Abstract
Background
Type 1 diabetes mellitus (T1DM) is disease caused by the destruction of β pancreatic cells. The activation of T-lymphocyte and proliferation inhibitor are induced by protein tyrosine phosphatase non-receptor type 22 (PTPN22). However, the link between PTPN22 C1858T gene polymorphism and T1DM is still controversy. This study aimed to analyse the C1858T gene polymorphism in Indonesian children with T1DM.
Materials and methods
This case-control study was conducted from March 2021 to May 2022 in the Endocrinology Outpatient Clinic at Dr. Soetomo Hospital and Tropical Disease Center Universitas Airlangga. Patients with controlled T1DM during the study period were included. The PTPN22 analysis used polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) method.
Results
Sixty-two children voluntarily participated in this study, and were equally divided into the T1DM and control groups. Most of the patients (94%, 58/62) are Javanese. This study revealed a more frequent CC genotype (9.4%) and allele-C (54.6%) polymorphism in the T1DM group, while more frequent CT genotype (100%) and allele-T (50%) polymorphism were in the control group. The C- and T-allele frequency was 54.6% and 45.4% in the T1DM group, respectively. The T1DM and control groups did not significantly differ (p= .2381).
Conclusions
PTPN22 homozygous genotype-CC and allele-C polymorphisms are more frequent in patients with T1DM. However, the PTPN22 C1858T gene polymorphism did not significantly correlate to T1DM children in this study.
The PTPN22 C1858T gene polymorphism does not significantly affect the susceptibility of T1DM in Indonesian children.
PTPN22 homozygous genotype-CC polymorphism was more observed in the T1DM group; thus, this genotype may play as a risk factor for T1DM children in the Indonesian population.
Key Messages:
Keywords:
Background
Diabetes mellitus (DM) is a metabolic disorder characterized by persistent hyperglycaemia. Two forms of DM frequently affect children; type 1 diabetes mellitus (T1DM) is commonly reported as an autoimmune illness due to β-pancreatic cell destruction, and type 2 DM (T2DM), also known as non-insulin dependent DM. Both types of diabetes are polygenic, which means they are linked to various genes [Citation1].
The prevalence of T1DM was 10% from diabetic patient [Citation2]. The prevalence of T1DM is <1% of the entire population, and its incidence rate rapidly increased globally, an estimated threefold increase in prevalence by 2040 [Citation3]. T1DM incidence is very different in some places. Some studies have found that the prevalence of T1DM was lower among Asians than Caucasians [Citation4–10]. China reported a T1DM annual incidence of 0.1/100,000, Japan with 1.4/100,000, and Finland with 43/100,000, while in Indonesia is approximately 0.3/100,000 [Citation2].
There are various causes of T1DM, such as genetic risk factors including β-pancreatic cell destruction induced by T-cell [Citation2,Citation11,Citation12]. In 2008, Ikegami et al. reported that Japanese and Korean populations had five new single-nucleotide polymorphisms (SNPs). The −1123G > C SNP was correlated with T1DM in both populations. The human leukocyte antigen (HLA), cytotoxic T lymphocyte antigen-4 (CTLA-4) and protein tyrosine phosphatase non-receptor type 22 (PTPN22) are the crucial genes associated with T1DM susceptibility [Citation13]. In Indonesian study, the HLA-DQA1 and HLA-DQB1 subtypes mainly found in Indonesian children with T1DM are HLA-DQA1 0101/0102 and HLA-DQB1 0301 [Citation14]. Furthermore, the CTLA-4 1822 C/T polymorphism might be a protective factor against T1DM [Citation15].
The PTPN22 gene has a significant role in preventing T cell activation and proliferation. The mutation of this gene can induce and maintain the autoimmunity [Citation16]. PTPN22 polymorphism varies among races. Studies have investigated the relationship between T1DM and the PTPN22 C1858T. There have been reports of the 1858T allele being linked to T1DM in several countries, including Italy, Germany, Spain, Ukraine and France [Citation13]. This topic has been studied since 12 years ago [Citation12,Citation17,Citation18]. However, PTPN22 studies in Indonesia is limited. Hence, this study aims to evaluate the association between children with PTPN22 C1858T gene polymorphism and T1DM.
Materials and methods
Study was conducted in the Pediatric Endocrine Outpatient Clinic at Dr. Soetomo Hospital and Tropical Disease Center (TDC) Universitas Airlangga from March 2021 to May 2022. Children aged 4–18 years, who are willing to join in this study, were included in the T1DM group. Type 1 DM diagnosis was based on classic symptoms, elevated blood glucose level, low C-peptide and positive antibodies (GAD-65 and ZnT8) [Citation19].
Meanwhile, children without T1DM, who visited the Pediatric Outpatient Clinic at Dr. Soetomo Hospital, in stable condition, and willing to join this study, belonged to the control group. Children due to severe illness and their parents who refused to join in the study were excluded. Consecutive random sampling was used for collecting samples and the sample size was determined by using the sample calculation formula for a cross-sectional study [Citation20]. This study was approved by the Clinical Research Unit at Dr. Soetomo Hospital, Surabaya, Indonesia with the number of 1889/KEPK/III/2020.
Genetic analysis
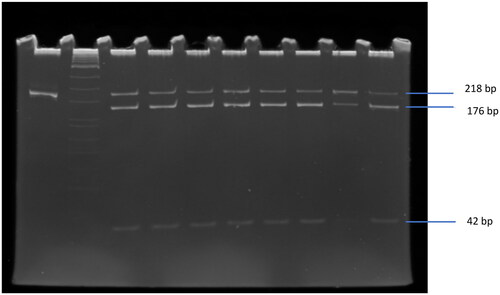
The QIAmp DNA Mini Kit was used to extract DNA from peripheral blood mononuclear cells according to standard procedure. The DNA fragment was 218 bp resulted from the amplification of forward primer: 5′-ACTGATAATGTTGCTTCAACGG-3′ and reverse primer: 5′-TCACCAGCTTCCTCAACCAC-3′ by polymerase chain reaction (PCR) (Applied BioSystems, Foster City, CA). The PCR mixture consisted of 10× PCR buffer, 250 ng template DNA, 20 pmol of each primer, 1.5 mM MgCl2, 0.2 mM of each of the deoxyribonucleotide triphosphates, and 1 U GoTaq DNA polymerase (Promega, Fitchburg, WI). The amplification consists of denaturation for 2 min at 94 °C followed by 35 cycles at 94 °C for 30 s, annealing 30 s at 60 °C and 30 s at 72 °C, and the final extension for 3 min at 72 °C. The polymorphism of C1858T was identified by restriction fragment length polymorphism (RFLP) as described by Bulut et al. [Citation21]. The PCR product was cut by RsaI enzyme (New England Biolabs, Ipswich, MA) at 37 °C for 4 h and resulted in two fragments, 176 bp and 42 bp that indicated the homozygous CC (wild type), whereas the heterozygous CT had three fragments at 218, 176 and 42 bp (see ). The mutant type (homozygous TT) was at 218 bp that cannot be cut by RsaI enzyme. The PCR product, which was not cut by RsaI enzyme, was used as the control of 218 bp position.
Statistical analysis
SPSS version 20.0 (IBM, Armonk, NY) was applied for data analysis. Either Fisher’s exact or Chi-square test was used as requisite for the genotypes polymorphisms and allele distribution comparison between T1DM and control groups. The significant difference or correlation was shown by p value (.05).
Results
Sixty-two children involved in this study were divided as follows: 31 in the T1DM and 31 in the control group. The median age of all subjects was 12.6 (2.7–16.8) years, with 27 males and 35 females, and mostly Javanese tribe (94%, 58/62). The onset of T1DM occurred mostly in the childhood under 12 year-old (90.3%, 28/31) (see ).
Table 1. Characteristics of subject.
The CC genotype was mostly found in T1DM group but not significantly different in the controls (p= .238 95% CI: 0 (0–NA)). The prevalence of CT genotype in T1DM children was 90.3% (28/31), while 100% (31/31) was in the control. The frequency of the C allele and T allele in children with T1DM was 54.8% and 45.2%, respectively ().
Table 2. Polymorphism of PTPN22 C1858T distribution.
Discussion
Genetics and environment contribute to the risk of T1DM. Although the major histocompatibility complex (MHC) is closely connected to the genetic susceptibility to T1DM, the non-HLA gene is also thought to be present to promote the disease. The non-HLA genes such as PTPN22 may confer the risk of T1DM through T-cell-mediated immune response [Citation22,Citation23].
The PTPN22 C1858T gene polymorphism in T1DM children was not significantly different in the control group. It seems that PTPN22 C1858T gene polymorphism is not associated with the occurrence of T1DM. Therefore, interestingly, the variant C allele was present homozygous in 9.7% (3/31) children with T1DM and not in the control group. This might play a role in T1DM susceptibility. Hence, it needs further study with a lot of samples.
Our study was in line with previous studies, which provided no significant association between PTPN22 C1858T polymorphism and T1DM [Citation22,Citation23]. A recent study from Azerbaijani also reported that polymorphisms of the PTPN22 gene (polymorphisms −1123 C/G and +2740 A/G) did not correlate with T1DM [Citation24]. The same result was shown in a study of Egyptian children with systemic lupus erythematosus [Citation25]. Furthermore, a meta-analysis study that described the PTPN22 C1858T polymorphism in Europe and the American population may play as a risk factor in T1DM. However, in contrast with this study, PTPN22 was associated with T1DM in the Colorado, Egyptian children and Asian population [Citation26–28].
This study found genotype-CC and allele-C more frequent in T1DM children. In contrast, another study showed that the TT-1858 genotype was more prevalent in children with T1DM (p= .038, OR: 3.16; 95% confidence interval (CI): 1.28–7.09). The study concludes that PTPN22 C1858T polymorphisms increased the risk factor of T1DM [Citation27]. Other studies also suggest that the prevalence of the PTPN22 variant (rs2542151), the G allele, may increase the risk of T1DM [Citation28].
The limitation is that this is a single-centre study, further multicentre research is necessary to gather additional information on Indonesian races.
Conclusions
The homozygous genotype-CC and allele-C are more often found in T1DM. This study shows that PTPN22 C1858T polymorphism did not significantly had a role in T1DM genetic susceptibility. Further studies, such as multicentre studies and studies in different ethnic groups, are needed for the influence of the PTPN22 C1858T polymorphism to the T1DM susceptibility.
Author contributions
Study concept and design: FA and MF. Acquisition of the data: FA. Analysed and interpreted the data: SB and NR. Drafted the manuscript: FA and NR. Critical revision of the manuscript: NR and MF. Supervised the study: NR and SB. All authors read and approved the final manuscript.
Ethics statement
This study was approved by Dr. Soetomo Hospital Clinical Research Unit with the reference number of 1889/KEPK/III/2020. The Declaration of Helsinki was followed by all research participants. The parents or legal guardians of each participant signed the informed consent form.
Acknowledgements
Thanks to the Dr. Soetomo General Hospital endocrinology staff and all of the study participants.
Disclosure statement
No potential conflict of interest was reported by the author(s)e.
Data availability statement
The corresponding author can provide you with the information needed to understand the results of this study.
Additional information
Funding
References
- Ali KI. Do second generation sequencing techniques identify documented genetic markers for neonatal diabetes mellitus? Heliyon. 2021;7(9):e07903.
- WHO. Diabetes and genetics. WHO; 2023. Available from: www.who.int/genomics/about/Diabetis-fin.pdf
- Yahaya T, Salisu T. Genes predisposing to type 1 diabetes mellitus and pathophysiology: a narrative review. Med J Indones. 2020;29(1):100–109.
- Rochmah N, Faizi M, Hisbiyah Y, et al. Quality of life differences in pre- and post-educational treatment in type 1 diabetes mellitus during COVID-19. Diabetes Metab Syndr Obes. 2021;14:2905–2911.
- Gomes KF, Semzezem C, Batista R, et al. Importance of zinc transporter 8 autoantibody in the diagnosis of type 1 diabetes in Latin Americans. Sci Rep. 2017;7(1):207.
- Bhatty A, Baig S, Fawwad A, et al. Association of zinc transporter-8 autoantibody (ZnT8A) with type 1 diabetes mellitus. Cureus. 2020;12(3):1–8.
- Braga de Souze AC, Felicio JS, Neto JF, et al. Health-related quality of life in people with type 1 diabetes mellitus: data from the Brazilian type 1 diabetes study group. Health Qual Life Outcomes. 2015;13:204.
- Kiani AK, John P, Bhatti A, et al. Association of 32 type 1 diabetes risk loci in Pakistani patients. Diabetes Res Clin Pract. 2015;108(1):137–142.
- Achenbach P, Lampasona V, Landherr U, et al. Autoantibodies to zinc transporter 8 and SLC30A8 genotype stratify type 1 diabetes risk. Diabetologia. 2009;52(9):1881–1888.
- Mattana TC, Santos AS, Fukui RT, et al. CD226 rs763361 is associated with the susceptibility to type 1 diabetes and greater frequency of GAD65 autoantibody in a Brazilian cohort. Mediators Inflamm. 2014;2014:1–7.
- Diet R, Niken PY, Novina A, et al. Buku ajar endokrinologi. Vol. 2. Jakarta: IDAI; 2018. p. 146–206.
- Jaya RI, Zettyana YR, Bakri A, et al. Polymorphisms associated with type 1 diabetes mellitus. Paediatr Indones. 2018;58(6):274–279.
- Giza S, Antonius G, Emmanouela G, et al. The role of PTPN22 C1858T gene polymorphism in diabetes mellitus type 1: first evaluation in Greek children and adolescents. BioMed Res Int. 2013;2013:721604.
- Soetjipto , Rochmah N, Faizi M, et al. HLA-DQA1 and HLA-DQB1 gene polymorphism in Indonesian children with type I diabetes mellitus. Appl Clin Genet. 2022;12(15):11–17.
- Rochmah N, Faizi M, Nova S, et al. CTLA-4 CT-60 A/G and CTLA-4 1822 C/T gene polymorphisms in Indonesians with type 1 diabetes mellitus. Appl Clin Genet. 2022;29(15):19–25.
- Cohen S, Dadi H, Shaoul E, et al. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93(6):2013–2024.
- Ozougwu JC, Obimba KC, Belonwu CD, et al. The pathogenesis and pathophysiology of type 1 and type 2 diabetes mellitus. J Physiol Pathophysiol. 2013;4(4):46–57.
- Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol. 2014;32(1):83–119.
- Mayer-Davis EJ, Kahkoska AR, Jefferies C, et al. ISPAD clinical practice consensus guidelines 2018: definition, epidemiology, and classification of diabetes in children and adolescents. Pediatr Diabetes. 2018;19(27):7–19.
- Pourhoseingholi MA, Vahedi M, Rahimzadeh M. Sample size calculation in medical studies. Gastroenterol Hepatol Bed Bench. 2013;6(1):14–17.
- Bulut F, Erol D, Elyas H, et al. Protein tyrosine phosphatase non-receptor 22 gene C1858T polymorphism in patients with coexistent type 2 diabetes and Hashimoto’s thyroiditis. Balkan Med J. 2014;31(1):37–42.
- Elsisi O, Kamal M, Madani H, et al. Association of protein tyrosine phosphatase non receptor type 22 (PTPN22) C1858T gene polymorphism with type 1 diabetes mellitus in Egyptian children cohort. Gaz Egypt Pediatr Assoc. 2015;69:75–79.
- Baniasadi V, Das SN. No evidence for association of PTPN22 R620W functional variant C1858T with type 1 diabetes in Asian Indian. J Cell Mol Med. 2008;12(3):1061–1062.
- Ahmadov G. Study of the PTPN22 gene in children with type 1 diabetes mellitus in the Azerbaijani population. Georgian Med News. 2017;271:45–49.
- Hamza RT, Awwad KS, Temsah KA, et al. R620W polymorphism of protein of protein tyrosine phosphatase PTPN22 in Egyptian children and adolescents with systemic lupus erythematosus: relation to thyroid autoimmunity. Int J Adolesc Med Health. 2013;25(2):143–149.
- Steck AK, Liu SY, McFann K, et al. Association of the PTPN22/LYP gene with type 1 diabetes. Pediatr Diabetes. 2006;7(5):274–278.
- Abou SS, Tawfik MA, Muhammed ZS, et al. PTPN22 gene and IL2RA rs11594656, rs2104286 gene variants: additional insights of polygenic single-nucleotide polymorphisms’ pattern among Egyptian children with type 1 diabetes. Gaz Egypt Pediatr Assoc. 2021;69:35.
- Tangjittipokin W, Teerawattanapong N, Tangjarusritaratorn T, et al. Immunogenetics of type 1 diabetes in Asian populations. Genomics Genet. 2022;15(1):16–33.