Abstract
Objectives
Chromosomal microarray analysis (CMA) has been widely applied to genetic diagnosis in miscarriages in clinical practice. However, the prognostic value of CMA testing of products of conception (POCs) after the first clinical miscarriage remains unknown. The aim of this study was to evaluate the reproductive outcomes after embryonic genetic testing by CMA in SM couples.
Methods
In this retrospective study, a total of 1142 SM couples referred for embryonic genetic testing by CMA, and 1022 couples were successfully followed up after CMA.
Results
Among 1130 cases without significant maternal cell contamination, pathogenic chromosomal abnormalities were detected in 680 cases (60.2%). The subsequent live birth rate did not differ significantly between couples with chromosomally abnormal and normal miscarriage (88.6% vs. 91.1%, p = .240), as well as the cumulative live birth rate (94.5% vs. 96.7%, p = .131). Couples with partial aneuploid miscarriage had a higher likelihood of spontaneous abortion both in the subsequent pregnancy (19.0% vs. 6.5%, p = .037) and cumulative pregnancies (19.0% vs. 6.8%, p = .044) when compared with couples with chromosomally normal miscarriage.
Conclusions
SM couples with chromosomally abnormal miscarriage manifested with a similar reproductive prognosis to couples with chromosomally normal miscarriage.
CMA testing of POCs could provide an accurate genetic diagnosis for couples with SM.
The live birth rate of couples with partial aneuploid miscarriage was as high as couples with chromosomally normal miscarriage, despite a higher risk of adverse pregnancy event.
Among couples with the most common single aneuploid miscarriage, the cumulative live birth rates of couples with trisomy 16, sex chromosomal abnormalities and trisomy 22 were 94.1%, 95.8% and 84.0%, respectively.
Key messages
Introduction
Sporadic miscarriage (SM), also known as pregnancy loss, is the most common complication in early pregnancy [Citation1]. Approximately 15.3% of all clinically recognized pregnancy ended in miscarriage in general population [Citation2]. Embryonic chromosomal abnormalities are the primary cause of pregnancy loss [Citation3]. Besides, parental chromosomal abnormalities, uterine malformations, thrombophilias, antiphospholipid syndrome, immunodeficiency, infection disorders, endocrine and metabolic disorders are potential aetiologies as well [Citation1].
Chromosomal microarray analysis (CMA) containing single-nucleotide polymorphism probes, which can simultaneously identify aneuploidies, copy number variations (CNVs), uniparental disomy (UPD), triploidies and maternal cell contamination (MCC), has replaced the application of conventional G-banding karyotype analysis in most paediatric and prenatal diagnosis as well as genetic analysis of miscarriage products [Citation4,Citation5]. Currently, genetic analysis of products of conception (POCs) was recommended in couples with recurrent miscarriages (RM) by guidelines [Citation3,Citation6]. However, it was not routinely recommended after the first pregnancy loss [Citation3,Citation6]. In spite of this, due to the anxiety of couples suffering from miscarriage, SM couples were also eager for embryonic genetic testing to uncover the cause of pregnancy loss and some SM couples underwent embryonic CMA testing in clinical practice. Furthermore, the importance of CMA testing of POCs after the first miscarriage has also been highlighted by several studies [Citation7,Citation8]. Therefore, genetic evaluation of POCs is equally necessary for SM couples. However, the prognostic value of CMA testing of POCs after the first clinical miscarriage remains unknown.
Hence, we conducted a retrospective study to assess the clinical value of CMA as a genetic testing tool for a cohort of 1142 POCs after SM. Furthermore, we investigated the long-term reproductive outcomes among SM couples with different types of chromosomally abnormal miscarriage.
Materials and methods
Study population
From August 2011 to December 2019, a total of 1142 SM couples referred to Nanjing Maternity and Child Health Care Hospital (Jiangsu, China) for embryonic genetic testing by CMA. Informed consent of genetic testing was obtained from all patients before CMA testing. Parental study was performed by karyotype analysis, fluorescence in situ hybridization (FISH) or CMA testing according to embryonic CMA results. Genetic counselling was offered to all individuals. Maternal age, gestational age of the index miscarriage (the first clinical miscarriage in our study) and previous reproductive history were recorded. The mean maternal age was 29.0 years (range 18.0 to 44.0) and the mean gestational age was 10.0 weeks (range 6.0 to 18.0) in this cohort of 1142 couples. Couples with live birth, biochemical miscarriage, ectopic pregnancy, therapeutic abortion or induced abortion before the index miscarriage were included in the current study. This study was approved by the Medicine Ethics Committee of Nanjing Maternity and Child Health Care Hospital (2021KY004).
Chromosomal microarray analysis
All fresh miscarriage specimens were rinsed with saline solution for three times. Chorionic villi were separated from maternal decidua according to the standardized technology [Citation9]. Samples where chorionic villi could not be clearly identified were excluded from this study. Genomic DNA was extracted from chorionic villi with the protocol of QIAamp DNA Mini Kit (Qiagen, Germany). Chromosomal abnormalities of POCs were detected by two CMA platforms in the current study, including CytoScan 750K array (Affymetrix, USA) and HumanCyto12-SNP array (Illumina, USA). SNP array experiments and molecular karyotype analysis for both platforms were performed as previously described [Citation10]. Quantitative fluorescent polymerase chain reaction (QF-PCR) was subsequently performed to identify the percentage of maternal and foetal DNA if MCC was detected by CMA. Significant MCC referred to the proportion of MCC exceeding 30%. Samples with significant MCC were excluded from our study.
The two platforms could detect CNVs at an effective minimal resolution of 100 kb and regions of allelic homozygosity (ROHs) at a threshold of 5 Mb. Mosaicism for aneuploidies or CNVs ≥ 5 Mb was reported when the detection threshold of 30% was exceeded. CNVs were further classified as partial aneuploidy (CNVs ≥ 10 Mb, large CNVs) and microdeletions/microduplications (CNVs < 10 Mb, submicroscopic CNVs) based on their sizes. Pathogenicity of detected CNVs were evaluated according to the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) [Citation11].
Reproductive outcomes
All eligible couples were followed up for at least 24 months after CMA testing. Each reproductive outcome was obtained by telephone interview or medical records. Cases identified as variants of uncertain significance (VOUS) in the index miscarriage were excluded from our follow-up study as pathogenicity of these CNVs remained ambiguous. Couples underwent preimplantation genetic testing (PGT) during the following pregnancies were excluded. We observed subsequent and cumulative reproductive outcomes after CMA testing. Subsequent outcome specifically referred to the first pregnancy outcome after the index miscarriage. Cumulative outcomes covered all pregnancy events after the index miscarriage. Live birth was our primary outcome, which was defined as the delivery of one or more viable infants after 28 weeks of gestation. The secondary outcomes referred to adverse pregnancy events, such as spontaneous abortion, biochemical miscarriage or ectopic pregnancy in our study.
Statistical analysis
Baseline characteristics were compared using the Kruskal–Wallis test for continuous variables and the Fisher’s exact test for categorical variables. Binary logistic regression analysis was performed to compare the reproductive outcomes between couples with different embryonic chromosomal abnormalities using the non-adjusted model and the adjusted model separately. The latter model adjusted for the maternal age of the index miscarriage. We performed additional sensitivity analysis to examine the robustness of our results. Firstly, we repeated binary logistic regression analysis adjusted for maternal age, gestational age of the index miscarriage and previous reproductive history simultaneously. Secondly, subgroup analysis was performed to further investigate the reproductive outcomes of couples with different maternal age of the index miscarriage (aged < 30 years, 30–34 years and ≥ 35 years). A p value < .050 was considered statistically significant. All statistical analyses were conducted with SPSS version 22.0.
Results
Embryonic chromosomal abnormalities detected by CMA
A total of 1142 cases of SM were analysed by CMA. We excluded 12 cases (1.1%) with significant MCC. Among the remaining 1130 cases with no or negligible MCC, chromosomal abnormalities were detected in 680 (60.2%) cases (). Normal results were obtained in 442 (39.1%) cases, and VOUS were obtained in 8 (0.7%) cases.
Table 1. Distribution and frequency of embryonic chromosomal abnormalities detected by chromosomal microarray analysis in SM.
Numerical chromosomal abnormalities were the most frequent abnormal finding, including 542 (48.0%) with aneuploidies and 87 (7.7%) with polyploidies. Among the cases with aneuploidies, 95.4% (517/542) of these cases were identified as single aneuploidies, and multiple aneuploidies composed the remaining 4.6% (25/542). With the exception of chromosomes 1 and 19, single aneuploidies were detected in all chromosomes. Trisomies represented the majority among the cases with single aneuploidies (420/517, 81.2%), and others were monosomies (97/517, 18.8%). With respect to trisomies, trisomy 16 was the most common (142/420, 33.8%), followed by trisomy 22 (64/420, 15.2%) and trisomy 21 (28/420, 6.7%). Monosomies were observed in chromosomes X (95/97, 97.9%), 8 (1/97, 1.0%) and 21 (1/97, 1.0%). Among the cases with polyploidies, 85 (97.7%) were triploidy and two (2.3%) were tetraploidy. In addition, four cases (0.4%) with whole-genome UPD were identified in our cohort.
In our study, 47 cases (4.2%) with pathogenic CNVs were identified, including 35 (3.1%) with partial aneuploidy (Table S1) and 12 (1.1%) with pathogenic microdeletions/microduplications (Table S2). Among cases with partial aneuploidies, multiple deletions and/or duplications accounted for the largest proportion (16/35, 45.7%). Seven cases (7/35, 20.0%) were identified as terminal deletion accompanied with terminal duplication, suggesting unbalanced translocation or recombinant inversion. After parental karyotype analysis, four couples were identified as balanced translocation carriers (three maternal inherited and one paternal inherited). Ten (28.6%) with terminal deletion/duplication and two (5.7%) with interstitial deletion/duplication were found in cases with partial aneuploidies. Of the cases with pathogenic submicroscopic CNVs, we detected five (41.7%) with terminal microdeletion/microduplication, six (50.0%) with interstitial microdeletion/microduplication and one (8.3%) with multiple microdeletions and/or microduplications. Theoretically, these 12 cases could not be identified by karyotype analysis. Ten of these 12 cases underwent parental study by FISH or CMA testing, and were considered to be de novo.
Table 2. Baseline characteristics of couples with sporadic miscarriage who were pregnant after chromosomal microarray analysis.
Baseline characteristics of couples with reproductive outcomes
Among 1130 cases with CMA results, eight cases with VOUS were not followed up. Among the remaining cases with normal or pathogenic findings, 100 couples were lost to follow-up. Of those couples successfully followed up, 83 decided not to conceive, 26 employed PGT during the following pregnancies and 66 failed to conceive. The above 283 cases were excluded from our study. The remaining 847 couples who conceived at least once after CMA testing were eventually included in our study. Details are seen in .
Figure 1. Flow chart of follow-up after embryonic chromosomal microarray analysis in couples with sporadic miscarriage.
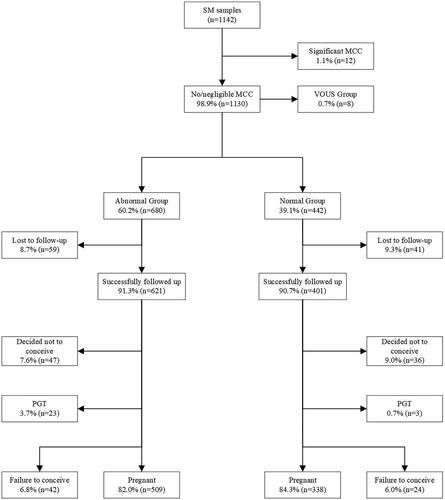
Baseline characteristics of couples with reproductive outcomes are illustrated in . We generally categorized cases into the following five subgroups: aneuploidy (n = 415), polyploidy (n = 62), partial aneuploidy (n = 21), pathogenic microdeletion/microduplication (n = 11) and chromosomally normal group (n = 338). The mean duration of follow-up was 49.0 (range 25.0 to 120.0) months.
Maternal age of the index miscarriage was found to vary significantly between five subgroups (p = .017). Women with chromosomally normal miscarriage in the index miscarriage were significantly younger than those with aneuploid miscarriage (p = .023). However, maternal age of the index miscarriage did not differ significantly between any remaining subgroups. Gestational age was similar in all subgroups. Besides, no significant differences were found in the incidence of live birth or adverse pregnancy events before the index miscarriage between subgroups.
Subsequent reproductive outcome
Among 847 couples pregnant during the follow-up period, 759 (89.6%) had a subsequent live birth. No significant difference was observed in the subsequent live birth rate between couples with chromosomally abnormal miscarriage and chromosomally normal miscarriage (88.6% vs. 91.1%, p = .240). The frequencies of subsequent spontaneous abortion, biochemical miscarriage and ectopic pregnancy were also similar between two groups and did not differ significantly. Detailed subsequent reproductive outcomes are summarized in .
Table 3. Subsequent reproductive outcome after embryonic chromosome analysis in couples with sporadic miscarriage.
In different subgroups, couples with polyploid miscarriage represented a higher live birth rate (95.2%) than those with other chromosomal abnormalities, whereas couples with partial aneuploid miscarriage presented a worse prognosis (81.0%) than others. The incidence of spontaneous abortion was significantly higher in couples with partial aneuploid miscarriage than those with chromosomally normal miscarriage (19.0% vs. 6.5%, p = .042), and the difference remained after adjusting for maternal age of the index miscarriage (p = .037). However, the rates in other three subgroups were similar to that in couples with chromosomally normal miscarriage. Compared with couples with chromosomally normal miscarriage, those with microdeletion/microduplication miscarriage displayed a significantly higher risk of ectopic pregnancy (9.1% vs. 0.9%, p = .044), while no statistical difference was found among the remaining three subgroups. When maternal age of the index miscarriage was used for adjustment, this difference was maintained (p = .046). The incidence of subsequent biochemical miscarriage was not associated with the types of chromosomal abnormalities of the index miscarriage. These statistically significant findings were consistent with the results after adjustment for maternal age, gestational age of the index miscarriage and previous reproductive history in the sensitivity analysis.
Among 67 women with a subsequent abortion, 16 (12 with single aneuploidy and four with normal results in the index miscarriage) underwent a subsequent CMA testing of POCs (Table S3). Among 12 cases with single aneuploidy in the index miscarriage, trisomies were detected in five cases, monosomy X in three cases, VOUS in one case and normal results in three cases. Among four cases with normal results, we found three cases with trisomy and one with normal result.
Cumulative reproductive outcomes
In total, the incidence of delivery of one or more viable infants in couples with chromosomally normal miscarriage did not differ significantly compared with chromosomally abnormal miscarriage (96.7% vs. 94.5%, p = .131). The frequencies of adverse pregnancy events were also similar between two groups. The cumulative outcomes are showed in .
Table 4. Cumulative reproductive outcomes after embryonic chromosomal analysis in couples with sporadic miscarriage.
In the subgroups of miscarriage, the proportion of couples giving birth to one or more viable infants during the follow-up period was 100% after partial aneuploid miscarriage or microdeletion/microduplication miscarriage, higher than that of couples with aneuploid miscarriage (93.7%), polyploid miscarriage (96.8%) and chromosomally normal miscarriage (96.7%). A significantly higher proportion of couples with partial aneuploid miscarriage than those with chromosomally normal miscarriage had one or more spontaneous abortions after CMA testing when we adjusted for maternal age of the index miscarriage (19.0% vs. 6.8%, p = .044). The presence of microdeletion/microduplication miscarriage led to a significantly higher risk of one or more ectopic pregnancies when compared with couples with chromosomally normal miscarriage (9.1% vs. 0.9%, p = .044). This difference also existed when maternal age of the index miscarriage was included in the logistic regression model (p = .046). The prevalence of one or more biochemical miscarriages during the follow-up period was similar between five subgroups. These statistically significant findings were maintained after adjustment for maternal age, gestational age of the index miscarriage and previous reproductive history in the sensitivity analysis.
Reproductive outcomes of couples classified by maternal age of the index miscarriage
Among women with different maternal ages (aged < 30 years, 30–34 years and ≥ 35 years), the live birth rates and the rates of adverse pregnancy events did not differ significantly between couples with chromosomally abnormal miscarriage and couples with chromosomally normal miscarriage (Tables S4, S5, S6 and S7).
In different subgroups, among women younger than 30 years, the subsequent incidence of spontaneous abortion in couples with partial aneuploid miscarriage was significantly higher than those with chromosomally normal miscarriage (23.1% vs. 6.5%, p = .041) (Table S5). However, the cumulative incidence of spontaneous abortion was similar between these two groups. There were also no significant differences in the rates of live birth, biochemical miscarriage and ectopic pregnancy between couples with different types of chromosomally abnormal miscarriage and couples with chromosomally normal miscarriage among women younger than 30 years. Among women aged 30–34 years, couples with microdeletion/microduplication miscarriage were more likely to experience ectopic pregnancy in the subsequent pregnancy when compared with couples with chromosomally normal miscarriage (25.0% vs. 1.9%, p = .035) (Table S7). This finding was maintained in the rate of cumulative ectopic pregnancy between these two groups (25.0% vs. 1.9%, p = .035). However, there were no significant differences in the rates of live birth, spontaneous abortion and biochemical miscarriage between couples with different types of chromosomally abnormal miscarriage and couples with chromosomally normal miscarriage among women aged 30–34 years. In addition, among women aged 35 years or older, the rates of live birth and adverse pregnancy events were similar between couples with different types of chromosomally abnormal miscarriage and couples with chromosomally normal miscarriage.
Reproductive outcomes of couples with different single aneuploid miscarriage
Pregnancy occurred at least once in 406 couples with single aneuploid miscarriage during the follow-up period. Trisomy 16 (118/406, 29.1%), sex chromosomal abnormalities (72/406, 17.7%) and trisomy 22 (50/406, 12.3%) were the most common aneuploidies (). The subsequent live birth rates of couples who suffered from miscarriage with these three aneuploidies (trisomy 16, sex chromosomal abnormalities and trisomy 22) were 87.3%, 95.8% and 70.0%, respectively (), and the cumulative live birth rates of these couples were 94.1%, 95.8% and 84.0%, respectively ().
Figure 2. Embryonic genetic findings and reproductive outcomes in couples with different single aneuploid miscarriage. (a) Distribution of embryonic single aneuploidies in the index miscarriage of the 406 couples with reproductive outcomes. (b) Subsequent reproductive outcome in couples with single aneuploid miscarriage. (c) Cumulative live birth rate in couples with single aneuploid miscarriage.
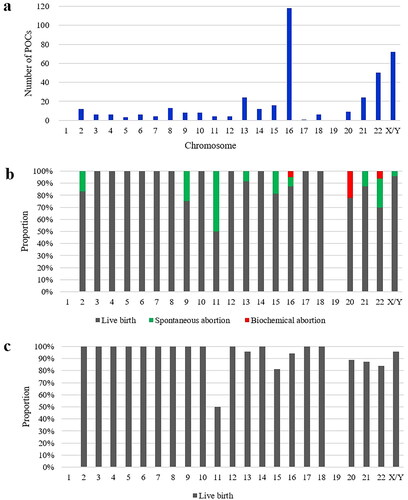
Besides, the live birth rates of couples with certain aneuploid miscarriage (chromosomes 3, 4, 5, 6, 7, 8, 10, 12, 14, 17 and 18) in the index miscarriage were 100% in the subsequent pregnancy. The cumulative live birth rate of couples suffering from miscarriage with trisomy 2 and 9 reached to 100% as well. Couples who suffered from miscarriage with trisomy 11 presented a worse prognosis, with a live birth rate of 50% in both the subsequent and cumulative pregnancies.
Discussion
In this retrospective study, we systematically evaluated the distribution and frequency of embryonic chromosomal abnormalities among couples with the first clinical pregnancy loss. Then, we conducted a follow-up study to compare the live birth rate among couples with different types of embryonic chromosomal abnormalities. The overall prognosis in couples with chromosomally abnormal miscarriage was similar to those with chromosomally normal miscarriage. These findings in our study could provide valuable information that guides genetic counselling for chance of live birth and estimation of risk for adverse pregnancy events in the following pregnancies.
Embryonic chromosomal abnormalities were the primary cause of SM. The overall prevalence of pathogenic chromosomal abnormalities in miscarriage specimens among SM couples was 60.2%, which is similar to the frequencies of SM (60.6%) and RM (61.2%) previously reported [Citation12]. Consistent with the findings of previous studies, aneuploidy accounted for the largest proportion in SM [Citation13,Citation14]. It is well accepted that large CNVs are causative of miscarriage. We totally detected 35 cases with partial aneuploidy at a prevalence of 3.1%, which is similar to the detection rate identified previously (3.0%) [Citation15]. Terminal deletion accompanied with terminal duplication, which is suggestive of recombinant inversion or unbalanced translocation, was detected in seven (0.6%) cases. Four couples were ultimately identified as balanced translocation carriers by karyotype analysis. These results suggested that segmental chromosomal abnormalities of miscarriages could be caused by parental chromosomal rearrangements, and genetic evaluation of POCs is necessary for these couples to reduce the risk of adverse pregnancy events. Moreover, the detection rate of pathogenic submicroscopic CNVs in SM was 1.1% in this study, which is consistent with the data previously reported (1.1%) [Citation16]. Our findings provide additional evidence that submicroscopic CNVs could be an important cause of miscarriage.
Nowadays, genetic counselling for SM couples about their risk of subsequent miscarriage or delivery of offspring with congenital defect is based on empirical evaluation that lacks objective and accurate data on reproductive outcome. In this study, we observed that couples with chromosomally abnormal miscarriage (88.6%) had a similar chance of live birth in the subsequent pregnancy compared with couples with chromosomally normal miscarriage (91.1%). This finding was consistent with one previous study [Citation17], which also found no statistical difference in the subsequent live birth rate among SM couples with aneuploid miscarriage compared with couples with euploid miscarriage (68% vs. 63%, p = .45). However, the overall live birth rate in our study was obviously higher than that in this previous study which focussed on infertile couples.
In this study, we observed that couples with partial aneuploid miscarriage (19.0%) showed a significantly higher risk of subsequent spontaneous abortion than couples with chromosomally normal miscarriage (6.5%). It is known that subsequent prognosis in couples carrying a reciprocal translocation (31.9%) was obviously poorer than that in non-carrier couples (71.7%) [Citation18]. Nevertheless, all couples with partial aneuploid miscarriage who had a second abortion in this study were identified as non-carrier couples. Accordingly, we suggest further investigation and treatment for these couples to determine the pathogenesis of a second miscarriage. Additionally, a significantly higher prevalence of ectopic pregnancy was found in couples with microdeletion/microduplication miscarriage (9.1%) than in couples with chromosomally normal miscarriage (0.9%). However, the currently available data are not robust enough to draw a solid conclusion due to a limited number of samples in this subgroup. It remains uncertain whether this finding could reflect the actual condition, and further large-scale research is required to investigate the reproductive outcomes of SM couples with microdeletion/microduplication miscarriage.
The long-term prognosis of SM couples after embryonic CMA testing remains uncertain. In our study, a similar chance of delivery of one or more viable infants was observed among couples with chromosomally normal miscarriage (96.7%) and couples with chromosomally abnormal miscarriage (94.5%) during the follow-up period. However, among women with RM, a significantly lower cumulative live birth rate was found in women with euploid miscarriage (44.7%) than women with aneuploid miscarriage (71.9%) (p = .02) [Citation19]. The possible reason could be that couples with euploid miscarriage may accompany with other systematic diseases which would reemerge in the following pregnancies. Our findings suggest that long-term reproductive prognosis in SM couples is better than that in RM couples.
At present, the effects of maternal age of the index miscarriage on following reproductive outcomes of SM couples with different types of chromosomally abnormal miscarriage remain unclear [Citation17]. Overall, the rates of live birth and adverse pregnancy events were similar between couples with chromosomally normal and abnormal miscarriage in all three maternal age cohorts (aged < 30 years, 30–34 years and ≥ 35 years). Among women younger than 30 years, couples with partial aneuploid miscarriage were more likely to experience spontaneous abortion in the subsequent pregnancy than those with chromosomally normal miscarriage. Among women aged 30–34 years, couples with microdeletion/microduplication miscarriage represented a significantly higher risk of ectopic pregnancy both in the subsequent pregnancy and cumulative pregnancies. However, the sample size in this subgroup was limited and we did not consider it as a robust conclusion currently. It is essential to further confirm this finding in a large cohort study.
Reproductive outcomes are mainly determined by the chromosome involved in the index aneuploid miscarriage. Our study revealed that reproductive prognosis would be optimistic to the majority of couples with single aneuploid miscarriage. Nevertheless, pregnancy loss was more likely to occur again in couples who suffered from miscarriage with trisomy 11; this could be due to the limited number of cases with trisomy 11 detected in our cohort. The subsequent live birth rate of couples who suffered from the most frequent aneuploid miscarriage (trisomy 16) was 87.3%, and the cumulative live birth rate reached to 94.1% in them. Overall, these findings may aid in evaluating reproductive prognosis of SM couples who suffered from single aneuploid miscarriage and provide risk assessment of adverse pregnancy events.
This was the first study to evaluate the reproductive outcomes in couples with first clinical pregnancy loss after embryonic genetic testing by CMA. A comprehensive and precise genetic counselling could be offered to SM couples to estimate the recurrent risk and take necessary measures to avoid subsequent miscarriage. The sensitivity analysis confirmed the robustness of our findings. Our research also exists some limitations. The sample size in some subgroups is not sufficient enough. In addition, about one-tenth of couples were lost to follow-up, and prognosis of these couples remains unknown.
In conclusion, our study demonstrated that CMA testing of POCs could provide an accurate genetic diagnosis for couples with SM. SM couples with chromosomally abnormal miscarriage manifested with a similar reproductive prognosis to couples with chromosomally normal miscarriage. The live birth rate of couples with partial aneuploid miscarriage was as high as couples with chromosomally normal miscarriage, despite a higher risk of adverse pregnancy event.
Authors contributions
All authors substantial contributed to the conception and design of the study. ZYX, RZ and YML acquired the data and drafted the manuscript. LLM, MTH, JXT, FCQ, HZ and PH analysed the data. QYZ, ZFX and YW conducted interpretation of the data. All authors revised the work critically for intellectual content, approved the final version to be published and agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (16.7 KB)Supplemental Material
Download MS Word (16.7 KB)Supplemental Material
Download MS Word (17.1 KB)Supplemental Material
Download MS Word (19.2 KB)Supplemental Material
Download MS Word (35 KB)Supplemental Material
Download MS Word (38 KB)Supplemental Material
Download MS Word (85.5 KB)Acknowledgements
We greatly appreciate all the participants for their cooperation in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Larsen EC, Christiansen OB, Kolte AM, et al. New insights into mechanisms behind miscarriage. BMC Med. 2013;11:154.
- Quenby S, Gallos ID, Dhillon-Smith RK, et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. 2021;397(10285):1658–1667.
- American College of Obstetricians and Gynecologists’ Committee. ACOG practice bulletin no. 200: early pregnancy loss. Obstet Gynecol. 2018;132(5):e197–e207.
- Sahoo T, Dzidic N, Strecker MN, et al. Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet Med. 2017;19(1):83–89.
- Zhang T, Sun Y, Chen Z, et al. Traditional and molecular chromosomal abnormality analysis of products of conception in spontaneous and recurrent miscarriage. BJOG. 2018;125(4):414–420.
- Huchon C, Deffieux X, Beucher G, et al. Pregnancy loss: French clinical practice guidelines. Eur J Obstet Gynecol Reprod Biol. 2016;201:18–26.
- Zhu X, Li J, Zhu Y, et al. Application of chromosomal microarray analysis in products of miscarriage. Mol Cytogenet. 2018;11:44.
- Lee JM, Shin SY, Kim GW, et al. Optimizing the diagnostic strategy to identify genetic abnormalities in miscarriage. Mol Diagn Ther. 2021;25(3):351–359.
- Lathi RB, Milki AA. Tissue sampling technique affects accuracy of karyotype from missed abortions. J Assist Reprod Genet. 2002;19(11):536–538.
- Wang Y, Li Y, Chen Y, et al. Systematic analysis of copy-number variations associated with early pregnancy loss. Ultrasound Obstet Gynecol. 2020;55(1):96–104.
- Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2):245–257.
- Finley J, Hay S, Oldzej J, et al. The genomic basis of sporadic and recurrent pregnancy loss: a comprehensive in-depth analysis of 24,900 miscarriages. Reprod Biomed Online. 2022;45(1):125–134.
- Choi TY, Lee HM, Park WK, et al. Spontaneous abortion and recurrent miscarriage: a comparison of cytogenetic diagnosis in 250 cases. Obstet Gynecol Sci. 2014;57(6):518–525.
- Nikitina TV, Sazhenova EA, Tolmacheva EN, et al. Comparative cytogenetic analysis of spontaneous abortions in recurrent and sporadic pregnancy losses. Biomed Hub. 2016;1(1):1–11.
- Gao J, Liu C, Yao F, et al. Array-based comparative genomic hybridization is more informative than conventional karyotyping and fluorescence in situ hybridization in the analysis of first-trimester spontaneous abortion. Mol Cytogenet. 2012;5(1):33.
- Robberecht C, Schuddinck V, Fryns JP, et al. Diagnosis of miscarriages by molecular karyotyping: benefits and pitfalls. Genet Med. 2009;11(9):646–654.
- Murugappan G, Leonard SA, Newman H, et al. Karyotype of first clinical miscarriage and prognosis of subsequent pregnancy outcome. Reprod Biomed Online. 2021;42(6):1196–1202.
- Sugiura-Ogasawara M, Ozaki Y, Sato T, et al. Poor prognosis of recurrent aborters with either maternal or paternal reciprocal translocations. Fertil Steril. 2004;81(2):367–373.
- Sugiura-Ogasawara M, Ozaki Y, Katano K, et al. Abnormal embryonic karyotype is the most frequent cause of recurrent miscarriage. Hum Reprod. 2012;27(8):2297–2303.
