Abstract
Background
Observational studies have suggested an association between inflammatory bowel disease [IBD] and psoriasis. However, the detailed genetic basis, causality, and direction of this association remain unclear.
Methods
Bidirectional two-sample Mendelian Randomization [MR] analysis was conducted using summary statistics from published genome-wide association studies. Bayesian Colocalization and multivariable MR [MVMR] analyses were performed to identify candidate variants and risk genes involved in the shared genetic basis between IBD, psoriasis, and their subtypes.
Results
Genetically predicted IBD and Crohn’s disease [CD] were associated with an increased risk of psoriasis, psoriasis vulgaris [PsV], and psoriatic arthritis [PsA] (IBD on psoriasis: pooled odds ratio [OR] 1.09, 95% confidence interval [CI] 1.04–1.14, p = .0001; CD on psoriasis: pooled OR 1.10, 95% CI 1.06–1.15, p < .0001) and vice versa (psoriasis on IBD: pooled OR 1.11, 95%CI 1.02–1.21), whereas CD only exhibited a unidirectional association with psoriasis. Colocalization analysis revealed eight candidate genetic variants and risk genes (including LINC00824, CDKAL1, IL10, IL23R, DNAJC27, LPP, RUNX3, and RGS14) associated with a shared genetic basis. Among these, IL23R, DNAJC27, LPP, and RGS14 were further validated by MVMR analysis.
Conclusion
Our findings indicated bidirectional causal associations between IBD and psoriasis (including PsV and PsA), which were attributed primarily to CD rather than Ulcerative colitis [UC]. Furthermore, we identified several candidate variants and risk genes involved in the shared genetic basis of IBD and psoriasis. Acquiring a better understanding of the shared genetic architecture underlying IBD and psoriasis would help improve clinical strategies.
Introduction
Inflammatory bowel disease (IBD) is a chronic immune-mediated inflammatory disorder of the gastrointestinal tract characterized by progressive or relapsing-and-remitting conditions [Citation1]; its subtypes include Crohn’s disease (CD) and ulcerative colitis (UC). IBD affected 6.8 million people worldwide in 2017 and was more common in developed countries. The incidence of IBD has rapidly increased in developing countries, with an annual increase of 11.1% for CD and 14.9% for UC [Citation2,Citation3]. Genetic susceptibility, abnormal immune responses to environmental predispositions, and microbial dysbiosis are thought to cause IBD [Citation4]. However, the pathophysiology of IBD remains unclear. Furthermore, it is generally accepted that IBD is associated with other immune-mediated disorders, particularly psoriasis, as they have been found to not only frequently co-occur but also share several genotypes, clinical courses, and immune features [Citation5]. Psoriasis is a chronic, relapsing, immune-mediated inflammatory skin disease. Its subtypes include psoriasis vulgaris (PsV), arthropathic psoriasis (PsA), guttate psoriasis (PsG), and unspecified psoriasis (PsN) [Citation6]. It affects approximately 2–4% of the population in Western countries [Citation7]. Both IBD and psoriasis are more likely to develop comorbidities, such as cardiovascular diseases, psychiatric disorders, and central nervous system diseases [Citation6]. Thus, IBD and psoriasis are major health problems that impose considerable economic burdens. Therefore, it is necessary to clarify the causal relationship and co-pathogenesis between IBD and psoriasis.
Some observational studies and meta-analyses have reported that psoriasis is a risk factor for IBD and vice versa. Specifically, in a nationwide Danish cohort with 75,209 cases of psoriasis, including 11,309 cases of CD and 30,310 cases of UC, psoriasis was associated with a higher risk of CD and UC [Citation8]. An analysis of data from 174,476 women in the Nurses’ Health Study revealed that psoriasis was associated with an increased risk of CD but not UC [Citation9]. In contrast, a study based on a national database in Taiwan (51,800 cases of psoriasis) showed that psoriasis was not associated with IBD development [Citation10]. However, the causal association between IBD and psoriasis remains unclear due to the bias of confounding variables and difficulty in identifying the causal direction in observational studies. Moreover, although genome-wide association studies (GWAS) have found shared genetic susceptibility regions 6p22, 20q13 and 19p13 [Citation11], many of them have not yet been demonstrated to be associated with the common pathogenesis of IBD and psoriasis.
Mendelian Randomization (MR) analysis has been proposed as a method to assess the causal relationships between traits by introducing genetic variants as instrumental variables. Bayesian colocalization analysis (COLOC) was used to evaluate the shared genetic architecture between two traits by applying a Bayesian statistical test to establish a logical regression model between the approximate Bayesian factor and p-value to assess whether the observed overlap or spatial proximity could be due to chance [Citation12,Citation13].
In this study, we performed bidirectional MR analyses to investigate the causal effects of IBD (including CD and UC) on psoriasis (PsV, PsA, guttate psoriasis, and unspecified psoriasis) and vice versa. We then applied COLOC analysis to detect causal variants and potential effector genes shared by IBD and psoriasis. Finally, we performed multivariable MR (MVMR) analysis to determine the direct association between IBD and psoriasis after adjusting for the expression of genes and inferred the effector genes mediating the causal relationship between IBD and psoriasis with more certainty [Citation14].
Materials and methods
Study design
We performed bidirectional two-sample MR to investigate the causal relationship between IBD (UC and CD) and psoriasis (including PsV, PsA, PsG, and PsN) based on GWAS summary statistics for IBD from the IBD Genetic Consortium (IBDGC) and European Bioinformatics Institute (EBI) databases, and psoriasis from the FinnGen database and UK Biobank (UKB). We then applied COLOC analysis at each locus of interest to identify shared genetic variants and potential risk genes in IBD and psoriasis. In the last step, we used MVMR analysis combining gene expression quantitative trait loci (eQTL) from the eQTLGen study with GWAS summary statistics to examine whether the putative causal relationship between IBD and psoriasis can be affected by genes identified by COLOC; in other words, to further identify the potential risk genes shared by IBD and psoriasis. A schematic of the study design is shown in . This study was based on large-scale meta-analysis data and publicly available GWAS summary data; therefore, no ethics committee approval or informed consent was required. This study was designed and reported in compliance with the STROBE MR statement [Citation15].
Figure 1. Schematic overview of the study design. Step-1. Bidirectional MR study between IBD and psoriasis: The independent SNPs associated with IBD or psoriasis as genetic instrumental variables were obtained from the GWAS summary statistics for IBD from the IBD Genetic Consortium (Liu et al.) and EBI database (de Lange et al.) and psoriasis from the FinnGen and UK Biobank databases. MR analyses were performed per outcome database and subsequently analyzed. Step-2. We extracted the genomic regions extending to 200 KB of lead SNPs for COLOC analysis as a training cohort, extracted the genomic regions within a 1MB window of the genes located in the COLOC analysis as the testing cohort, and identified seven potential risk genes shared in IBD and psoriasis. In the last step, we applied univariable and multivariable MR on respective trait pairs combining genetic variants from eQTLGen and GWAS summary statistics to detect direct causal effects between IBD and psoriasis after correcting for the effects of risk genes.
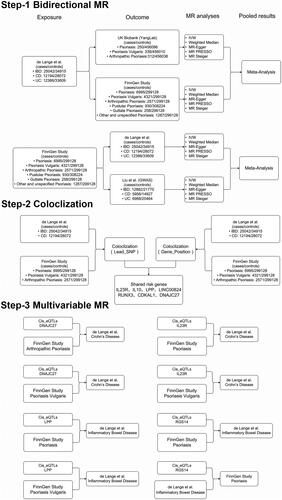
Data sources
We extracted the European-only GWAS meta-analysis for IBD from the IBDGC (Liu et al.) [Citation16] (cases/controls for IBD:12,882/21,770; CD:5956/14,927; UC:6968/20,464) and EBI databases [Citation17] (cases/controls for IBD:25,042/34,915; CD:12,194/28,072; UC:12,366/33,609). The diagnosis of IBD was based on accepted radiological, endoscopic, and histopathological evaluations, and all included cases fulfilled the clinical diagnostic criteria for IBD.
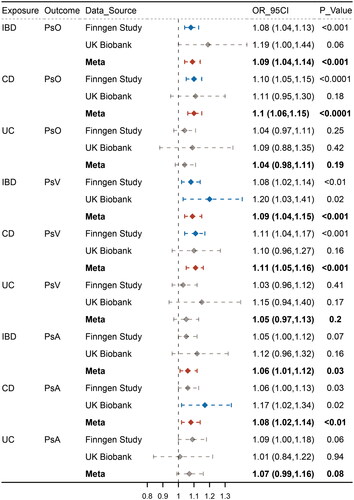
Figure 2. Causal estimates for effect of IBD and subentities on psoriasis, psoriasis vulgaris and Psoriatic arthritis. Causal estimates for the effect of inflammatory bowel disease (IBD) and its subtypes, Crohn’s disease (CD) and ulcerative colitis (UC), on psoriasis (PsO), psoriasis vulgaris (PsV), and psoriatic arthritis (PsA). Estimated ORs represent the effect of per log-OR increase in IBD, CD, and UC on PsO, PsV, and PsA, obtained from an inverse-variance weighted analysis and combined over Finngen and UKB databases using meta-analyses.
Summary statistics of genetic variants for psoriasis were acquired from the FinnGen database (R7 version) (cases/controls for psoriasis:6995/29,9128; PsV:4321/29,9128; PsA:2571/29,9128; PsG:258/29,9128; PsN:1287/29,9128) [Citation18] and UK Biobank cohort (cases/controls for psoriasis:250/45,6098; PsV:338/45,6010; PsA:312/45,6036). The phenotype definitions of psoriasis and its subtypes were based on the International Statistical Classification of Diseases and Related Health Problems (ICD) codes [Citation19]. Further information regarding phenotypes, participants, and summary statistics are accessible on the FinnGen website (https://www.finngen.fi/en/) and the fastGWA data portal (http://fastgwa.info/ukbimpbin) [Citation20].
Expression quantitative trait loci (eQTL) are defined as loci associated with genetic variants that alter gene expression levels [Citation21]. Considering that cis-eQTLs were more proximal to the gene of interest in drug development studies, we obtained blood-based cis-eQTL summary statistics of genes identified after COLOC tests from the eQTLGen Consortium [Citation22]. The eQTLGen cohort comprised 31,684 whole blood and peripheral blood mononuclear cell samples from 37 datasets. Detailed information on the participants, gene expression measurements, and genotyping has been previously described [Citation23]. We selected the cis genetic variants for analyses that were located within 1 megabase (Mb) of the target gene regions whose expression was robustly associated with (based on p < 5 × 10−8, r2< 0.1 within 1000 kb), as previously described by Westra et al. [Citation24]
All of the databases in the current study were open available. Therefore, ethical approval was not applicable.
Selection of genetic instrumental variables
To ensure the relevance assumption of MR analysis, the correlation p-value should be less than 5 × 10−8 in the corresponding GWAS study. To avoid weak instrument bias, instrumental variables with an F-statistic value of >10 were used in subsequent analyses. To ensure SNP independence, we excluded SNPs with r2> 0.001 and <10,000 kb to eliminate the linkage disequilibrium (LD) effects. Based on the key assumption of MR analyses, genetic variants must not be associated with outcomes and known confounding factors (including ever and current smoking); thus, we performed a searched the PhenoScanner website (http://wwwcam.ac.uk) and excluded SNPs with pleiotropic effects (p > 1 × 10−5). The excluded SNPs are listed in Supplementary eTable 2.
Statistical analysis
The statistical analysis consisted of three steps. First, a bidirectional two-sample MR was performed to investigate the causal effects between IBD and psoriasis and their subtypes. Second, we performed a COLOC analysis for diseases with bidirectional causality to identify shared genetic variants and potential risk genes. Third, MVMR analysis was conducted to examine the direct causal relationship between IBD and psoriasis, after correcting for risk genes.
We used inverse-variance weighted (IVW) meta-analysis as the main analysis method to combine each SNP-specific Wald ratio estimate [Citation25]. The heterogeneity in the IVW meta-analysis was measured using Cochran’s Q-test. In addition, we applied a series of sensitivity analyses to assess the pleiotropy and potential outliers [Citation26]. The weighted median estimator is more robust than the IVW when individual genetic variants with strongly outlying causal estimates are included [Citation27]. In the MR-Egger regression model, the intercept deviating from zero indicates pleiotropy, and the slope provides a causal estimate correcting horizontal pleiotropic effects, albeit with insufficient statistical efficiency [Citation28]. To further detect significant outliers and eliminate potential horizontal pleiotropy, we performed Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) analysis [Citation29]. Leave-one-out analysis was performed by the sequential exclusion of each SNP to assess the potential influence of a particular variant on the estimates. The IVW method was then applied to the remaining SNPs and was used as the final estimate. In addition, a pooled meta-analysis was performed to combine individual estimates using a fixed-effects model for each direction. As all outcome variables in the current analyses were binary, the final effect estimates were interpreted as ORs. MR results were presented as ORs with the corresponding 95% confidence intervals (95% CI).
Bayesian colocalization analysis
Bayesian colocalization analysis can evaluate the shared, local genetic architecture between two traits and is valuable in further identifying MR associations caused by LD confounding [Citation12]. We applied the COLOC package [Citation13] to detect shared causal variants responsible for significant bidirectional associations between IBD and psoriasis. For each trait pair, we used the genetic variants involved in the final bidirectional MR analysis as lead SNPs, extracted the genomic regions extending to 200 KB of lead SNPs for COLOC analysis, and used them as a training cohort. Moreover, we also extracted the genomic regions within a 1MB window of the genes where the Lead SNP was located for COLOC analysis as the testing cohort. The posterior probability of hypothesis 4 [PPH4] > 0.75 both in the training and testing cohorts, was deemed as powerful evidence for colocalization [Citation13].
Univariable and multivariable MR
To further investigate the role of the genes identified by COLOC analyses, we conducted univariable and multivariable MR analyses [Citation30] on respective trait pairs combining cis genetic variants from eQTLGen and GWAS summary statistics. The IVW method was used as the primary result. Genes that showed significant association with the respective traits in univariable MR were selected for further MVMR analysis. As the associations for each genetic instrument of each exposure were conditional on one another in MVMR, a direct causal effect between psoriasis and IBD could be shown after correcting for the effect of specific genes. In other words, we could further identify the common effector genes of trait pairs by MVMR as further evidence of colocalization analysis.
Analyses were performed using R (v. 4.2.1) statistical software. The two-sample MR and MVMR analysis were performed using the package ‘TwoSampleMR’ (0.5.6) and ‘MRPRESSO’ (1.0). Meta-analyses were conducted using the ‘meta’ package (5.5.0) [Citation31]. The colocalization analyses were conducted with the ‘coloc’ package (5.1.0.1). Regional association plots were generated using the R package ‘LocusCompareR’ (5.1.0) [Citation32].
Results
Bidirectional Mendelian Randomization
When we investigated the causal effect of IBD on psoriasis, we identified one GWAS summary statistic for IBD from the EBI database and two GWAS summary statistics for psoriasis from the FinnGen database and UK Biobank. We identified 16 SNPs associated with psoriasis, its subtypes, and known confounders by searching the PhenoScanner database. In the other direction, we used the FinnGen database for psoriasis and IBDGC and EBI databases for IBD. We also detected eight SNPs associated with IBD, its subtypes, and known confounders in the PhenoScanner database. To improve the power statistics, we conducted a meta-analysis of the results. Causal estimates and sensitivity analyses for each MR study are presented in Supplementary eTable 1. The eliminated SNPs associated with the outcomes and known confounders are listed in Supplementary eTable 2. Detailed information on the SNPs for each exposure and outcome is listed in Supplementary eTable 3. The differences between IBD and guttate psoriasis and unspecified psoriasis are provided in the Supplementary Results section.
IBD and psoriasis
Meta-analyses of estimates from IVW suggested that IBD and CD were associated with an increased risk of psoriasis, whereas UC did not show a significant causal effect (IBD: OR 1.09, 95% CI 1.04-1.14, p = .0001; CD: OR 1.10, 95% CI 1.06–1.15, p < .0001; UC: OR 1.04, 95% CI 0.98–1.11, p = .186). In the other direction, the meta-analysis indicated that psoriasis was associated with an increased risk of IBD, but did not show significant causal effects with CD and UC (IBD: OR 1.11, 95% CI 1.02–1.21, p = 1.96 × 10−2; CD: OR 1.08, 95% CI 0.98–1.18, p = .115; UC: OR 1.00, 95% CI 0.87–1.15, p = .980) ().
IBD and psoriasis vulgaris
Meta-analyses of estimates from IVW suggested that IBD and CD were associated with an increased risk of PsV, whereas UC did not show a significant causal effect (IBD: OR 1.09, 95% CI 1.04–1.15, p = .0006; CD: OR 1.11, 95% CI 1.05–1.16, p = .0002; UC: OR 1.05, 95% CI 0.97–1.13, p = .204). On the other hand, the meta-analysis indicated that PsV was associated with an increased risk of IBD and CD but was not significantly associated with UC (IBD: OR 1.08, 95% CI 1.02–1.15, p = .0147; CD: OR 1.12, 95% CI 1.02–1.23, p = .0136; UC: OR 0.94, 95% CI 0.87–1.02, p = .126) ().
IBD and arthropathic psoriasis
Meta-analyses of estimates from IVW suggested that IBD and CD were associated with an increased risk of PsA, whereas UC did not show a significant causal effect (IBD: OR 1.06, 95% CI 1.01–1.12, p = .0297; CD: OR 1.08, 95% CI 1.02–1.14, p = .0045; UC: OR 1.07, 95% CI 0.99–1.16, p = .083). On the other hand, the meta-analysis indicated that PsA was associated with an increased risk of IBD and CD but did not show significant causal effects with UC (IBD: OR 1.12, 95% CI 1.07–1.16, p < .0001; CD: OR 1.17, 95% CI 1.11–1.22, p < .0001; UC: OR 1.00, 95% CI 0.96–1.05, p = .973) ().
Validation and sensitivity analyses
The results of the investigation of the bidirectional causal relationships between IBD and psoriasis and their subtypes were well replicated in the independent datasets for validation and summarized by meta-analysis. Although some degree of heterogeneity remained after removing outliers, there was little evidence to support the horizontal pleiotropy effect across the genetic predictors detected by MR-Egger intercept analysis (Supplementary eTable 1). Furthermore, the results of the weighted-median and MR-Egger analyses were generally consistent with the results of the IVW analyses for each database, suggesting that underlying heterogeneity did not greatly bias our results. Leave-one-out analyses suggested that the associations were unlikely to be driven by certain extreme SNPs, which indicated the robustness of the results.
Bayesian colocalization analysis
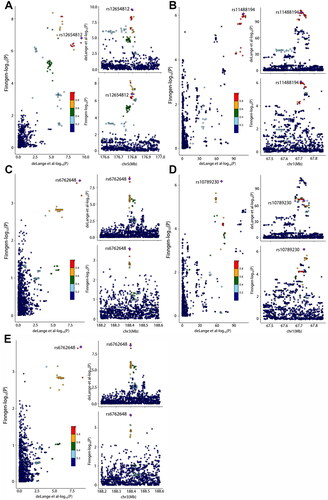
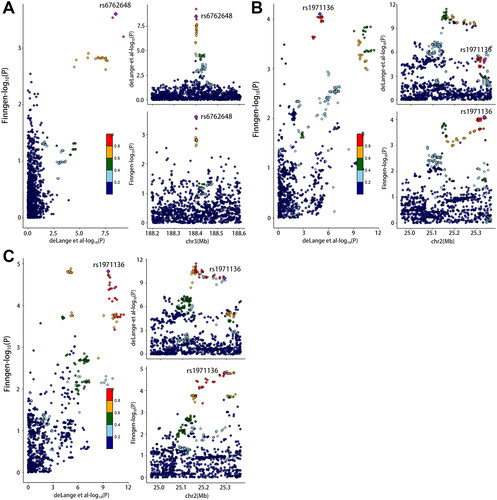
We found evidence to support the presence of shared causal variants, using COLOC analysis in the training and testing cohorts (PPH4 > 0.75) for the association between IBD and psoriasis (rs11209013, IL23R; rs13153019, RGS14; rs56116661, LPP), IBD and PsV (rs11209013, IL23R; rs56116661, LPP), IBD and PsA (rs3024493, IL10), CD and psoriasis (rs56116661, LPP; rs7542079, RUNX3), CD and PsV (rs4343432, DNAJC27; rs7542079, RUNX3; rs938650, LINC00824), and CD and PsA (rs1012636, CDKAL1; rs4343432, DNAJC27). The regional colocalization plots for these associations are shown in and . The detailed results of the shared genetic variants and located genes are listed in Supplementary eTable 4 and eTable 5.
Figure 3. Causal estimates for effect of psoriasis, psoriasis vulgaris and Psoriatic arthritis on IBD and its subentities. Causal estimates for the effects of psoriasis (PsO), psoriasis vulgaris (PsV), and psoriatic arthritis (PsA) on inflammatory bowel disease (IBD) and its subtypes, Crohn’s disease (CD), and ulcerative colitis (UC). Estimated ORs represent the effect of per log-OR increase in PsO, PsV, and PsA on IBD, CD, and UC, obtained from an inverse-variance weighted analysis and combined over IBDGC and EBI databases using meta-analyses.
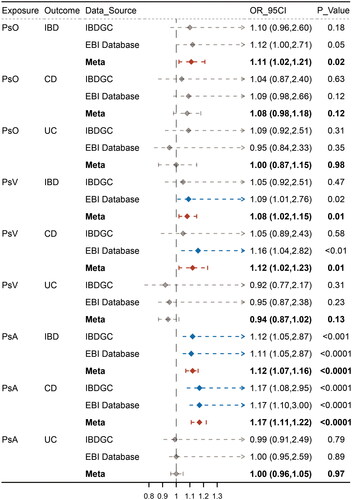
Figure 4. Colocalization of genetic IBD and psoriasis, psoriasis vulgaris and psoriatic arthritis in genomic regions extending to 200 KB of lead SNPs. (A): rs13153019 (IBD-PsO) (B): rs11209013 (IBD-PsO), (C): rs56116661 (IBD-PsO), (D): rs11209013 (IBD-PsV) and (E): rs56116661 (IBD-PsV). For each illustration, each dot represented a genetic variant with the candidate causal variant, shown as a purple diamond. The color of other variants indicated their linkage disequilibrium (r2). The left panel showed − log10 p values for SNP associations with IBD from EBI database on the x-axes, and − log10 p values for associations with psoriasis from Finngen database on the y-axes. The right panel illustrated genomic positions based on GRCh37 on the x-axes and − log10 p values of IBD GWAS (upper panel) and − log10 p values of psoriasis (below panel) on the y-axes.
Univariable and multivariable MR analyses
Given the genes identified by COLOC analyses, we obtained cis-eQTL summary statistics from the eQTLGen Consortium and conducted univariable MR analysis. We observed that increased IL23R had a positive effect on the occurrence of CD (OR:3.13, 95% CI 2.12–4.62, p < .0001) and IBD (OR:2.07, 95% CI 1.55–2.77, p < .0001), whereas increased LPP had a negative effect on the occurrence of IBD (OR:0.93, 95% CI 0.88–0.99, p = .026). Meanwhile, increased DNAJC27 had a positive effect on the occurrence of both IBD (OR:1.11, 95% CI 1.04–1.17, p = .0007) and CD (OR:1.13, 95% CI 1.05–1.21, p = .0005), and increased RGS14 had a positive effect on psoriasis (OR:1.42, 95% CI 1.22–1.66, p < .0001) (Supplementary eTable 6).
We performed MVMR imaging to detect a direct association between IBD and psoriasis. After reciprocal adjustment, both psoriasis and PsV showed insignificant causal effects on IBD after correcting for the effect of IL23R (psoriasis: OR,1.05; 95% CI 0.77–1.42, p = .776; PsV: OR,1.07; 95% CI 0.85–1.35; p = .553). Similarly, psoriasis and PsV showed insignificant causal effects in IBD after correcting for the effect of LPP (psoriasis: OR:1.08, 95% CI 0.97–1.21, p = .178; PsV: OR:1.02, 95% CI 0.94–1.10, p = .671), while PsV and PsA continued to have a significant causal effect on CD after correcting for the effect of DNAJC27 (PsV: OR:1.13, 95% CI 1.03–1.25, p = .012; PsA: OR:1.16, 95% CI 1.06–1.26, p = .0006). Interestingly, after correcting for the effect of RGS14, the causal effect of psoriasis on IBD became insignificant, whereas the effect of IBD on psoriasis remained significant (psoriasis: OR:1.09, 95% CI 0.94–1.26, p = .262; PsA: OR:1.09, 95% CI 1.04–1.14, p = .0005) (Supplementary eTable 7).
Discussion
Based on GWAS summary statistics, we used bidirectional MR analyses to investigate the potential causal associations between IBD and psoriasis as well as their respective subtypes. The findings indicated positive bidirectional causal associations between IBD and psoriasis (including PsV and PsA), which were primarily attributed to CD rather than UC. These bidirectional associations were similar among the sensitivity analyses and were supported by colocalization analyses. Univariable and multivariable MR analyses of colocalized genes (IL23R, LPP, and RGS14) also provided further evidence. Interestingly, UC did not show any causal association with psoriasis or its subtype. Additionally, we found a positive unidirectional association between IBD (including CD) and unspecified psoriasis. These unidirectional associations were consistent with sensitivity analyses but were not supported by colocalization analyses.
Figure 5. Colocalization of genetic crohn’s disease and psoriasis, psoriasis vulgaris and psoriatic arthritis in genomic regions extending to 200 KB of lead SNPs.
(A): rs56116661 (CD-PsO) (B): rs4343432 (CD-PsV) and (C): rs4343432 (CD-PsA). For each illustration, each dot represented a genetic variant with the candidate causal variant, shown as a purple diamond. The color of other variants indicated their linkage disequilibrium (r2). The left panel showed − log10 p values for SNP associations with CD from EBI database on the x-axes, and − log10 p values for associations with psoriasis from Finngen database on the y-axes. The right panel illustrated genomic positions based on GRCh37 on the x-axes and − log10 p values of CD GWAS (upper panel) and − log10 p values of psoriasis (below panel) on the y-axes.
To date, no consensus has been reached concerning the role of psoriasis in the etiology of IBD and vice versa. Compared with previous studies, this study inferred a bidirectional causal association between IBD, in particular the CD subtype, and psoriasis (including PsV and PsA), which is not only an affirmation of previous studies, but also a further explanation and extension of their contradictions. The pathophysiological mechanisms underlying this relationship have not been fully elucidated; therefore, we performed a co-localization analysis to identify nine causal variants and localized them to their respective genes. Further MVMR analysis of cis-eQTL data provided more evidence for the four potential effector genes. Current evidence suggests that IL23R involvement in the IL23/Th17 axis is involved in the pathogenesis of CD and psoriasis. The IL-23R chain forms a heterodimeric complex with IL-12Rβ1 to form the IL-23 receptor [Citation33]. The activation of the IL-23 receptor and downstream pathways is essential for the differentiation and activation of Th17 cells, which play a key role in chronic organotrophic inflammatory processes such as psoriasis and CD [Citation34,Citation35]. Previous GWASs identified four common psoriasis and CD risk loci (IL23R, IL12B, REL, and TYK2). In clinical trials, antibodies against IL-23 receptor were also effective for both CD and psoriasis [Citation36].
The other detected genes also play a role in the pathogenesis of IBD or psoriasis, although they have not been identified in large GWAS studies. IL-10 is a potent anti-inflammatory and immunosuppressive cytokine produced by both innate and adaptive immune cells. The deregulation of IL-10 plays a role in the development of a large number of inflammatory diseases such as IBD, psoriasis, and rheumatoid arthritis [Citation37]. S1PR5, a member of the S1P receptor family, is implicated in the pathophysiology of immune-mediated diseases. Research on S1PR modulators has led to the approval of safe and effective treatments for immune-mediating diseases, including multiple sclerosis, IBD, rheumatoid arthritis, and psoriasis [Citation38,Citation39]. RUNX3 is crucial for the promotion of Th1 and Th17 polarization, which is important in the pathogenesis of psoriasis and IBD, particularly CD [Citation40,Citation41]. Furthermore, the RUNX3 rs2236851 CT genotype was found to be associated with susceptibility to UC and the risk of early onset in the Chinese population. Twelve genes involved in IBD were identified as the downstream target genes of RUNX3 [Citation42]. A previous study mapped trans-eQTLs in a monocyte dataset stimulated with LPS and discovered that a risk variant for IBD, rs17622517, located in C5ORF56, affects the expression of IRF1 as an enhancer [Citation43]. Smith et al. identified CDKAL1 as a candidate biomarker of the efficacy of tumor necrosis factor inhibitors in psoriasis. It was also found to be involved in the immune cellular crosstalk implicated in psoriasis pathogenesis, including the activation and differentiation of Th17 cells and the positive regulation of NF-κB [Citation44]. The remaining genes, LPP and OR2W6P, have not been reported to be involved in the pathogenesis of IBD and psoriasis.
Environmental factors have also been implicated in the pathogenesis of IBD and psoriasis. Both skin and gut have large microbial populations and robust immune responses [Citation45]. Furthermore, microbiota can trigger abnormal immune activation, combined with genetic predisposition [Citation46]. There increasing evidence supports the gut-skin axis hypothesis of a close association between intestinal dysbiosis and cutaneous manifestations.
Strengths and limitations
The primary strength of our study was its design. We first applied bidirectional MR to estimate the causal association between IBD and psoriasis and their subtypes. We then conducted Bayesian colocalization analysis and MVMR to determine the causal variant and potential effector genes, proposing a possible biological basis and providing confidence in result robustness and strengthening causal inference. Another strength is the large sample size and abundant GWAS summary statistics, including the IBD Genetics Consortium, EBI database, FinnGen database, and UK Biobank. We pooled the above datasets by meta-analysis to improve statistical power and enhance the reliability and robustness of the results. To minimize bias, we strictly confined the study population to those of European ancestry. We strictly screened SNPs using multiple methods, such as MR-Egger and MR-PRESSO analyses, to minimize the impact of confounding factors and reverse causality. More importantly, we obtained cis-SNPs of colocalized genes for univariable and multivariable MR analyses, which were closer to the genes of interest in drug development studies.
This study had some limitations. First, the GWAS statistics were summary level rather than the individual level, which may have introduced interstudy heterogeneity. Second, although we investigated bidirectional causal effects between IBD and psoriasis, the evidence of estimation from psoriasis to IBD and the statistical power were relatively weak because fewer SNPs in the FinnGen database and UK Biobank met the selection criteria. Third, the participants were of European ancestry, which hampered the extrapolation of our findings to other ancestries. It is worth mentioning that there were potential overlapping participants in the exposure and outcome studies but it is difficult to estimate the exact overlap ratio. However, the F statistics (almost all of the IVs over 30) in this study is high enough to avoid the bias of potential participants overlap. Fourth, although we screened the SNPs by searching in PhenoScanner and eliminated SNPs associated with known confounders, potential pleiotropies could not be fully eliminated because confounders for IBD and psoriasis have not been fully elucidated. Finally, the effector genes identified by COLOC and MVMR analyses are still at a theoretical level, and biological experiments and clinical trials are underway.
Clinical implications
Combined with the above results and prior findings, the causal association between IBD and psoriasis is considered reliable. However, the coexistence of IBD and psoriasis poses new diagnostic and therapeutic challenges. First, patients with IBD, especially CD, should be informed of the risk of psoriasis and extraintestinal manifestations, such as skin lesions and arthritis. Similarly, patients with psoriasis, especially PsV and PsA, should be informed about the increased risk of CD, and gastroenterological consultation is indicated for patients with psoriasis presenting with bowel symptoms. Second, the risk factors shared between IBD and psoriasis (e.g. smoking) should be emphasized. More detailed risk factors for various subtypes of IBD and psoriasis warrant further investigation. Notably, a series of important comorbidities, such as cardiovascular disease, obesity, diabetes, hypertension, dyslipidemia, metabolic syndrome, nonalcoholic fatty liver disease, cancer, and anxiety/depression, shared in both psoriasis and IBD also need more attention. Additionally, the paradoxical treatment of IBD and psoriasis requires further investigation. For instance, monoclonal antibodies against IL-17 exhibited exceptional efficacy in the treatment of moderate-to-severe psoriasis, but failed or even had detrimental effects in moderate-to-severe CD.
In this study, we identified several potential effects of variants and genes shared between psoriasis and IBD. Among these, IL23R is a recognized gene that affects the development of IBD and psoriasis by mediating the IL23/Th17 pathway and has been clinically targeted by drugs. However, studies on other genes, such as RUNX3, S1PR5, and IL10, in IBD and psoriasis are still lacking. They may be potential effector genes mediating the causal effect of IBD and psoriasis; however, further functional validation is needed.
Conclusion
In conclusion, our study identified bidirectional causal associations between IBD (including CD) and psoriasis (including PsV and PsA); however, CD exhibited only a unidirectional association with psoriasis. In addition, we identified several candidate variants and risk genes involved in the shared genetic basis of IBD and psoriasis. Acquiring a better understanding of the shared genetic architecture underlying IBD and psoriasis would help improve clinical strategies.
Authors contributions
YJ, MJS, and CJT designed the study and reviewed the relevant literature. CYK and CDL performed data analyses and wrote the manuscript for this study. XSY, JX, and WDH contributed to the writing of the original draft. BWW contributed to literature review. All authors have read and approved the final manuscript for submission.
Consent form
The authors confirm that the work has not been published before nor elsewhere.
Supplemental Material
Download Zip (305.2 KB)Acknowledgements
We wish to acknowledge the participants and investigators of the IBDGC, EBI, FinnGen, and UK Biobank studies.
Disclosure statement
The authors declare that they have no competing interests.
We are currently conducting a study on bidirectional and have encountered some confusion. I want to figure out whether bidirectional MR analysis will make extra alpha errors and lead to overestimation of causal effects? In other words, do reverse causal effects affect the estimation of forward causal effects. If so, can we use a Bonferonni-corrected threshold to mitigate this effect. I have tried my best to searched in recently published articles about bidirectional MR analysis. However, there are no relevant descriptions about the disadvantages of bidirectional MR analysis. We would be grateful if you provide some professional insights.
I sincerely value and appreciate your contributions to the development of the MR field and look forward to your reply!
Best regards,
Siyuan Xie & Delong Chen
Additional information
Funding
References
- Rosen MJ, Dhawan A, Saeed SA. Inflammatory bowel disease in children and adolescents. JAMA Pediatr. 2015;169(11):1–13. doi: 10.1001/jamapediatrics.2015.1982.
- Kaplan GG, Windsor JW. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2021;18(1):56–66. doi: 10.1038/s41575-020-00360-x.
- Ghoneim HE, Fan YP, Moustaki A, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170(1):142–157.e19. doi: 10.1016/j.cell.2017.06.007.
- Huang H, Fang M, Jostins L, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547(7662):173–178. doi: 10.1038/nature22969.
- Peppas S, Piovani D, Peyrin-Biroulet L, et al. Statins and inflammatory bowel disease: where do we stand?. Eur J Intern Med. 2020;75:10–14. doi: 10.1016/j.ejim.2020.02.017.
- Griffiths CEM, Armstrong AW, Gudjonsson JE, et al. Psoriasis. Lancet. 2021;397(10281):1301–1315. doi: 10.1016/s0140-6736(20)32549-6.
- Parisi R, Symmons DP, Griffiths CE, et al. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol. 2013;133(2):377–385. doi: 10.1038/jid.2012.339.
- Egeberg A, Mallbris L, Warren RB, et al. Association between psoriasis and inflammatory bowel disease: a Danish nationwide cohort study. Br J Dermatol. 2016;175(3):487–492. doi: 10.1111/bjd.14528.
- Li WQ, Han JL, Chan AT, et al. Psoriasis, psoriatic arthritis and increased risk of incident Crohn’s disease in US women. Ann Rheum Dis. 2013;72(7):1200–1205. doi: 10.1136/annrheumdis-2012-202143.
- Tsai TF, Wang TS, Hung ST, et al. Epidemiology and comorbidities of psoriasis patients in a national database in Taiwan. J Dermatol Sci. 2011;63(1):40–46. doi: 10.1016/j.jdermsci.2011.03.002.
- Wolf N, Quaranta M, Prescott NJ, et al. Psoriasis is associated with pleiotropic susceptibility loci identified in type II diabetes and Crohn disease. J Med Genet. 2008;45(2):114–116. doi: 10.1136/jmg.2007.053595.
- Kanduri C, Bock C, Gundersen S, et al. Colocalization analyses of genomic elements: approaches, recommendations and challenges. Bioinformatics. 2019;35(9):1615–1624. doi: 10.1093/bioinformatics/bty835.
- Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLOS Genet. 2014;10(5):e1004383. doi: 10.1371/journal.pgen.1004383.
- Giambartolomei C, Zhenli Liu J, Zhang W, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34(15):2538–2545. doi: 10.1093/bioinformatics/bty147.
- Skrivankova VW, Richmond RC, Woolf BaR, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. doi: 10.1136/bmj.n2233.
- Liu JZ, Van Sommeren S, Huang HL, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–986. doi: 10.1038/ng.3359.
- De Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49(2):256–261. doi: 10.1038/ng.3760.
- Kurki MI, Karjalainen J, Palta P, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. medRxiv. 2022. doi: 10.1101/2022.03.03.22271360.
- The Lancet. Icd. 11. Lancet. 2019;393(10188):2275–2275.
- Jiang LD, Zheng ZL, Fang HL, et al. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53(11):1616–1621. doi: 10.1038/s41588-021-00954-4.
- Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2011;27(2):72–79. doi: 10.1016/j.tig.2010.10.006.
- Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. doi: 10.1038/ng.3538.
- Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. doi: 10.1038/s41588-021-00913-z.
- Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–1243. doi: 10.1038/ng.2756.
- Bowden J, Del Greco M F, Minelli C, et al. A framework for the investigation of pleiotropy in two-sample summary data mendelian randomization. Stat Med. 2017;36(11):1783–1802. doi: 10.1002/sim.7221.
- Burgess S, Bowden J, Fall T, et al. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30–42. doi: 10.1097/EDE.0000000000000559.
- Verbanck M, Chen C, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Eur J Hum Genet. 2019;27:854–855.
- Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080.
- Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7.
- Burgess S, Dudbridge F, Thompson SG. Re: “multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects”. Am J Epidemiol. 2015;181(4):290–291. doi: 10.1093/aje/kwv017.
- Balduzzi S, Rucker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. 2019;22(4):153–160. doi: 10.1136/ebmental-2019-300117.
- Liu BX, Gloudemans MJ, Rao AS, et al. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet. 2019;51(5):768–769. doi: 10.1038/s41588-019-0404-0.
- Parham C, Chirica M, Timans J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12R beta 1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168(11):5699–5708. doi: 10.4049/jimmunol.168.11.5699.
- Cargill M, Schrodi SJ, Chang M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80(2):273–290. doi: 10.1086/511051.
- Nair RP, Duffin KC, Helms C, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappa B pathways. Nat Genet. 2009;41(2):199–204. doi: 10.1038/ng.311.
- Krueger GG, Langley RG, Leonardi C, et al. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med. 2007;356(6):580–592. doi: 10.1056/NEJMoa062382.
- Mollazadeh H, Cicero AFG, Blesso CN, et al. Immune modulation by curcumin: the role of interleukin-10. Crit Rev Food Sci Nutr. 2019;59(1):89–101. doi: 10.1080/10408398.2017.1358139.
- Chun J, Giovannoni G, Hunter SF. Sphingosine 1-phosphate receptor modulator therapy for multiple sclerosis: differential downstream receptor signalling and clinical profile effects. Drugs. 2021;81(2):207–231. doi: 10.1007/s40265-020-01431-8.
- Stepanovska B, Huwiler A. Targeting the S1P receptor signaling pathways as a promising approach for treatment of autoimmune and inflammatory diseases. Pharmacol Res. 2020;154:104170. doi: 10.1016/j.phrs.2019.02.009.
- Djuretic IM, Levanon D, Negreanu V, et al. Transcription factors T-bet and Runx3 cooperate to activate lfng and silence ll4 in T helper type 1 cells. Nat Immunol. 2007;8(2):145–153. doi: 10.1038/ni1424.
- Wong WF, Kohu K, Chiba T, et al. Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology. 2011;132(2):157–164. doi: 10.1111/j.1365-2567.2010.03381.x.
- Guo CC, Yao FF, Wu KC, et al. Chromatin immunoprecipitation and association study revealed a possible role of runt-related transcription factor 3 in the ulcerative colitis of Chinese population. Clin Immunol. 2010;135(3):483–489. doi: 10.1016/j.clim.2010.01.004.
- Brandt M, Kim-Hellmuth S, Ziosi M, et al. An autoimmune disease risk variant: a trans master regulatory effect mediated by IRF1 under immune stimulation?. PLOS Genet. 2021;17(7):e1009684. doi: 10.1371/journal.pgen.1009684.
- Corbett M, Ramessur R, Marshall D, et al. Biomarkers of systemic treatment response in people with psoriasis: a scoping review. Br J Dermatol. 2022;187(4):494–506. doi: 10.1111/bjd.21677.
- Thrash B, Patel M, Shah KR, et al. Cutaneous manifestations of gastrointestinal disease part II. J Am Acad Dermatol. 2013;68(2):211.e1–211.e33. doi: 10.1016/j.jaad.2012.10.036.
- Zakostelska Z, Malkova J, Klimesova K, et al. Intestinal microbiota promotes Psoriasis-Like skin inflammation by enhancing Th17 response. PLoS One. 2016;11(7):e0159539. doi: 10.1371/journal.pone.0159539.
