Abstract
Objective
Accumulating evidence suggests that differentially expressed circular RNAs (circRNAs) play critical roles in immune cells of systemic lupus erythematosus (SLE) patients. Hsa_circ_0000479 has been studied in the field of cancer and infection, whereas seldom studied in autoimmune diseases. The aim of this study was to investigate the role and clinical value of neutrophil hsa_circ_0000479 in SLE.
Methods
The expression levels of hsa_circ_0000479 in both healthy individuals and SLE patients’ neutrophils were detected by qPCR and compared with those in peripheral blood mononuclear cells (PBMCs) . In addition, the correlation of hsa_circ_0000479 levels in neutrophils with the clinical and immunological features of SLE patients was also analysed.
Results
The expression levels of hsa_circ_0000479 in the patients with SLE were significantly higher in neutrophils than that of PBMCs, and also significantly higher than that in healthy controls (HCs). Moreover, the expression levels of hsa_circ_0000479 in neutrophils were negatively associated with absolute neutrophil count and complement 3 (C3), whereas positively correlated with anti-dsDNA and anti-nucleosome antibodies in SLE. In addition, SLE patients with higher levels of hsa_circ_0000479 demonstrated more several clinical manifestations, including Raynaud’s phenomenon, alopecia and leucopenia.
Conclusions
Hsa_circ_0000479 is up-regulated in neutrophils of SLE patients, and is also associated with several important laboratory indicators and clinical manifestations, suggesting that hsa_circ_0000479 in neutrophils was one of probable factors involved in the pathogenesis of SLE with potential clinical value.
Key messages
Hsa_circ_0000479 was expressed in neutrophils and was considerably higher than that of PBMCs in SLE patients.
The neutrophil hsa_circ_0000479 was correlated with laboratory parameters, including NEUT, C3, anti-dsDNA antibodies and AnuA, in addition to being associated with Raynaud’s phenomenon, alopecia, and leucopenia in patients with SLE.
Hsa_circ_0000479 in neutrophils may play an influential role in SLE patients and will be important to understand the pathogenesis, stratification and treatment in SLE.
1. Introduction
Systemic lupus erythematosus (SLE) is a typical autoimmune disease characterized by the presence of autoimmune complex and diverse organ damage [Citation1]. The exact mechanism of SLE pathogenesis incompletely understood. Dysfunction of neutrophils, monocytes/macrophages, B cells and T cells result in extensive production of pathogenic autoantibodies and inflammatory cytokines, leading to severe clinical manifestations [Citation2–4].
Neutrophils play a key role in defense against infection and in the activation and immunoregulation [Citation5]. It is well known that the proportion and number of low-density neutrophils are markedly increased in SLE patients [Citation6,Citation7]. The studies have confirmed that increased neutrophils apoptosis, abnormal formation of neutrophil extracellular traps (NETs) and increased ferroptosis may over-activate the immune response and promote the development in SLE through exposure to extensive autoantigen [Citation8–10].
Circular RNAs (circRNAs) belong to a newly discovered type of noncoding RNAs, consisting of transcripts from the exons that are formed by non-colinear reverse splicing, with covalently closed single-stranded structures lacking 5′–3′ polarity and a polyadenine tail [Citation11,Citation12]. The special structure protects circRNAs from degradation by RNA exonuclease, which makes them more stable and suitable as potential biomarkers than other types of RNA [Citation13,Citation14].
CircRNAs often have tissue-restricted and cell-specific expression patterns, and exert important biological functions by acting as microRNA or protein inhibitors (known as sponges), by regulating the protein function or by being translated themselves [Citation15,Citation16]. Furthermore, circRNAs have been implicated in diseases, such as neurological disorders [Citation17], cardiovascular diseases [Citation18], diabetes mellitus [Citation19] and cancer [Citation20], and have clinical potential as biomarkers, therapeutic agents and drug targets.
Recently, an increasing number of studies have been conducted to clarify the role of circRNAs in SLE, such as circGARS [Citation21], circIBTK [Citation22] and hsa_circ_0123190 [Citation23]. Interestingly, a meta-analysis has been performed based on the pooled data from published studies that focused on the association between circRNAs and SLE [Citation24]. Two studies showed that hsa_circ_0000479 in peripheral blood mononuclear cells (PBMCs) from SLE patients may serve as a potential biomarker for SLE patients, respectively [Citation25,Citation26]. Thus, hsa_circ_0000479 may be an essential molecule in some subsets of immune cells in SLE.
The number of neutrophils in PBMCs is significantly higher than in healthy individuals, which is a prominent feature in SLE patients [Citation7]. Given the crucial role of neutrophils and circRNAs in SLE, this study aims to compare the expression of hsa_circ_0000479 in PBMCs and homologous neutrophils between SLE patients and HCs and further investigate the expression and clinical value of hsa_circ_0000479 in SLE neutrophils in an attempt to provide potential clues for the disease understanding.
2. Materials and methods
2.1. Patients and controls
A total of 125 participants were enrolled in this study, including 80 SLE patients and 45 healthy controls (HCs). The information is displayed in . All SLE patients fulfilled the American College of Rheumatology 1997 revised classification criteria [Citation27] and were recruited at the Department of Rheumatology and Immunology, Peking University People’s Hospital between 2021 and 2022. The HC with matched age and sex were recruited from the physical examination center simultaneously. Clinical information of SLE patients, including clinical symptoms and signs, sociological characteristics and laboratory parameters, was collected from the patient’s medical records system. The study was approved by the ethics committee of Peking University People’s Hospital. All participants had signed informed consent forms.
Table 1. Clinical characteristics of SLE patients and healthy controls.
2.2. Clinical and laboratory data
The Clinical data of patients with SLE were recorded, including gender, age, disease duration, clinical manifestations. The laboratory parameters including white blood cells (WBC), red blood cells (RBC), hemoglobin (Hb), platelets (PLT), the urine protein of 24 h, Albumin/Globulin, erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), complement 3 (C3), complement component 4 (C4), immunoglobulin (Ig) A, IgG, IgM, anti-dsDNA antibody (anti-dsDNA Ab). Disease activity was assessed using the Systemic Lupus Erythematosus Disease Activity Index-2000 (SLEDAI-2K) [Citation28] at the same time of blood collection. As the standard of classification, SLE patients with SLEDAI-2K ≥ 5 were defined as a middle and high disease activity.
2.3. Isolation and identification of neutrophils
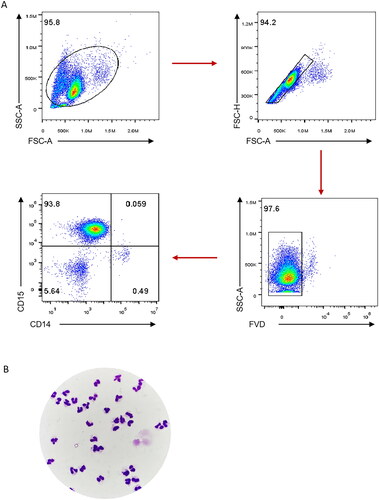
A total of 4 mL of blood with ethylenediamine tetraacetic acid (EDTA) was obtained from each subject. The blood was centrifuged at 3500 rpm for 10 min at room temperature. Then the cells of the white membrane obtained were re-suspended with 3 mL of sterile PBS buffer (pH = 7.2 − 7.4). The PBMCs and neutrophils were separated by Percoll solution (Cat No. 40501ES60, Yeasen, Shanghai, China) with the concentrations of 60% and 75%, respectively. Finally, the neutrophils were detected by staining with fixable viability dye (FVD), anti-CD15 antibody and anti-CD14 antibody, which were purchased from BioLegend (San Diego, CA). The purity and viability of neutrophils were 93.8% and 97.6%, respectively (). The appearance of neutrophils was predominant under microscope ().
Figure 1. Purity and viability of neutrophils isolated from human peripheral whole blood. Neutrophils isolated from 4 mL of peripheral EDTA anticoagulant whole blood were treated by percoll discontinuous density gradient centrifugation. The neutrophils were detected by staining with fixable viability dye (FVD), anti-CD15 antibody and anti-CD14 antibody. FACS gating strategy for identifying the viability and purity of neutrophils were shown (A). The hyper-segmented neutrophils were mainly observed by Wright’s staining and 1000× oil microscope (B).
2.4. RT-qPCR
Total RNA was extracted from PBMCs and neutrophils by an RNAsimple Total RNA Extraction Kit (Cat No. DP419; Tiangen, Beijing, China) following manufacturer’s recommendations. The quantity and quality of RNA were assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Waltham, MA) by measuring the absorbance ratios of A260/A280 and A260/A230.
Reverse transcription (RT) was carried out using Hifair III 1st Strand cDNA Synthesis SuperMix for quantitative real-time PCR (qPCR) (Cat No. 11141ES10; Yeasen, Shanghai, China) following manufacturer’s recommendations. Of 1 μg of total RNA was converted to cDNA in 20 μL reactions. To determine the levels of hsa_circ_0000479, qPCR was performed following cDNA generation.
Briefly, 1 μL of cDNA was used as template in a 10 μL qPCR reaction by adding Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (Cat No. 11184ES03; Yeasen, Shanghai, China) and 0.2 μL pre-designed primers for target genes. The primers were synthesized by Tsingke Biological Technology (Beijing, China), and the sequences were as follows: hsa_circ_0000479 forward: 5′-AAGAGAAGAATCTGTAAGAATCA-3′,
reverse: 5′-TGGTGCTATCAAGGTGTA-3′ [Citation25];
β-Actin forward: 5′-CGGAGTCAACGGATTTGGTC-3′;
reverse: 5′-CGGTGCCATGGAATTTGCCA-3′.
For the relative quantification analysis of hsa_circ_0000479 expression, data were normalized to the geometric mean of β-Actin, the comparative threshold cycle value (Ct) method was used and the results were calculated by the 2−ΔΔCt.
2.5. Statistical analysis
Statistical analysis and graphic presentation were carried out with GraphPad Prism version 9.0 (GraphPad Software, San Diego, CA) and SPSS version 26.0 (SPSS Inc., Chicago, IL). Data were presented as the mean ± standard deviation (SD) or median with interquartile range (IQR) or with a range (P25, P75). The Student’s t-test, Mann–Whitney U-test, Spearman’s rank correlation test and/or Benjamini–Hochberg multiple testing correction were used where appropriate. P values < 0.05 were considered statistically significant: *p < 0.05, **p < 0.01 and ***p < 0.001, ns: not significance.
3. Results
3.1. Demographic and clinical characteristics of SLE patients
A total of 125 participants were recruited for this study. 8 SLE patients and 8 HCs were allocated randomly into a training cohort, and 45 healthy subjects and 80 SLE patients were served as a validation cohort. The ratio of females to males was 16:1 (75 females and 5 males), the age ranged between 16 and 78 years old with a mean of 34.5 years old. The median disease duration was 2 years (IQR: 0.29–8). The median score of SLEDAI-2K score was 10 (IQR: 6–15.75) and 85% SLE patients were under middle/high disease activity (SLEDAI-2K ≥ 5). No statistically significant difference was observed between the training and validation cohorts in the demographic and clinical characteristics of SLE patients, as shown in .
3.2. Basic information of hsa_circ_0000479 and PCR product identification in neutrophils
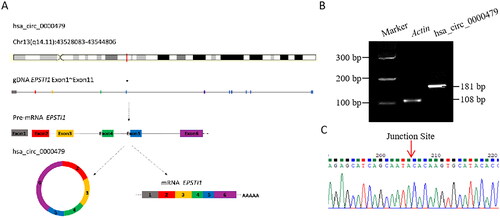
According to human reference genome (GRCh37/hg19), hsa_circ_0000479 was a circular RNA structure formed by reverse splicing from the exon 2 to exon 6 of the Epithelial Stromal Interaction 1 (EPSTI1) gene, which is located on chromosome 13q14 with a total length of 375 bp (). The length of the PCR verification product was 181 bp, which was amplified by divergent primers (). The sequence of hsa_circ_0000479 was confirmed by Sanger sequencing, with the junction site especially included (). These results indicated that hsa_circ_0000479 was indeed expressed in human peripheral blood neutrophils, with a consistent sequence as reported in PBMCs [Citation25].
Figure 2. Basic information of hsa_circ_0000479 and PCR product identification in neutrophils. (A) The schematic diagram of the genomic position of hsa_circ_0000479 is from EPSTI1 gDNA. (B) Percoll solution sorted neutrophils (1 × 10^6) from human peripheral blood was set to detect the expression of hsa_circ_0000479 by PCR. The transcript of human β-actin gene was used as an amplification control. The length of the product detected by PCR agarose gel electrophoresis was 181 bp. (C) PCR products were connected to T-easy vector and then sequenced by Beijing TsingKe Biotech Co., ltd with Sanger sequencing methods. The results of blast sequence alignment were consistent and validated the specific reverse splicing junction of hsa_circ_0000479.
3.3. Upregulated expressions of hsa_circ_0000479 in neutrophils of SLE
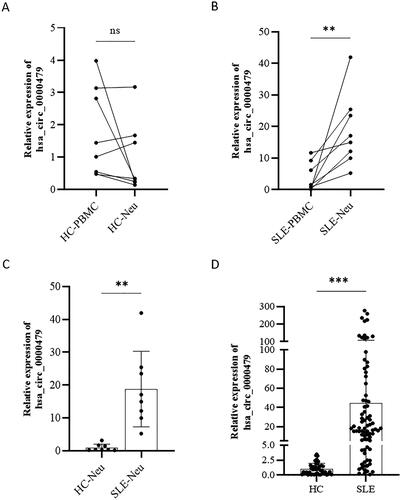
This study is the first to investigate the differential expression of hsa_circ_0000479 in PBMCs and neutrophils. Eight HCs and eight SLE patients were enrolled as a training cohort. The results showed that there was no significant difference expression of hsa_circ_0000479 between PBMCs (1.73 ± 1.38) and neutrophils (0.94 ± 1.08) in HCs (t = 1.272, p = 0.224) (). However, the expression levels of hsa_circ_0000479 in neutrophils (18.78 ± 11.50) were much higher than that of PBMCs (3.83 ± 4.56) in SLE (t = 3.416, p = 0.008) (). Notably, the hsa_circ_0000479 expression levels in neutrophils of SLE patients (18.78 ± 11.50) were significantly higher than those in HCs (0.94 ± 1.08) (t = 4.366, p = 0.003) (). In order to confirm these results, 80 SLE patients and 45 HCs were further enrolled as the validation cohort. As expected, the expression of hsa_circ_0000479 in neutrophils of SLE [18.03 (4.89, 51.18)] was significantly higher than that in HCs [0.93 (0.23, 1.47)] (Z = 263, p < 0.001) ().
Figure 3. Up-regulated expressions of hsa_circ_0000479 in neutrophils of SLE. (A, B) Expression of hsa_circ_0000479 in the PBMCs and neutrophils of healthy controls (n = 8) (A) and SLE patients (n = 8) (B). (C) Expression of hsa_circ_0000479 in the neutrophils of healthy controls (n = 8) and SLE patients (n = 8). (D) Statistical results of hsa_circ_0000479 detected in the neutrophils of healthy controls (n = 45) and SLE patients (n = 80) by qPCR compared with internal reference β-Actin; Ns: no significance; **p < 0.01, ***p < 0.001 (Mann–Whitney U-test, A–D).
3.4. Correlation of hsa_circ_0000479 with SLE patients’ laboratory features
The relationship between hsa_circ_0000479 levels in neutrophils and laboratory features of SLE patients was analysed (). The results showed that the expression of hsa_circ_0000479 in neutrophils was negatively correlated with age (r = −0.294, padj =0.023), WBC (r = −0.313, padj = 0.016), absolute neutrophils count (r = −0.323, padj = 0.015) and hypo-complementaemia (r = −0.347, padj = 0.010) in SLE patients (). In addition, the expression of hsa_circ_0000479 was positively correlated with serum IgA (r = 0.350, padj = 0.013), IgG (r = 0.337, padj = 0.013) and IgM (r = 0.299, padj = 0.021) in SLE (). Importantly, the levels of hsa_circ_0000479 were also positively correlated with the levels of autoantibodies, including anti-dsDNA antibody and anti-nucleosome antibody (AnuA) (). Moreover, SLE patients with positivity of ANA, anti-dsDNA antibody, anti-U1RNP or AnuA showed higher levels of hsa_circ_0000479 than those negative patients ().
Figure 4. Correlation of hsa_circ_0000479 with SLE patients’ clinical and immunological features. The correlation of hsa_circ_0000479 levels with SLE patients’ age (A), white blood cell (WBC) (B), absolute neutrophil count (NEUT) (C), C3 (D), IgA level (E), IgG (F), IgM (G), anti-dsDNA antibodies (H) and anti-nucleosome antibodies (AnuA) (I) were analysed. *p < 0.05; **p < 0.01 (Spearman’s rank correlation test and benjaminiand hochberg correction, A–I).
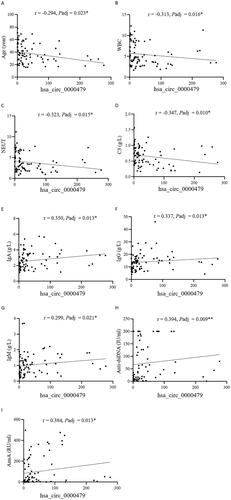
Figure 5. The expression of hsa_circRNA_000479 was increased in neutrophils of patients with positive autoantibodies. The levels of hsa_circRNA_000479 in SLE patients with or without autoantibodies. Sub-group analyses in SLE patients were performed according to the negative or positive of ANA (A), anti-dsDNA antibodies (B), anti-U1RNP (C), and anti-AnuA (D). *p < 0.05; **p < 0.01; ***p < 0.001 (Mann–Whitney U-test, A–D).
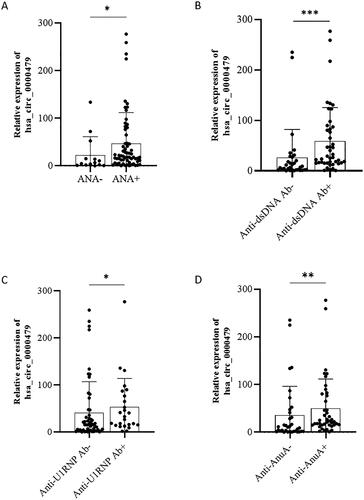
Table 2. Correlation of hsa_circ_0000479 with SLE patients’ clinical and immunological features.
3.5. Association of hsa_circ_0000479 with SLE clinical features
The relationship between hsa_circ_0000479 levels in neutrophils and the clinical features of SLE patients was also analysed (). Totally 80 SLE patients were divided into with or without specific clinical manifestations. The expression of hsa_circ_0000479 in neutrophils of SLE patients with Raynold’s phenomenon, alopecia or leucopenia was higher than those without. However, there was no significant correlation between hsa_circ_0000479 levels and SLEDAI-2K score in SLE ().
Table 3. Association of hsa_circ_0000479 with SLE clinical features.
3.6. Correlation between hsa_circRNA_000479 in neutrophils and immune cells in SLE patients
The relationship between the levels of hsa_circ_0000479 in neutrophils and the immune cell profile of SLE patients was further analysed (). The results showed that the levels of hsa_circ_0000479 were positively correlated with Foxp3+ regulatory T cells (Treg, ) and negatively correlated with effector T cells (Teff, ). However, no correlation was found between hsa_circ_0000479 and the other T cell subsets, as well as the B and NK cell subsets.
Figure 6. The expression level of hsa_circRNA_000479 in neutrophils is closely related to Treg and Teff cell subsets in SLE patients. The correlation of hsa_circ_0000479 levels with SLE patient Foxp3+ Treg cells (A) and Teff cells (B) were analysed. *p < 0.05; **p < 0.01 (Spearman’s rank correlation test, A,B).
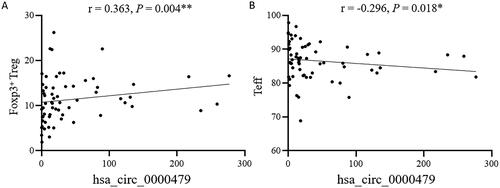
Table 4. Correlation between hsa_circRNA_000479 in neutrophils and immune cells in SLE patients.
4. Discussion
In this study, we reported that hsa_circ_0000479 was expressed in neutrophils and was significantly increased in patients with SLE. Moreover, the neutrophils hsa_circ_0000479 was correlated with laboratory parameters, including absolute neutrophils count, C3, anti-dsDNA and anti-nucleosome antibodies, and was also with specific clinical manifestations, including Raynaud’s phenomenon, alopecia and leucopenia in SLE patients in this study. These findings suggest a potential role of hsa_circ_0000479 in the pathogenesis and clinical features of SLE.
It is well known that neutrophils, as one of the important innate immune cells, are the most abundant nucleated cells in the peripheral circulation of the human and play an important role as regulators of adaptive immune responses [Citation29,Citation30]. Dysregulation of neutrophils contributes to the occurrence and progression of SLE [Citation8,Citation10]. In this study, neutrophils with high expression of hsa_circ_0000479 may be more prone to cell death, resulting in a reduction in absolute neutrophil count, leading to the release of a large amount of DAMPs that bind to receptors on immune cells [Citation31]. This triggers an inflammatory response and recruits immune cells. Furthermore, the activation of auto-reactive B cells results in the production of IgA/G/M, such as anti-dsDNA and AnuA autoantibodies, forming immune complexes that eventually deposit in the skin tissues and activate the complement pathway, leading to lower levels of C3. This process may lead to clinical manifestations in SLE patients, including Raynaud’s phenomenon and alopecia. It is suggested that hsa_circ_0000479 of neutrophils may directly or indirectly regulate innate and adaptive immune responses in SLE. However, further research is needed to validate the relationship between neutrophils and conditions, such as alopecia and Raynaud’s phenomenon. Additionally, exploring the expression profile of circRNAs and identifying key circRNAs in peripheral neutrophils of SLE patients would provide valuable insights and merits in-depth investigation.
Prior to this study, the prominent presence of hsa_circ_0000479 in PBMCs of SLE had been noted. After comprehensively comparing of the circRNAs microarray data of PBMCs samples from SLE patients tested by different researches, hsa_circ_0000479 was elevated in the data from two laboratories [Citation25,Citation26], and the change fold of both was very significant, but hsa_circ_0000479 was not focus on in the other groups [Citation32–34], which was also a question of our concern. Presumably, the complex pathology and physiology of SLE with significant heterogeneity may contribute to the overall difference. For another reason, we speculated that the number of low-density neutrophils mixed in PBMCs isolated in different laboratories may directly influence the final experimental results.
Additionally, hsa_circ_0000479, also called circEPSTI1, shares the base sequence from exon 2 to exon 6 with EPSTI1. Previous research suggested that EPSTI1 promoted B cells activation and participated in the development and progression of primary Sjögren’s syndrome (pSS) via NF-κB signaling pathway [Citation35]. Besides, based on the microarray and Gene Expression Omnibus (GEO) database analysis, EPSTI1 expression was up-regulated in peripheral blood of SLE patients [Citation36,Citation37]. This study specifically analysed the expression of hsa_circ_0000479 for neutrophils in SLE patients with or without secondary Sjogren’s syndrome. However, the difference in hsa_circ_0000479 between the two groups was not statistically significant. In addition, hsa_circ_0000479 is a potential biomarker and play a vital role in cancer, such as breast cancer [Citation38–40] and ovarian cancer [Citation41]. Notably, hsa_circ_0000479 was detected in non-invasive biological samples such as urine and bile, according to the Exorbase Database (http://www.exorbase.org/exoRBaseV2/circRna/toResultBySymbolOrId), and deserve more attention.
Lacking functional validation and mechanism studies is the main deficiency of our study. By summing up what has been reported, we recognize that hsa_circ_0000479 play a role mainly via the competing endogenous RNAs (ceRNA) mechanism, regulating the expression and function of miRNA-targeted mRNA. In recent studies, it has been reported that the miRNA/mRNA axis of hsa_circ_0000479 action, including miR-149-5p/RIG-I/IL-6 axis could play a role in regulating the immune response against SARS-CoV-2 [Citation42], miR-4753/6809-BCL11A axis affected the proliferation and apoptosis of triple-negative breast cancer [Citation43], miR-375/409-3P/515-5p-SLC7A11 axis was relative to ferroptosis in cervical cancer, miR-942-5p/LTBP2 axis regulated oral squamous cell carcinoma (OSCC) cells proliferation and invasion via the acceleration of epithelial-mesenchymal transition (EMT) and the phosphorylation of PI3K/Akt/mTOR signaling pathway components [Citation44]. Moreover, hsa_circ_0000479 may regulate SLE progression by modulating metabolic pathways and the Wnt signaling pathway by bioinformatics analysis [Citation25]. And more imperative, it is worth noting that hsa_circ_0000479 exhibits significantly higher expression in neutrophils compared to PBMCs in SLE patients, highlighting the need for further elucidation of its function and mechanism in neutrophils during the pathogenesis of SLE.
In summary, hsa_circ_0000479 expression is significantly dysregulated in neutrophils and associated with various clinical features of SLE, suggesting that it may play a crucial role in neutrophils, which will be of essential for understanding the pathogenesis, stratification and treatment in SLE.
Author contributions
RY performed the experiments, analysed the data and prepared the manuscript. YS and FH conceived and designed the study, reviewed and edited the manuscript. LX, GC, ZW, RL, WP, LC, YJ and HY contributed to reagents/materials/analysis.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
Data supporting the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Fava A, Petri M. Systemic lupus erythematosus: diagnosis and clinical management. J Autoimmun. 2019;96:1–12. doi: 10.1016/j.jaut.2018.11.001.
- Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47(12):1457–1464. doi: 10.1038/ng.3434.
- Orme J, Mohan C. Macrophages and neutrophils in SLE-An online molecular catalog. Autoimmun Rev. 2012;11(5):365–372. doi: 10.1016/j.autrev.2011.10.010.
- Sharabi A, Tsokos GC. T cell metabolism: new insights in systemic lupus erythematosus pathogenesis and therapy. Nat Rev Rheumatol. 2020;16(2):100–112. doi: 10.1038/s41584-019-0356-x.
- Mantovani A, Cassatella MA, Costantini C, et al. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024.
- Hacbarth E, Kajdacsy-Balla A. Low density neutrophils in patients with systemic lupus erythematosus, rheumatoid arthritis, and acute rheumatic fever. Arthritis Rheum. 1986;29(11):1334–1342. doi: 10.1002/art.1780291105.
- Tay SH, Celhar T, Fairhurst AM. Low-density neutrophils in systemic lupus erythematosus. Arthritis Rheumatol. 2020;72(10):1587–1595. doi: 10.1002/art.41395.
- Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nat Rev Rheumatol. 2011;7(12):691–699. doi: 10.1038/nrrheum.2011.132.
- Li P, Jiang M, Li K, et al. Glutathione peroxidase 4-regulated neutrophil ferroptosis induces systemic autoimmunity. Nat Immunol. 2021;22(9):1107–1117. doi: 10.1038/s41590-021-00993-3.
- Gupta S, Kaplan MJ. Bite of the wolf: innate immune responses propagate autoimmunity in lupus. J Clin Invest. 2021;131(3):e144918. doi: 10.1172/JCI144918.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003.
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017.
- Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036.
- Kristensen LS, Jakobsen T, Hager H, et al. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi: 10.1038/s41571-021-00585-y.
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi: 10.1186/s12943-020-01286-3.
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y.
- Dube U, Del-Aguila JL, Li Z, et al. An atlas of cortical circular RNA expression in alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22(11):1903–1912. doi: 10.1038/s41593-019-0501-5.
- Lu D, Chatterjee S, Xiao K, et al. A circular RNA derived from the insulin receptor locus protects against doxorubicin-induced cardiotoxicity. Eur Heart J. 2022;43(42):4496–4511. doi: 10.1093/eurheartj/ehac337.
- Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004.
- Wang Y, Wu C, Du Y, et al. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol Cancer. 2022;21(1):13. doi: 10.1186/s12943-021-01484-7.
- Zhao X, Dong R, Zhang L, et al. N6-methyladenosine-dependent modification of circGARS acts as a new player that promotes SLE progression through the NF-κB/A20 axis. Arthritis Res Ther. 2022;24(1):37. doi: 10.1186/s13075-022-02732-x.
- Wang X, Zhang C, Wu Z, et al. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Res Ther. 2018;20(1):118. doi: 10.1186/s13075-018-1618-8.
- Zhang C, Gao C, Di X, et al. Hsa_circ_0123190 acts as a competitive endogenous RNA to regulate APLNR expression by sponging hsa-miR-483-3p in lupus nephritis. Arthritis Res Ther. 2021;23(1):24. doi: 10.1186/s13075-020-02404-8.
- Wang L, Lu M, Li W, et al. Significance of circRNAs as biomarkers for systemic lupus erythematosus: a systematic review and meta-analysis. J Int Med Res. 2022;50(7):3000605221103546. doi: 10.1177/03000605221103546.
- Guo G, Wang H, Ye L, et al. Hsa_circ_0000479 as a novel diagnostic biomarker of systemic lupus erythematosus. Front Immunol. 2019;10:2281. doi: 10.3389/fimmu.2019.02281.
- Luo Q, Zhang L, Fang L, et al. Circular RNAs hsa_circ_0000479 in peripheral blood mononuclear cells as novel biomarkers for systemic lupus erythematosus. Autoimmunity. 2020;53(3):167–176. doi: 10.1080/08916934.2020.1728529.
- Hochberg MC. Updating the American college of rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725–1725. doi: 10.1002/art.1780400928.
- Bombardier C, Gladman DD, Urowitz MB, et al. Derivation of the SLEDAI. A disease activity index for lupus patients. The committee on prognosis studies in SLE. Arthritis Rheum. 1992;35(6):630–640. doi: 10.1002/art.1780350606.
- Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol. 2014;15(7):602–611. doi: 10.1038/ni.2921.
- Gupta S, Kaplan MJ. The role of neutrophils and NETosis in autoimmune and renal diseases. Nat Rev Nephrol. 2016;12(7):402–413. doi: 10.1038/nrneph.2016.71.
- Kang N, Liu X, Haneef K, et al. Old and new damage-associated molecular patterns (DAMPs) in autoimmune diseases. Rheumatol Autoimmun. 2022;2(4):185–197. doi: 10.1002/rai2.12046.
- Song W, Qiu J, Yin L, et al. Integrated analysis of competing endogenous RNA networks in peripheral blood mononuclear cells of systemic lupus erythematosus. J Transl Med. 2021;19(1):362. doi: 10.1186/s12967-021-03033-8.
- Zhang MY, Wang JB, Zhu ZW, et al. Differentially expressed circular RNAs in systemic lupus erythematosus and their clinical significance. Biomed Pharmacother. 2018;107:1720–1727. doi: 10.1016/j.biopha.2018.08.161.
- Luo Q, Zhang L, Li X, et al. Identification of circular RNAs hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus. Int J Mol Med. 2019;44(4):1462–1472. doi: 10.3892/ijmm.2019.4302.
- Sun JL, Zhang HZ, Liu SY, et al. Elevated EPSTI1 promote B cell hyperactivation through NF-κB signalling in patients with primary sjögren’s syndrome. Ann Rheum Dis. 2020;79(4):518–524. doi: 10.1136/annrheumdis-2019-216428.
- Ishii T, Onda H, Tanigawa A, et al. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 2005;12(6):429–439. doi: 10.1093/dnares/dsi020.
- Zhao X, Zhang L, Wang J, et al. Identification of key biomarkers and immune infiltration in systemic lupus erythematosus by integrated bioinformatics analysis. J Transl Med. 2021;19(1):35. doi: 10.1186/s12967-020-02698-x.
- Raval AP, Desai UN, Joshi JS, et al. Role of epithelial - Stromal interaction protein-1 expression in breast cancer. Indian J Pathol Microbiol. 2020;63(3):382–387. doi: 10.4103/IJPM.IJPM_672_19.
- Li T, Lu H, Shen C, et al. Identification of epithelial stromal interaction 1 as a novel effector downstream of krüppel-like factor 8 in breast cancer invasion and metastasis. Oncogene. 2014;33(39):4746–4755. doi: 10.1038/onc.2013.415.
- Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of Triple-Negative breast cancer progression. Theranostics. 2018;8(14):4003–4015. doi: 10.7150/thno.24106.
- Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi: 10.1111/jcmm.14260.
- Firoozi Z, Mohammadisoleimani E, Shahi A, et al. Hsa_circ_0000479/hsa-miR-149-5p/RIG-I, IL-6 axis: a potential novel pathway to regulate immune response against COVID-19. Can J Infect Dis Med Microbiol. 2022;2022:2762582. [q]
- Wu P, Li C, Ye DM, et al. Circular RNA circEPSTI1 accelerates cervical cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging (Albany NY). 2021;13(3):4663–4673. doi: 10.18632/aging.202518.
- Wang J, Jiang C, Li N, et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis. 2020;11(8):682. doi: 10.1038/s41419-020-02851-w.
