Abstract
Objective
To screen and identify microRNAs (miRNAs) associated with the prognosis of lung adenocarcinoma (LUAD) using clinical samples and construct a prediction model for the prognosis of LUAD.
Methods
160 patient samples were used to screen and identify miRNAs associated with the prognosis of LUAD. Differentially expressed miRNAs were analyzed using gene chip technology. The selected miRNAs were validated using samples from the validation sample group. Cox proportional hazards regression was used to construct the model and Kaplan-Meier was used to plot survival curves. Model power was assessed by testing the prognosis of the constructed model using real-time polymerase chain reaction (RT-PCR) data.
Results
The data showed that miR-1260b, miR-21-3p and miR-92a-3p were highly expressed in the early recurrence and metastasis group, while miR-2467-3p, miR-4659a-3p, miR-4514, miR-1471 and miR-3621 were lowly expressed. It was further confirmed that miR-21-3p was significantly highly expressed in the early recurrence and metastasis group (p = 0.02). Receiver operating characteristic (ROC) curve results showed cut-off point value of 0.0172, sensitivity of 88.2% and specificity of 100%. The predictive results of the constructed model were in good agreement with the actual prognosis of patients by using the validation sample test (Kappa = 0.426, p < 0.001), with a model sensitivity of 74.4%, a specificity of 68.3%, and an accuracy of 71.3%.
Conclusion
miRNAs associated with the prognosis of patients with stage I LUAD were screened and validated, and a risk model for predicting the prognosis of patients was constructed. This model has good consistency with the actual prognosis of patients.
Introduction
Lung cancer is currently one of the leading causes of cancer-related death in the world [Citation1]. It is the most prevalent cancer in men and the second most prevalent cancer in women [Citation2]. Histologically, lung cancer can be divided into small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC) [Citation3]. About 80% of these lung cancer types are NSCLC [Citation4]. Lung adenocarcinoma (LUAD) is the most common histological subtype of NSCLC, accounting for approximately 40% of all cases [Citation5]. Despite improvements in molecular diagnosis and treatment, the prognosis of LUAD remains poor and the risk of metastasis and recurrence remains high [Citation6]. The main reason is that early lung adenocarcinoma is difficult to diagnose, most LUAD patients are diagnosed at an advanced stage, and effective treatments are lacking [Citation7]. Therefore, the 5-year survival rate of lung adenocarcinoma is poor. Identifying biomarkers and potential therapeutic agents for the diagnosis and prognosis of LUAD is important to improve survival in patients with lung adenocarcinoma [Citation8].
MicroRNAs (miRNAs) are small non-coding RNAs ranging from 18 nucleotides to 25 nucleotides in length [Citation9]. MiRNAs interfere with mRNA translation by base-pairing with the 3 ‘untranslated region (UTR) of target mRNAs, resulting in mRNA degradation or translational repression [Citation10]. Several studies have shown that miRNAs have multiple cellular regulatory roles and functions associated with cancer initiation and progression [Citation11–13]. MiRNAs can therefore currently be an effective biological marker for detecting, classifying, and predicting a variety of cancers [Citation14]. Li et al. identified eight miRNA signatures as potential biomarkers for predicting survival status in LUAD patients [Citation15]. Although considerable progress has been made in systematically evaluating cancer-related miRNAs and molecular markers to predict overall survival (OS) or response to immunotherapy in LUAD patients, professional in-depth clinical evaluation still requires more diagnostic and therapeutic miRNA biomarkers to support personalized treatment in lung cancer patients, which needs to be confirmed by randomized multicenter clinical trials [Citation16–18]. Expression profiling using high-throughput microarrays has become a widely used technique. It can measure the expression of thousands of genes at a time and identify new cancer biomarkers [Citation19]. In this study, miRNA expression microarrays and clinical patient samples were used to screen miRNAs associated with the prognosis of LUAD, and a risk prediction model was constructed in the hope of finding new therapeutic targets and biomarkers for LUAD.
Methods and materials
Patient information and tissue samples
Samples of tumor tissue were collected from 160 patients with lung adenocarcinoma who received a surgical resection between January 2004 and December 2012 at the Department of Thoracic Surgery, Cancer Hospital, Chinese Academy of Medical Sciences. All tissue samples were fixed and paraffin-confirmed (FFPE) and were confirmed by pathological diagnosis. All samples were primary lung adenocarcinoma. Clinicopathological features of all patients were collected (). All patients were followed up until April 2019. This study complied with the approved guidelines of the Cancer Hospital Chinese Academy of Medical Sciences and informed consent was obtained from all patients.
Table 1. Basic clinical characteristics of initial sample group.
MiRNA expression microarray hybridization
After fluorescently labeling total RNA, according to the miRNA expression microarray hybridization step, 10 × blockers, hybridization samples, hybridization cassettes were prepared successively, and a clean gasket slide was placed in the hybridization cassette chamber, and all specimens of fluorescently labeled RNA were slowly added to gasket well of gasket slide, respectively, to avoid bubbles as much as possible, and then covered. In this process, Agilent faces down on the side labeled with number and covers the cover of SureHyb chamber on the Microarray slide, slides the clamp to the chamber, tightens the Hybridization chamber, turns in the vertical direction for 3 weeks, and the fused large bubbles rotate accordingly, in normal state. Following this, they were placed in a hybridization oven at 55 °C with a rotation speed of 20 rpm for 20 h of hybridization.
MiRNA microarray data extraction and analysis
Quality reports were generated after data extraction with Agilent Feature Extraction software. The quality of chip hybridization in the process of labeling, hybridization and cleaning is shown from the aspects of Grid selection, chip cleaning, chip signal distribution range, chip background noise, and chip fluorescence signal distribution. Normalized miRNA microarray data were obtained by GeneSpring GX software analysis. Further principal component analysis, differential miRNA screening based on p-value and Fold Change, differential miRNA screening based on p-value and Fold Change and survival analysis, and cluster analysis were subsequently performed.
Real-time polymerase chain reaction (RT-PCR)
RNA content was extracted from tumor tissues using miRNeasy FFPE Kit (217504, Qiagen, Shanghai, China) and RNA quality and concentration were measured with a Nanodrop2000 Micro UV spectrophotometer (Thermo, Waltham, MA, USA). MiRNAs were reverse transcribed into cDNA according to TaqMan® MicroRNA Reverse Transcription Kit (4366596, Invitrogen, Waltham, MA, USA). PCR was performed with TaqMan® microRNA Assay and TaqMan™ Universal Master Mix II, no UNG (4440049, Invitrogen, Waltham, MA, USA). Fold change calculations were performed using the 2-ΔΔCT method using miR-191 as an internal reference for miRNAs.
Statistical analysis
Cox proportional hazards regression was used to construct the model. The prognostic index (PI) calculated by the equation was used for receiver operating characteristic (ROC) curve analysis. Kaplan-Meier was used to draw the survival curve. Chi-square test (McNemer test and Kappa agreement test) was performed for paired design between PI prediction results and the actual prognosis of patients. The R programming language was used for all statistical analyses (version 3.6). p < 0.05 was considered to indicate a statistically significant difference.
Results
Patient basic information in different groups
Eighty LUAD patients’ samples served as the initial sample group for the screening of miRNAs associated with patient prognosis. Forty patients in the initial sample group developed recurrence and metastasis within 2 years after surgery and were classified as the early recurrence and metastasis group. The other 40 patients did not develop recurrence and metastasis more than 5 years after surgery and were classified as the group without recurrence and metastasis. The statistical analysis results showed that there was no significant difference in age, gender, smoking status, surgical methods, tumor location, tumor differentiation, pleural invasion, and postoperative adjuvant therapy between the two groups (p ≥ 0.05) (). This suggests that the balance of demographic and clinical characteristics between the two groups of patients in the initial sample group meets the requirements of univariate analysis and can be used for the correlation study between miRNAs and recurrence and metastasis of stage I LUAD.
An additional sample of 80 lung adenocarcinoma patients was subsequently used as a validation sample group for validation of the selected miRNAs. In the validation sample group, 39 patients developed recurrence and metastasis within 2 years after surgery and were classified as the early recurrence and metastasis group. The other 41 patients did not develop recurrence and metastasis more than 5 years after surgery and were classified as the non-recurrence and metastasis group. The statistical analysis results showed that there was no significant difference in age, gender, smoking status, surgical methods, tumor location, tumor differentiation, pleural invasion, and postoperative adjuvant therapy between the two groups (p ≥ 0.05) (). This suggests that the balance of demographic and clinical characteristics between the two groups of patients with the validation sample group meets the requirements of univariate analysis and can be used to verify the association between candidate differentially expressed miRNAs and recurrence and metastasis of stage I LUAD.
Table 2. Basic clinical characteristics of validation sample group.
MiRNAs expression microarray data quality analysis
The miRNA expression profiles of the two groups of patients were preliminarily obtained using gene chip technology (miRNA expression microarray) with the initial sample group of research materials. The miRNA expression microarray hybridization process met the operating requirements and the microarray data quality was good (). Principal component analysis showed that miRNA expression profiles were similar between patients with early recurrence and metastasis and those without recurrence and metastasis in the initial sample group ().
Figure 1. miRNA microarray quality control images. A. Reflects correct grid selection. B: Reflects clean chip cleaning. C. Reflects chip signal distribution range. D. Reflects background noise of chip. E. Reflects fluorescence signal distribution. F. Quality Control Metrics-plot of miRNA microarray data. Indicators are as follows: Any color PrcntFeatPopnOL: percentage content of Feature escape value; gTotalSignal75pctiIe: 75% value of all gene signals; Add Error Estimate Green: additional error value; gNon Ctrl Med Prcnt CVBG Sub Sig: coefficient of variation value to remove background signal. G. Each point represents one case. Red: early recurrence metastasis group. Blue: no recurrence metastasis group.
Screening of candidate miRNAs
Normalized microarray data were filtered according to Flags values, and then probes with no detected signal in more than 50% of samples in both groups were removed. And 14 miRNAs that were significantly differentially expressed in the two groups of samples based on p-values as well as Fold Change were obtained as candidate miRNAs using p < 0.05 and Fold Change > 1.5 as screening criteria ().
Table 3. Differentially expressed miRNAs in the initial sample group (based on p value and Fold change).
For the above 14 candidate miRNAs, the initial sample group was divided into low and high miRNA expression groups using the median of their expression in the samples as the cut-off point, and the survival curves were plotted using the Kaplan-Meier method, and the log-rank method was used to compare the survival rates between the two groups. Combined with the results of survival analysis of patients (), a total of 8 candidate differentially expressed miRNAs associated with postoperative recurrence and metastasis of stage I lung adenocarcinoma were obtained (). Among them, miR-1260b, miR-21-3p, and miR-92a-3p were highly expressed in the early recurrence and metastasis group, while miR-2467-3p, miR-4659a-3p, miR-4514, miR-1471, and miR-3621 were lowly expressed compared with the non-recurrence and metastasis group.
Figure 2. Relationship between expression levels of eight candidate differentially expressed miRNAs and progression-free survival (PFS) in the initial sample group. A. Relationship between miR-1260b expression and prognosis of patients, p = 0.001. B. Relationship between miR-21-3b expression and prognosis of patients, p = 0.02. C. Relationship between miR-4514 expression and prognosis of patients, p = 0.024. D. Relationship between miR-4659a-3p expression and prognosis of patients, p = 0.009. E. Relationship between miR-2467-3p expression and prognosis of patients, p < 0.001. F. Relationship between miR-3621 expression and prognosis of patients, p = 0.026. G. Relationship between miR-1471 expression and prognosis of patients, p = 0.013. H. Relationship between miR-92a-3p expression and prognosis of patients, p = 0.011. I. Heatmap of eight candidate miRNAs. The abscissa is the case and the ordinate is eight candidate miRNAs.
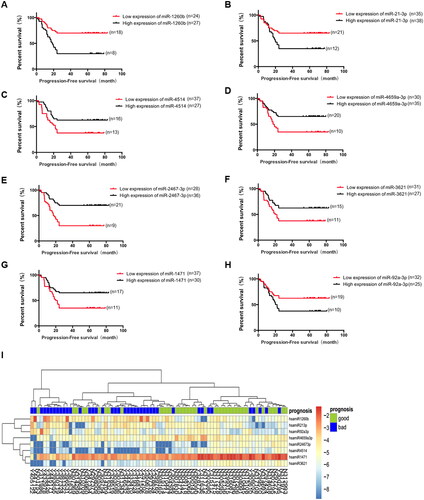
Table 4. Differentially expressed miRNAs in the initial sample group (based on p value, Fold change and patient survival analysis).
By cluster analysis, the candidate miRNAs could better distinguish between lung adenocarcinoma cases with early postoperative recurrence and metastasis and those without recurrence and metastasis compared with principal component analysis (PCA) of miRNA expression profiles of tumor tissues from an initial sample group of 80 patients ().
Validation of candidate miRNAs
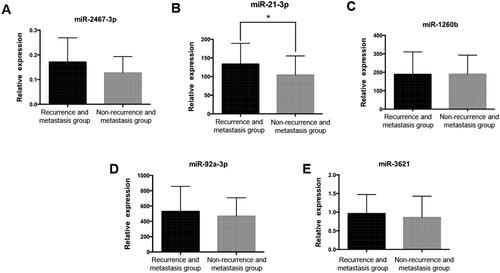
For the above 8 candidates differentially expressed miRNAs associated with postoperative recurrence and metastasis of stage I lung adenocarcinoma, 5 miRNAs (miR-21-3p, miR-92a-3p, miR-1260b, miR-2467-3p, miR-3621) were finally selected by referring to the literature [Citation20–24], and the differential expression of these 5 miRNAs between the two groups of early recurrence and metastasis and no recurrence and metastasis was validated using RT-PCR with the validation sample group as the research material (). Among them, miR-21-3p was significantly highly expressed in the early recurrence and metastasis group (p = 0.02), while the expression of miR-2467-3p, miR-92a-3p, miR-1260b and miR-3621 was not significantly different between the two groups (p > 0.05).
Figure 3. Real-time PCR validation of candidate miRNAs in validation sample group. A. miR-2467-3p expression level in validation sample group, p > 0.05. B. miR-21-3p expression level in validation sample group, *: p = 0.02. C. miR-1260b expression level in validation sample group, p > 0.05. D. miR-92a-3p expression level in validation sample group, p > 0.05. E. miR-3621 expression level in validation sample group, p > 0.05.
Construction and validation of prognostic evaluation model
Cox proportional hazards regression was used to construct the model, and the results yielded that miR-1260b, miR-2467-3p, miR-92a-3p, and tumor grade entered the hazard function expression equation (). The expression equation of the risk function was: PI = ZmiR-1260b × 0.499 - ZmiR-2467-3p × 0.853 + ZmiR-92a-3p × 0.641 - Zgrade × 0.496. The PI calculated by the above equation was used for ROC curve analysis to distinguish the early recurrence and metastasis group from the recurrence-free metastasis group. And the cut-off point value of ROC curve was obtained as PI = 0.0172, with a sensitivity of 88.2% and a specificity of 100% ().
Figure 4. The ROC curve for the early recurrence and metastasis group and the non-recurrence and metastasis group. A. The area under the ROC curve was 0.917, p < 0.001. B. Prognostic index in the initial sample group and progression-free survival in patients with lung adenocarcinoma, p < 0.001. C. Prognostic index in the validation sample group and progression-free survival in patients with lung adenocarcinoma, p < 0.001.
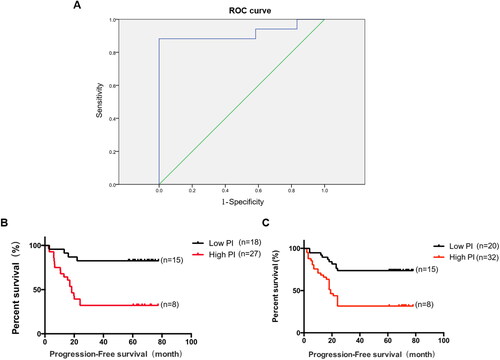
Table 5. Multivariate Cox regression analysis results for the initial sample group.
PI was divided into high and low groups by the cut-off point values obtained above, with poor prognosis in the high PI group and good prognosis in the low PI group, and survival curves were plotted with Kaplan-Meier (). The mean progression-free survival (PFS) of patients in the high PI group was significantly lower than that of patients in the low PI group, and the difference was statistically significant (p < 0.001). Pairwise design chi-square test (McNemer test and Kappa agreement test) was performed between PI prediction results and actual patient prognosis. The results showed that the percentage of poor prognosis in PI prediction results was 51.0%, which was not significantly different from the percentage of poor actual patient prognosis (45.1%) (McNemer test, p = 0.549), and the consistency between PI prediction results and actual patient prognosis was good (Kappa = 0.569, p < 0.001) (). The sensitivity, specificity and accuracy of the model were 82.6%, 75.0% and 78.4%, respectively.
Table 6. Chi-square test for paired design between PI prediction results of initial sample group and actual patient survival.
RT-PCR analysis of the validation sample group was used as the test data to test the efficacy of the prognostic evaluation model for stage I LUAD. Survival curves were plotted with Kaplan-Meier (). The mean PFS of patients in the high PI group was significantly lower than that of patients in the low PI group, and the difference was statistically significant (p < 0.001). Chi-square test of paired design data (McNemer test and Kappa agreement test) was performed between PI prediction results and actual prognosis of patients. The results showed that the percentage of poor prognosis predicted by PI was 52.5%, which was not significantly different from the percentage of poor prognosis predicted by PI (50%) (McNemer test, p = 0.678), and the consistency between PI prediction results and actual prognosis of patients was good (Kappa = 0.426, p < 0.001) (). The model sensitivity was 74.4%, specificity was 68.3% and accuracy was 71.3%.
Table 7. Chi-square test for paired design between PI prediction results of validation sample group and actual patient survival.
Discussion
Early extensive metastasis and easy recurrence are the main features of LUAD. Most patients with lung adenocarcinoma have distant metastases at the time of diagnosis and have a poor prognosis [Citation25,Citation26]. At the molecular level, the pathogenesis of LUAD is still unclear and early diagnosis is difficult [Citation27]. Therefore, there is an urgent need to find more effective biomarkers for early diagnosis and risk prediction of prognosis. Microarray technology is an effective method for analyzing biomarkers and can be used to investigate gene expression profiles in LUAD. MiRNAs can influence the development and metastasis of LUAD by downregulating or upregulating mRNA expression levels [Citation28]. In this study, we screened miRNAs associated with the prognosis of lung adenocarcinoma using patient samples and constructed a prognostic evaluation model, and also validated the selected miRNAs and prognostic evaluation models using clinical patient samples, providing new biomarkers for the prognosis of lung adenocarcinoma and new strategies for the treatment of lung adenocarcinoma.
Many studies have found that miRNAs are associated with many biological functions, such as proliferation, development, differentiation, apoptosis, metabolism and other physiological processes [Citation29–31]. Studies have shown that abnormal miRNA expression is significantly associated with the development of a variety of tumors [Citation32,Citation33]. Adenocarcinoma is the most common subtype of lung cancer and is associated with high morbidity and mortality [Citation34]. Several studies have shown that miRNAs have crucial functions in the prevention and prognosis of LUAD [Citation35,Citation36]. MiRNAs are complex combinations of gene expression and pathway regulatory systems and prognostic markers and therapeutic targets in a variety of cancers, including lung cancer [Citation37]. Multiple miRNAs play a key role in the development of lung cancer and regulate the process of lung cancer deterioration. Some studies reported miRNAs associated with prognostic value, such as miR-221 [Citation38], miR-372 [Citation39], miR-429 [Citation40], miR-486 [Citation41], and miR-137 [Citation42]. However, many miRNAs have not been identified in LUAD, and the role of miRNAs in the prognosis of lung adenocarcinoma remains to be explored.
In this study, 160 clinical samples of lung adenocarcinoma were used and divided into an initial sample group and a validation sample group. 80 samples in each group. Patients in each group were divided into a recurrence group and non-recurrence group according to their prognosis. The statistical analysis showed that there was no significant difference in age, gender, smoking status, surgical methods, tumor location, tumor differentiation, pleural invasion, and postoperative adjuvant therapy between the two groups (p ≥ 0.05) ( and ). This suggests that the balance of demographic and clinical characteristics between the two groups of patients meets the requirements of univariate analysis and can be used for the correlation study between miRNAs and recurrence and metastasis of stage I lung adenocarcinoma as well as validation studies. By using gene chip technology, miRNA expression profiles were obtained in two groups of patients in the relapse and non-relapse groups. Fourteen miRNAs based on p-value as well as Fold Change that were significantly differentially expressed in both sets of samples were obtained as candidate miRNAs using p < 0.05 and Fold Change > 1.5 as screening criteria (). A total of eight candidate differentially expressed miRNAs associated with postoperative recurrence and metastasis of stage I lung adenocarcinoma were subsequently obtained in combination with the survival curves of the patients (). Among them, miR-1260b, miR-21-3p, and miR-92a-3p were highly expressed in the early recurrence and metastasis group, while miR-2467-3p, miR-4659a-3p, miR-4514, miR-1471, and miR-3621 were lowly expressed compared with the non-recurrence and metastasis group. Cluster analysis showed that 8 candidates with differentially expressed miRNAs could better distinguish early postoperative recurrence and metastasis from lung adenocarcinoma cases without recurrence and metastasis ().
We subsequently validated the candidate miRNAs obtained from the screening using samples from the validation panel. By consulting the literature, five miRNAs (miR-21-3p, miR-92a-3p, miR-1260b, miR-2467-3p, and miR-3621) were finally selected for validation. MiR-21-3p was significantly highly expressed in the early recurrence and metastasis group as determined by RT-PCR results (p = 0.02), while miR-2467-3p, miR-92a-3p, miR-1260b, and miR-3621 were not significantly different between the two groups (p > 0.05) (). Luo et al. reported that two machine learning algorithms confirmed that miR-615-3p, miR-4652-5p, miR-450a-5p, hsa-miR-196a-5p, miR-21-3p, miR-139-5p, and miR-424-5p were critical diagnostic factors in laryngeal squamous cell carcinoma [Citation43]. Xie et al. [Citation44] reported that miR-21-3p upregulated the expression of PI3K-AKT pathway factors, CDK1, CCND1 but decreased RAF1 expression in hepatocellular cancer cells. And Zhang et al. [Citation45] also reported that miR‑21‑3p promoted chemo‑resistance via negatively regulating FOXO3a. The above reports are consistent with our conclusion that miR‑21‑3p is a biomarker of poor prognosis.
After identifying candidate miRNAs, we constructed a prognostic evaluation model for stage I lung adenocarcinoma using Cox proportional hazards regression based on the resulting miRNA data. According to the results in , miR-1260b, miR-2467-3p, miR-92a-3p, and tumor grade all entered the risk function expression equation. The expression equation of the risk function was: PI = ZmiR-1260b × 0.499 - ZmiR-2467-3p × 0.853 + ZmiR-92a-3p × 0.641 - Zgrade × 0.496. The PI calculated by the above equation was used for ROC curve analysis to distinguish the early recurrence and metastasis group from the recurrence-free metastasis group. The high PI group had a poor prognosis, and the low PI group had a good prognosis. The validation of this assessment model was subsequently performed using clinical data from the validation panel. PI prediction results were in good agreement with the actual prognosis of patients (Kappa = 0.426, p < 0.001) (), with a model sensitivity of 74.4%, specificity of 68.3%, and accuracy of 71.3%. This risk estimation model can be used to predict the prognosis of patients in subsequent clinical applications.
Conclusion
MiRNAs associated with the prognosis of lung adenocarcinoma were screened and validated by using clinical samples of lung adenocarcinoma. MiR-1260b, miR-21-3p and miR-92a-3p were found to be highly expressed in the early recurrence and metastasis group, while miR-2467-3p, miR-4659a-3p, miR-4514, miR-1471 and miR-3621 were lowly expressed. It was verified that miR-21-3p was significantly highly expressed in the early recurrence and metastasis group. The prognostic risk of lung adenocarcinoma patients can be predicted by constructing a prognostic evaluation model for lung adenocarcinoma. It provides a new biomarker for the prognosis detection of lung adenocarcinoma in clinical practice, and at the same time, it can provide a new target for the treatment of lung adenocarcinoma.
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of Cancer Hospital Chinese Academy of Medical Sciences. We obtained signed informed consent from the participants/legal guardians in this study.
Authors’ contributions
DHS and GYN were involved in the conception and design; YYK and CH were involved in the acquisition of patient information and tissue samples. DHS was involved in the analysis and interpretation of the data; DHS, LL, and GYN were involved in the drafting of the paper; DX and MYS were involved in revising it critically for intellectual content. All authors were involved in the final approval of the version to be published and agreed to be accountable for all aspects of the work.
Acknowledgements
Not applicable.
Disclosure statement
No potential conflict of interest was reported by the author(s). All of the authors had no personal, financial, commercial, or academic conflicts of interest separately.
Data availability statement
All data generated or analyzed during this study are included in this article.
Additional information
Funding
References
- Nugent WC, Edney MT, Hammerness PG, et al. Non-small cell lung cancer at the extremes of age: impact on diagnosis and treatment. Ann Thorac Surg. 1997; 63(1):1–12. doi: 10.1016/s0003-4975(96)00745-x.PMID: 8993264.
- Mederos N, Friedlaender A, Peters S, et al. Gender-specific aspects of epidemiology, molecular genetics and outcome: lung cancer. ESMO Open. 2020; 5(Suppl 4):e000796. doi: 10.1136/esmoopen-2020-000796.
- Ding Y, Zhang L, Guo L, et al. Comparative study on the mutational profile of adenocarcinoma and squamous cell carcinoma predominant histologic subtypes in Chinese non-small cell lung cancer patients. Thorac Cancer. 2020; 11(1):103–112. doi: 10.1111/1759-7714.13208.
- Gyoba J, Shan S, Roa W, et al. Diagnosing lung cancers through examination of Micro-RNA biomarkers in blood, plasma, serum and sputum: a review and summary of current literature. Int J Mol Sci. 2016;17(4):494. doi: 10.3390/ijms17040494.
- Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25(5):561–570. PMID: 17290066. doi: 10.1200/JCO.2006.06.8015.
- Xie D, Marks R, Zhang M, et al. Nomograms predict overall survival for patients with Small-Cell lung cancer incorporating pretreatment peripheral blood markers. J Thorac Oncol. 2015;10(8):1213–1220. PMID: 26200277. doi: 10.1097/JTO.0000000000000585.
- Li RZ, Fan XX, Duan FG, et al. Proscillaridin a induces apoptosis and suppresses non-small-cell lung cancer tumor growth via calcium-induced DR4 upregulation. Cell Death Dis. 2018;9(6):696. doi: 10.1038/s41419-018-0733-4.
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. 2019;322(8):764–774. PMID: 31454018. doi: 10.1001/jama.2019.11058.
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. PMID: 15105502. doi: 10.1126/science.1097434.
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. Epub 2007 Nov 29. PMID: 18048652. doi: 10.1126/science.1149460.
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. PMID: 16557279. doi: 10.1038/nrc1840.
- Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med. 2016;374(23):2287–2289. PMID: 27276568. doi: 10.1056/NEJMcibr1603785.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20.
- Lopez-Rincon A, Martinez-Archundia M, Martinez-Ruiz GU, et al. Automatic discovery of 100-miRNA signature for cancer classification using ensemble feature selection. BMC Bioinformatics. 2019;20(1):480. doi: 10.1186/s12859-019-3050-8.
- Li X, Shi Y, Yin Z, et al. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12:159. doi: 10.1186/1479-5876-12-159.
- Wu ZH, Zhong Y, Zhou T, et al. miRNA biomarkers for predicting overall survival outcomes for head and neck squamous cell carcinoma. Genomics. 2021;113(1 Pt 1):135–141. Epub 2020 Dec 3. PMID: 33279650. doi: 10.1016/j.ygeno.2020.12.002.
- Bishop JA, Benjamin H, Cholakh H, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010; 16(2):610–619. Epub 2010 Jan 12. PMID: 20068099. doi: 10.1158/1078-0432.CCR-09-2638.
- Zhong S, Golpon H, Zardo P, et al. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl Res. 2021;230:164–196. Epub 2020 Nov 28. PMID: 33253979. doi: 10.1016/j.trsl.2020.11.012.
- Takikita M, Chung JY, Hewitt SM. Tissue microarrays enabling high-throughput molecular pathology. Curr Opin Biotechnol. 2007;18(4):318–325. Epub 2007 Jul 20. PMID: 17643281. doi: 10.1016/j.copbio.2007.05.007.
- Gao Z, Liu H, Shi Y, et al. Identification of cancer stem cell molecular markers and effects of Hsa-Mir-21-3p on stemness in esophageal squamous cell carcinoma. Cancers. 2019;11(4): 518. doi: 10.3390/cancers11040518.
- Casadei L, Calore F, Creighton CJ, et al. Exosome-derived mir-25-3p and mir-92a-3p stimulate liposarcoma progression. Cancer Res. 2017;77(14):3846–3856. doi: 10.1158/0008-5472.CAN-16-2984.
- Kim DH, Park H, Choi YJ, et al. Exosomal mir-1260b derived from non-small cell lung cancer promotes tumor metastasis through the inhibition of Hipk2. Cell Death Dis. 2021;12(8):747. doi: 10.1038/s41419-021-04024-9.
- Gao S-L, Fan Y, Liu X-D, et al. Circ_0089153 exacerbates breast cancer cells proliferation and metastasis via sponging mir-2467-3p/E2f6. Environ Toxicol. 2022;37(6):1458–1471. PMID: 35225430. doi: 10.1002/tox.23498.
- Jung CK, Jung S-H, Yim S-H, et al. Predictive micrornas for lymph node metastasis in endoscopically resectable submucosal colorectal cancer. Oncotarget. 2016;7(22):32902–32915. doi: 10.18632/oncotarget.8766.
- Koinis F, Kotsakis A, Georgoulias V. Small cell lung cancer (SCLC): no treatment advances in recent years. Transl Lung Cancer Res. 2016;5(1):39–50. doi: 10.3978/j.issn.2218-6751.2016.01.03.
- Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol. 2012; 25 Suppl 1:S18–S30. PMID: 22214967. doi: 10.1038/modpathol.2011.150.
- Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene. 2003;22(42):6497–6507. PMID: 14528274. doi: 10.1038/sj.onc.1206865.[Mismatc]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222.
- Zhang X, He X, Liu Y, et al. MiR-101-3p inhibits the growth and metastasis of non-small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT-1. Biomed Pharmacother. 2017;93:1065–1073. Epub 2017 Jul 18. PMID: 28738500. doi: 10.1016/j.biopha.2017.07.005.
- Duca RB, Massillo C, Dalton GN, et al. MiR-19b-3p and miR-101-3p as potential biomarkers for prostate cancer diagnosis and prognosis. Am J Cancer Res. 2021; 11(6):2802–2820.
- Cao S, Lin L, Xia X, et al. lncRNA SPRY4-IT1 regulates cell proliferation and migration by sponging miR-101-3p and regulating AMPK expression in gastric cancer. Mol Ther Nucleic Acids. 2019;17:455–464. doi: 10.1016/j.omtn.2019.04.030.
- Chava S, Reynolds CP, Pathania AS, et al. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol Oncol. 2020;14(1):180–196. doi: 10.1002/1878-0261.12588.
- Luo J, Pan J, Jin Y, et al. MiR-195-5p inhibits proliferation and induces apoptosis of non-small cell lung cancer cells by targeting CEP55. Onco Targets Ther. 2019;12:11465–11474. doi: 10.2147/OTT.S226921.
- Yan F, Zhao W, Xu X, et al. LncRNA DHRS4-AS1 inhibits the stemness of NSCLC cells by sponging miR-224-3p and upregulating TP53 and TET1. Front Cell Dev Biol. 2020;8:585251. doi: 10.3389/fcell.2020.585251.
- Peterson SM, Thompson JA, Ufkin ML, et al. Common features of microRNA target prediction tools. Front Genet. 2014; 5:23. doi: 10.3389/fgene.2014.00023.
- Jiang N, Zou C, Zhu Y, et al. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and notch signaling. Theranostics. 2020; 10(6):2553–2570. doi: 10.7150/thno.41120.
- Wu X, Sui Z, Zhang H, et al. Integrated analysis of lncRNA-Mediated ceRNA network in lung adenocarcinoma. Front Oncol. 2020;10:554759. doi: 10.3389/fonc.2020.554759.
- Guo Y, Wang G, Wang Z, et al. Reck-notch1 signaling mediates miR-221/222 regulation of lung cancer stem cells in NSCLC. Front Cell Dev Biol. 2021;9:663279. doi: 10.3389/fcell.2021.663279.
- He H, Song X, Yang Z, et al. Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12-mediated autophagy via miR-372-3p. Cell Death Dis. 2020;11(10):883. doi: 10.1038/s41419-020-03083-8.
- Guo CM, Liu SQ, Sun MZ. miR-429 as biomarker for diagnosis, treatment and prognosis of cancers and its potential action mechanisms: a systematic literature review. Neoplasma. 2020;67(2):215–228. doi: 10.4149/neo_2019_190401N282.
- Wang A, Zhu J, Li J, et al. Downregulation of KIAA1199 by miR-486-5p suppresses tumorigenesis in lung cancer. Cancer Med. 2020;9(15):5570–5586. doi: 10.1002/cam4.3210.
- Xue M, Hong W, Jiang J, et al. Circular RNA circ-LDLRAD3 serves as an oncogene to promote non-small cell lung cancer progression by upregulating SLC1A5 through sponging miR-137. RNA Biol. 2020;17(12):1811–1822. doi: 10.1080/15476286.2020.1789819.
- Luo K, Zhao Y, Liu H, et al. Identification of critical miRNAs as novel diagnostic markers for laryngeal squamous cell carcinoma. Dis Markers. 2022;2022:6858411. doi: 10.1155/2022/6858411.
- Xie H, Jing R, Liao X, et al. Arecoline promotes proliferation and migration of human HepG2 cells through activation of the PI3K/AKT/mTOR pathway. Hereditas. 2022; 159(1):29. doi: 10.1186/s41065-022-00241-0.
- Zhang YQ, Chen RL, Shang LQ, et al. Nicotine-induced miR-21-3p promotes chemoresistance in lung cancer by negatively regulating FOXO3a. Oncol Lett. 2022; 24(2):260. doi: 10.3892/ol.2022.13380.
