepsy according to this study outcomes. We also propose that within the SCN2A gene, the C/CC of rs4667485 as well as the G/GG of rs1469649 may act as increased risk factor for epilepsy development and progression in Saudis. Moreover, The GATGCTCGGTTTCGCTACGCA haplotype of SCN2A gene was significantly related to epilepsy increased risk.
The other part of the current study explored the association between different clinical features of epilepsy and SCN genes variants. We found that these variants, rs1965757, rs7581811, rs7592445 rs3943809, rs12614399 of SCN2A and rs7598098 of SCN3A imply variation of epilepsy risk depending on the individuals’ gender. It has been indicated that men are more vulnerable to localization-related symptomatic epilepsies, whereas cryptogenic localization-related epilepsies were more frequent in women [Citation29]. We also infer that SCN2A genetic variants, rs10194956 and rs4667869, may affect the duration of first seizure. Vitamin B12 deficiency may result in neurological problems (Benbir et al. 2006). In this regard, the concentration of vitamin B12 found to be associated with the rs303778 of SCN8A in epilepsy patients in this study. History of febrile seizure of epilepsy can be posed as an essential prognostic factor that can predict epilepsy development. We propose that the rs2169312, rs13405797, rs2162600 of SCN1A and rs353139 of SCN2A are risk factor for epilepsy among individual with history of febrile seizure. Furthermore, the rs13405797 of SCN1A and rs2119067 of SCN3A can be considered as epilepsy risk biomarkers within individuals have a family history of epilepsy.
Epileptic spasms are a type of seizure that last 1 to 2 s and spasms occur predominantly in infants. The prognosis of epileptic spasms is generally connected with deprived neurodevelopmental and behavioural outcomes [Citation30]. In this study, we found that SCN2A gene variants (rs4667485, rs1469649, rs353112 and rs10182570) may be a risk factor for periodic epilepsy. Several Biomarkers of SCN1A gene have been proposed as risk factor to drug response and drug resistant of epilepsy. This prognosis clinical factor is important for clinical practice and medication enhancement [Citation12]. In light of our finding, it seems more likely that the rs353139, rs16850331, rs12614399 of SCN2A found to influence the drug responsiveness to epilepsy. Otherwise, Citation31 have explored the clinical response of SCN1A variations to sodium channel blocking drugs in Chinese epilepsy patients and found no correlation. Another clinical parameter that was examined in this study is the drug level and was significantly associated with rs10194956, rs13383628, rs11690959 of SCN1A, rs7596422 of SCN3A and rs11169883 of SCN8A. We deduce that these variants may affect the demanded dose of AED for epilepsy. Classification of epilepsy and seizure types is significant for the primary diagnostic testing. We found that the rs10182570 of SCN2A was correlated to Epilepsy classification, whereas rs935403 of the same gene was found in association with Seizure classification.
In conclusion, within (SCN1A) gene the rs3812718, rs10194956, rs13383628, rs6432861 and rs1542484 may impact the risk of epilepsy. In addition, both rs4667485 and rs1469649 of SCN2A were significantly associated with epilepsy risk. For SCN3A, rs16850186 and the rs72550243 of SCN1B, we declare them as genetics factors that may influence the development and progression of epilepsy in Saudi population. Considering the clinical impact of epilepsy prognosis features, we propose voltage sodium channels as predictor factors for the clinical outcome of epilepsy. Molecular and Pharmacogenomics studies are highly recommended to elucidate the inter-individuals variability in response to AED in the light of different prognosis parameters.
The main limitations of this case-control study were gender and the relatively small sample number that could lead to selection bias. However other potential limitations were avoided such as bias was caused by population stratification, as the Saudi Arab population is a relatively homogenous population. Moreover, there were no significant differences between case and control groups in terms of basic demographic characteristics.
Author contributions
The work presented here was carried out in collaboration among all authors. MAA, LNA, AA, EME, MYO, DMR, and NFA were involved in the conception and design of this study. MAA, AA, SAA, MSA, MA, RSA, AAA, AHS, MYO, and NFA participated in the collection of relevant data. MAA, LNA, DMR, SAA, MSA, MA, RSA, AAA, AHS, and EME were involved in the analysis and interpretation of the data. All authors were involved in drafting the manuscript and revising it critically for intellectual content. All authors reviewed and approved the final manuscript and agree to be held accountable for all aspects of the work.
Acknowledgments
The authors express their gratitude to King Khalid University, Saudi Arabia, for providing administrative and technical support.
Disclosure statement
Authors declare no conflict of interest.
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Steinlein OK. Genetics and epilepsy. Dialogues Clin Neurosci. 2008;10(1):29–38.
- Anwar H, Khan QU, Nadeem N, et al. Epileptic seizures. Discoveries (Craiova). 2020;8(2):e110.
- Valton L, Benaiteau M, Denuelle M, et al. Etiological assessment of status epilepticus. Rev Neurol (Paris). 2020;176(6):408–426.
- Barela AJ, Waddy SP, Lickfett JG, et al. An epilepsy mutation in the sodium channel SCN1A that decreases channel excitability. J Neurosci. 2006;26(10):2714–2723.
- Baulac S, Huberfeld G, Gourfinkel-An I, et al. First genetic evidence of GABAA receptor dysfunction in epilepsy: a mutation in the γ2-subunit gene. Nat Genet. 2001;28(1):46–48.
- Benbir G, Uysal S, Saltik S, et al. Seizures during treatment of vitamin B12 deficiency. Seizure. 2007;16(1):69–73.
- Andavan GSB, Lemmens-Gruber R. Voltage-gated sodium channels: mutations, channelopathies and targets. Curr Med Chem. 2011;18(3):377–397.
- Widmark J, Sundstrom G, Ocampo Daza D, et al. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 2011;28(1):859–871.
- Mulley JC, Scheffer IE, Petrou S, et al. SCN1Amutations and epilepsy. Hum Mutat. 2005;25(6):535–542.
- Esterhuizen AI, Mefford HC, Ramesar RS, et al. Dravet syndrome in South African infants: tools for an early diagnosis. Seizure. 2018;62:99–105.
- Escayg A, Goldin AL. Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia. 2010;51(9):1650–1658.
- Margari L, Legrottaglie AR, Vincenti A, et al. Association between SCN1A gene polymorphisms and drug resistant epilepsy in pediatric patients. Seizure. 2018;55:30–35.
- Patino GA, Claes LR, Lopez-Santiago LF, et al. A functional null mutation of SCN1B in a patient with dravet syndrome. J Neurosci. 2009;29(34):10764–10778.
- Ramadan W, Patel N, Anazi S, et al. Confirming the recessive inheritance of SCN1B mutations in developmental epileptic encephalopathy. Clin Genet. 2017;92(3):327–331.
- Haerian BS, Baum L, Kwan P, et al. SCN1A, SCN2A and SCN3A gene polymorphisms and responsiveness to antiepileptic drugs: a multicenter cohort study and meta-analysis. Pharmacogenomics. 2013;14(10):1153–1166.
- Hammer MF, Wagnon JL, Mefford HC, et al. SCN8A-Related epilepsy with encephalopathy. 2016. In: Adam MP, Ardinger HH, Pagon RA editors. GeneReviews® [internet]. Seattle (WA): University of Washington, Seattle; 1993–2022. Available from: https://www.ncbi.nlm.nih.gov/books/NBK379665/
- Howell KB, McMahon JM, Carvill GL, et al. SCN2Aencephalopathy. Neurology. 2015;85(11):958–966.
- Lu Y, Su Q, Li M, et al. Association of SCN1A, SCN2A, and UGT2B7 polymorphisms with responsiveness to valproic acid in the treatment of epilepsy. Biomed Res Int. 2020;2020:8096235–8096238.
- Lamar T, Vanoye CG, Calhoun J, et al. SCN3A deficiency associated with increased seizure susceptibility. Neurobiol Dis. 2017;102:38–48.
- Zaman T, Helbig I, Božović IB, et al. Mutations in SCN3A cause early infantile epileptic encephalopathy. Ann Neurol. 2018;83(4):703–717.
- Voros NS, Antonopoulos C. Cyberphysical systems for epilepsy and related brain disorders. In: Multi-parametric monitoring and analysis for diagnosis and optimal disease management; 2015.
- Gong J, Liao H, Long H, et al. SCN1B and SCN2B gene variants analysis in dravet syndrome patients. Medicine (Baltimore). 2019;98(13):e14974.
- Bender AC, Morse RP, Scott RC, et al. SCN1A mutations in dravet syndrome: Impact of interneuron dysfunction on neural networks and cognitive outcome. Epilepsy Behav. 2012;23(3):177–186.
- Shneker BF, Fountain NB. Epilepsy. Dis Mon. 2003;49(7):426–478.
- Sokka A, Olsen P, Kirjavainen J, et al. Etiology, syndrome diagnosis, and cognition in childhood-onset epilepsy: a population-based study. Epilepsia Open. 2017;2(1):76–83.
- Ma QL, Wang B, Chen GF, et al. Association between SCN1A rs3812718 polymorphism and generalized epilepsy with febrile seizures plus. Zhongguo Dang Dai er ke za Zhi = Chinese Journal of Contemporary Pediatrics. 2018;20(2):130–133.
- Makoff A, Lai T, Barratt C, et al. High-density SNP screen of sodium channel genes by haplotype tagging and DNA pooling for association with idiopathic generalized epilepsy. Epilepsia. 2010;51(4):694–698.
- Wang ZJ, Chen J, Chen HL, et al. Association between SCN1A polymorphism RS3812718 and valproic acid resistance in epilepsy children: a case–control study and Meta-analysis. Bioscience Reports. 2018;38(6):1–8.
- Christensen J, Kjeldsen MJ, Andersen H, et al. Gender differences in epilepsy. Epilepsia. 2005;46(6):956–960.
- Liao J, Huang T, Srour M, et al. Status epilepticus manifested as continuous epileptic spasms. Front Neurol. 2020;11:65.
- Kwan P, Poon WS, Ng H, et al. Multidrug resistance in epilepsy and polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, and SCN3A: Correlation among phenotype, genotype, and mRNA expression. Pharmacogenet Genomics. 2008;18(11):989–998.
