Abstract
Background. Adenosine triphosphate (ATP)‐binding cassette transporter A3 (ABCA3) gene mutations cause fatal respiratory failure in term infants, but common ABCA3 polymorphisms have remained uncharacterized at the population level.
Aim. To define a subset of tagging single‐nucleotide polymorphisms (tSNPs) which capture most of the variation within the ABCA3 gene, and to assess ABCA3 as a novel candidate gene for susceptibility to respiratory distress syndrome (RDS) in preterm infants.
Methods. Based on an initial screen, nine tSNPs were selected. These 9 tSNPs and a length variation, representing >90% of haplotypic variation of the gene, and 5 nonsynonymous coding SNPs were genotyped in 267 preterm infants. SNP rs13332514 was genotyped in an additional 48 infants.
Results. The fourth common haplotype was overrepresented in very premature infants with RDS, being accounted for by SNP rs13332514 (F353F), with an increased minor allele frequency in RDS. Furthermore, rs13332514 associated significantly with chronic lung disease defined as a requirement for supplemental O2 at 28 postnatal days in very premature infants.
Conclusions. The results are suggestive of an association of a synonymous SNP in the ABCA3 gene with a prolonged course of respiratory distress syndrome in very premature infants and serve as a reference for further population‐based studies of ABCA3.
| Abbreviations | ||
| ABCA3 | = | ATP‐binding cassette transporter A3 |
| ATP | = | adenosine triphosphate |
| BPD | = | bronchopulmonary dysplasia |
| GA | = | gestational age |
| LD | = | linkage disequilibrium |
| MAF | = | minor allele frequency |
| OR | = | odds ratio |
| RDS | = | respiratory distress syndrome |
| SNP | = | single‐nucleotide polymorphism |
| SP | = | surfactant protein |
| TDI‐FP | = | template‐directed dye‐terminator incorporation with fluorescence polarization detection |
| tSNP | = | tagging single‐nucleotide polymorphism |
Introduction
Adenosine triphosphate (ATP)‐binding cassette A3 transporter (ABCA3) is a member of the evolutionarily highly conserved family of ABC transporters. They are transmembrane proteins which use the hydrolysis of ATP to drive the transport of a wide variety of substrates across cellular membranes Citation1. ABC A subfamily consists of 12 structurally related transporters involved in lipid transport, and several of them have been linked to unrelated human genetic diseases, including ABCA1 in Tangiers disease, ABCA12 in lamellar ichthyosis, and others Citation2. ABCA3 is predominantly expressed in lung alveolar type II cells localizing to the limiting membrane of lamellar bodies, which are intracellular organelles responsible for the storage and regulated secretion of pulmonary surfactant Citation3. Surfactant, a mixture of lipids and proteins covering the distal epithelial surface of the lung, is required for normal lung function. Its main function is to reduce surface tension at the air‐liquid interface, thus preventing alveolar collapse Citation4.
Deficiency of surfactant due to prematurity is the main cause of respiratory distress syndrome (RDS), a multifactorial disease of newborn infants characterized by respiratory failure and deficient gas exchange Citation5. RDS is the most common cause of serious morbidity in prematurely born infants. It is often followed by bronchopulmonary dysplasia (BPD), a chronic form of pulmonary dysfunction, in infants born very preterm Citation6. According to current understanding, genetic susceptibility to RDS and even BPD is at least partly related to surfactant metabolism Citation7–9. Allelic variants of the genes encoding surfactant proteins (SP)‐A, ‐B, and ‐C have been shown to be predisposing factors to RDS Citation8. The genes associated with heritable susceptibility to BPD include those encoding SP‐A and ‐B, angiotensin‐converting enzyme (ACE), glutathione S transferase (GST), and tumor necrosis factor α (TNFα) Citation9. Very preterm infants born before 32 weeks of pregnancy with a high incidence of RDS and a moderate risk of BPD have different genetic risk factors of respiratory failure from those of less preterm infants born after 32 weeks of gestation with a moderate risk of RDS and a very low risk of BPD Citation10–12.
Several in vitro and in vivo studies have shown that ABCA3 is essential for normal surfactant homeostasis Citation3, Citation13–21, presumably through its potential role in the transport of surfactant lipids. Multiple missense, splice‐site and frameshift mutations in the gene encoding ABCA3 have been identified in term newborns with fatal surfactant deficiency or unexplained severe respiratory distress Citation13, Citation16 and in older children with interstitial lung disease (ILD), a heterogeneous group of poorly known chronic respiratory diseases characterized by pulmonary fibrosis and inflammation of the lung Citation22. As yet uncharacterized in population level studies, common ABCA3 polymorphisms, which could have a minor effect on ABCA3 protein expression or function, can be regarded as novel candidate risk factors for severe multifactorial respiratory diseases.
Haplotype analysis, i.e. simultaneous analysis of combinations of single‐nucleotide polymorphisms (SNPs), summarizes the information of several SNPs and allows a search for disease‐predisposing alterations at the gene‐wide level Citation23. The present study was designed to identify a set of informative markers for characterizing common variation within the ABCA3 gene to assess whether ABCA3 haplotypes or individual polymorphisms play a role in the susceptibility to respiratory distress. The results implicate a novel role for ABCA3 as a genetic determinant of susceptibility to multifactorial respiratory disease in preterm infants.
Key messages
Loss‐of‐function mutations involving the ATP‐binding cassette transporter A3 (ABCA3) gene result in fatal respiratory failure. Here we propose that common ABCA3 gene variation plays a role in genetic susceptibility to multifactorial pulmonary disease.
A synonymous SNP (F353F) in the ABCA3 gene associates with a prolonged course of respiratory distress syndrome in very premature infants.
Materials and methods
Diagnostic criteria for RDS and mild BPD
The diagnosis of RDS was established using clinical (severe respiratory distress, including requirement for supplemental O2 for at least 48 hours or requirement for surfactant therapy for established RDS) and radiographic criteria (diffuse haziness, edema, or a reticulogranular pattern with air bronchograms). None of the infants received surfactant prior to the diagnosis of RDS. The infants not recovering from RDS and developing chronic lung disease had requirement for supplemental oxygen or ventilation due to diffuse lung disease at the postnatal age of 28 days (also termed mild BPD).
Study population and DNA sample preparation
The present study population was originally enrolled for and included in our previous studies of association between SP genes and RDS or BPD Citation10, Citation11, Citation24, Citation25. These samples were included in the present study according to the inclusion criteria, whenever an adequate amount of DNA was still available to perform the ABCA3 genotyping. The study population consisted altogether of 407 infants of Finnish ancestry. A total of 359 infants were sampled prospectively, using blood specimens, and 48 retrospectively, using buccal cells. The prospectively sampled population consisted of 267 preterm infants born at 25–35 weeks of gestation in Oulu University Hospital, Seinäjoki Central Hospital, and Tampere University Hospital during 1996–2002 and 92 healthy term infants (>37 weeks of gestation) born in Oulu in 1998. To confirm the detected allelic association after an analysis of the prospectively collected samples, a retrospectively collected set of very preterm infants with gestational age (GA) of 25–31 weeks born in Oulu during 1991–1996 was analyzed. Written informed consent was obtained from the parents. The study was approved by the ethical committees of the participating centers. The clinical characteristics of the preterm infants are shown in Table
Table I. Clinical characteristics of preterm infants.
Due to the well established profound effect modification by the length of gestation on candidate gene associations observed in our earlier studies (with 32 weeks as a stratifying GA threshold to restrict e.g. particular SP‐A gene variants as high‐risk alleles in the lower but not in the higher GA group, as shown statistically by a homogeneity of odds ratios test) Citation10, Citation12, Citation24, Citation26, the preterm infants were divided into two GA‐matched groups for the analyses (Table ). All prospectively sampled infants with mild BPD (n = 25) came from the group of very premature infants with RDS. The retrospectively collected samples were from children born very premature with (n = 41) or without RDS (n = 7). Of these, 26 had both RDS and mild BPD, while 2 did not have either. The infants born at term served as reference for population allele and haplotype frequencies.
Genomic DNA was extracted from whole blood using the Puregene DNA Isolation kit (Gentra Systems, Minneapolis, MN, USA) and from buccal smears using Chelex 100 (Bio‐Rad, Hercules, CA, USA).
Genotyping
SNP genotyping was performed by template‐directed dye‐terminator incorporation with fluorescence polarization detection (TDI‐FP Citation27) using AcycloPrimeTM II SNP Detection Kits (Perkin Elmer Life Sciences, Boston, USA). In addition to the SNPs, a previously unknown length polymorphism consisting of 1–4 repeats of 80 bp (European Molecular Biology Laboratory EMBL accession numbers AM691772, AM691773, and AM691774) in intron 15 of the ABCA3 gene was genotyped using polymerase chain reaction. Primers were purchased from Oligomer Oy (Helsinki, Finland). Primer sequences and a detailed description of genotyping are available on request.
Selection of SNPs, haplotype prediction, and data analysis
Intronic SNPs with a validated minor allele frequency (MAF) ⩾0.1 among Caucasians and all coding SNPs (cSNPs) indicated in the dbSNP database (http://ncbi.nih.gov/SNP/) within the ABCA3 gene in the NCBI entry NT_037887, a novel intronic SNP (320‐17G>A) and 875A>T (E292V) were initially screened. Nine tagging SNPs (tSNPs) were selected from the first screen of SNPs listed in Table using a method based on pairwise r2 linkage disequilibrium (LD) statistics Citation28 with r2 set to a threshold of 0.8. The intron‐15 length variation was treated as one of the tSNPs and included in the haplotypes. The data analysis and statistical tests for haplotype and allele frequencies, including calculations of pairwise LD (D´) measures and Hardy‐Weinberg equilibrium, were performed using Haploview v. 3.2 Citation29. Haploview uses an accelerated expectation maximization (EM) algorithm to create haplotype frequency estimates from unphased genotype data. All the P‐values for SNP frequencies were permutation‐corrected for multiple testing with 100,000 replicates, and a P‐value <0.05 was considered significant.
Table II. ABCA3 SNPs genotyped in 176 infants to determine the linkage disequilibrium structure for tagging SNP (tSNP) selection, and their frequencies in the population.
Results
Haplotype structure of the ABCA3 gene and tagging SNP selection
The 23 SNPs selected for genotyping are shown in Table . We selected SNPs with a known MAF of ⩾0.1 among the Caucasian population from dbSNP database for this study, including all cSNPs, of which three were synonymous and four nonsynonymous. Of the 176 SNPs indicated in dbSNP for the ABCA3 gene and the flanking regions, 18 intronic SNPs and 1 cSNP had MAF ⩾0.1 at the time of the database search. No promoter SNPs fulfilling the selection criteria were identified in the database. Four intronic SNPs were omitted due to genotyping failure. Additionally, a novel SNP (320‐17G>A) and a single‐base change (875A>T) corresponding to a glutamine‐to‐valine substitution at amino acid position 292 (E292V) were included in the analysis, as this position has been reported to be frequently mutated in infants suffering from ILD Citation22. Only three cSNPs (rs13332514, rs323043, and rs149532, corresponding to residues F353F, P585P, and S1372S, respectively), all synonymous, were polymorphic (MAF>0.01) in our population. In addition to the SNPs, a novel length variation in intron 15 was analyzed. The most frequent allele of the length variation consisted of four repeats and accounted for ∼55% of the alleles, and the allele with one repeat accounted for ∼45%. A three‐repeat variant was only detected in two samples (0.03% of the alleles), and it was pooled together with the one‐repeat variant. No instances of a two‐repeat variant were detected.
We aimed to identify a set of informative markers for characterizing the population level variation in the genomic region of the ABCA3 gene. Thus, the entire ABCA3 gene locus was treated as a single block for tSNP selection. To characterize the LD patterns of the ABCA3 gene in the study population, the selected polymorphisms were genotyped in an initial population screen of 176 infants consisting of 92 term (GA>37 weeks) and 84 preterm (GA 25–35 weeks) infants. SNP rs149532 (corresponding to residue S1372S) was rare (MAF = 0.02) in our population, and it was thus excluded from further analysis, together with the nonpolymorphic cSNPs (rs28936412, 875A>T, rs13332760, rs28936690, and rs28936691, corresponding to residues L101P, E292V, V839F, L1552P, and Q1591P, respectively). The average intermarker distance was ∼2.9 kb, the largest distance between adjacent SNPs being ∼7.4 kb and the smallest ∼0.9 kb. None of the SNPs showed deviation from Hardy‐Weinberg equilibrium.
SNPs that capture >90% of the haplotypic variation were selected based on pairwise r2 LD statistics Citation28 with r2 initially set to a threshold of 0.8, resulting in a set of nine tSNPs, which are indicated in Table . The tSNPs are positioned in intron 5, exon 10, and the introns 10, 11, 12, 21, 22, 26, and 29. The intron‐15 length variation was included in the haplotypes and treated similarly to the tSNPs. Altogether 46 haplotypes were predicted (data not shown). Of these, 11 haplotypes occurred at a frequency ⩾0.01. Four common haplotypes with a frequency >0.05 were detected, accounting for ∼75% of all haplotypes. The locations of the polymorphisms are presented in Figure , the six most common haplotypes along with their frequencies in Table , and pairwise LD values for tSNPs in Figure .
Table III. Estimated ABCA3 haplotypes and frequencies.a
Figure 1 Map of theABCA3 gene showing the positions of exons (black bars) and polymorphisms (short and long lines) genotyped in the initial population screen of 176 infants. The positions of tagging single nucleotide polymorphisms (tSNPs) and intron‐15 length variation (i15) are indicated by long lines.
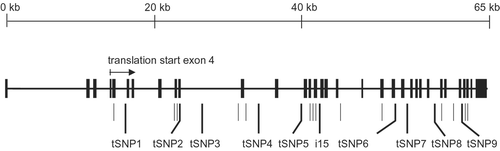
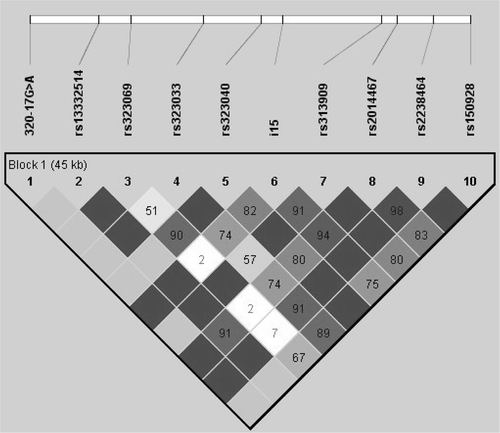
Figure 2 Haploview display of the pairwise linkage disequilibrium(D′) values for ABCA3 tagging single nucleotide polymorphisms and intron‐15 length variation (i15). Names and relative positions of the polymorphism are shown on top. The number in the square is the D′ value for each pairwise comparison. When no number is shown, linkage disequilibrium is complete (D′ = 1).
Association of ABCA3 haplotypes and polymorphisms with RDS and mild BPD
To assess the contribution of single polymorphisms and haplotypes to RDS, the nine tSNPs, the intron‐15 length variation, and the nonsynonymous cSNPs were genotyped in a panel of 267 preterm infants sampled prospectively. Comparisons of the haplotype, genotype, and allele frequencies in the RDS patients were performed in two groups consisting of very premature (GA 25–31 weeks) and late‐preterm infants (GA 32–35 weeks). In addition, retrospectively collected samples (n = 48) were analyzed for SNP rs13332514.
The six most common haplotypes and the comparisons of their frequencies between the cases and the controls are listed in Table . Haplotype 4 was overrepresented in the RDS infants compared to the controls among the very premature infants, whereas no haplotypic associations were observed among the late‐preterm infants. The allele frequencies of each polymorphism were also analyzed for potential associations with RDS (Table ). Among the very premature infants, the frequency of the minor (T) allele of a synonymous cSNP (rs13332514 corresponding to residue F353F) located in exon 10 was significantly higher in the infants with RDS compared to the controls (Table ). rs13332514 discriminated haplotype 4 from the other haplotypes and thus explained the detected nominal haplotypic association. As expected based on the relatively low MAF of rs13332514, the proportion of TT homozygotes was very low (none of 105 very premature infants, 2 of 158 late‐preterm infants). Similarly to allele frequencies, the genotype frequencies differed significantly between the RDS and control infants (data not shown). No other allelic associations were observed in either of the GA groups. Due to their potential effect on protein structure, we also screened for nonsynonymous cSNPs in 267 preterm infants. None of the five nonsynonymous cSNPs (rs28936412, 875A>T, rs13332760, rs28936690, and rs28936691, corresponding to residues L101P, E292V, V839F, L1552P, and Q1591P, respectively) were detected in any individuals.
Table IV. Frequencies of the six most common ABCA3 haplotypes.
Table V. Minor allele frequencies of the ABCA3 tSNPs and intron‐15 length variation (i15).
Since very preterm infants are at risk of developing BPD, a chronic lung disease affecting mainly very premature infants with RDS, we further analyzed rs13332514 for a potential association with mild BPD defined as a requirement for supplemental O2 at 28 postnatal days. All the BPD infants included in the analysis came from the group of very premature infants with RDS. Some of the very premature infants could not be included in the BPD allele frequency comparisons due to early death or missing information concerning their long‐term status. The minor (T) allele was significantly overrepresented in the infants with mild BPD compared to the control groups with and without RDS, suggesting that it could be associated with BPD rather than RDS (Table ). When the two control groups were combined, the association was even more evident (Table ). Genotype frequencies also differed significantly in all comparisons (data not shown).
Table VI. rs13332514 minor allele frequency comparisons in very premature infants with RDS or mild BPD.a
To confirm the detected, initially observed association, a set of retrospectively collected samples of very premature infants were analyzed for rs13332514, and the results were combined with those of the prospective samples. The T allele was significantly overrepresented in RDS infants, being restricted to infants with mild BPD (Table ). The differences in the genotype frequencies were significant in all comparisons (data not shown). Since the primary aim of this work was to investigate ABCA3 variation in relation to RDS, infants included in the mild BPD group were restricted to those with both RDS and prolonged O2 dependence. Altogether 11 infants had no RDS and developed mild BPD. Of these infants, which were excluded from the BPD analyses, six came from the prospectively (rs13332514 genotypes 5 CC and 1 CT) and five from the retrospectively sampled population (5 CC). In the infants with mild BPD, the mean GA of minor (T) allele carriers tended to be higher than in the noncarriers (mean±SD: 199.5±10.8 versus 189.8±11.1 days, respectively). rs133332514 had no detectable association with the degree of prematurity per se in the whole population of very premature infants (data not shown). The present study population was not large enough to perform an association study for more strict diagnosis of new BPD as defined at 36 weeks postmenstrual age, nor was it adequate for a multiparameter analysis to consider the effects of factors such as singleton versus multiple birth and severity of RDS.
Discussion
In the present study, the impact of variation in the ABCA3 gene on heritable susceptibility to RDS, a complex disorder of premature infants, and to mild BPD as its prolonged course was evaluated. The main cause of RDS is deficiency in pulmonary surfactant, which leads to respiratory failure and deficient gas exchange in the newborn infant. ABCA3 encodes a lamellar body membrane protein important for surfactant homeostasis. In term infants, loss‐of‐function mutations in the ABCA3 gene were recently identified as a cause of fatal respiratory distress Citation13, Citation16. ABCA3 mutations have also been identified in interstitial lung disease (ILD), a group of less severe respiratory diseases of unknown cause Citation22. Our data provide further evidence for the role of ABCA3 in the pathogenesis of nonlethal multifactorial pediatric lung diseases.
To date, the role of ABCA3 in relation to RDS or BPD in preterm infants has not been evaluated. Population level polymorphism and haplotype data of the ABCA3 gene have not been previously described. We used a tagging single‐nucleotide polymorphism (tSNP) approach which is a method to maximize the amount of genetic variation captured for a candidate gene using a minimal number of SNPs. Our aim was to define a subset of SNPs (tSNPs) which capture most of the variation (>90%) within the ABCA3 gene thereby serving as informative markers for association studies, and subsequently to investigate whether ABCA3 haplotypes are associated with heritable susceptibility to RDS in preterm infants. While selecting the tSNPs, a length polymorphism was detected in intron 15 and included in the analysis as one of the tSNPs. Four frequent haplotypes (frequency >5%) were identified in our population, with the most common haplotype occurring at an estimated frequency of 28.7%. We found a nominally significant association between the fourth common haplotype (H4) and RDS in the very premature infants born before 32 weeks of gestation (Table ), whereas no association was detected for any of the haplotypes among the late‐preterm infants born at 32–35 weeks of gestation. The process of testing the tSNPs individually for associations with RDS revealed a significant association of a single synonymous cSNP (rs13332514 corresponding to residue F353F) with RDS in the group of very premature infants (Table ). The nominal association of haplotype H4 with RDS in the very premature infants appears to be dependent solely on this SNP, since removal of the SNP from the haplotypes causes all evidence of association to be lost (data not shown).
Since infants with RDS are at a high risk of developing BPD, a chronic lung disease affecting almost exclusively infants with RDS born very prematurely, we also tested rs13332514 for an association with BPD in the group of very premature RDS infants. An association between this SNP and prolonged course of RDS or mild BPD (defined as O2 dependency at the postnatal age of 28 days) was detected, since the RDS cases that did not develop chronic lung disease had a significantly lower frequency of the minor (T) allele than the infants with mild BPD (Table ). Therefore, the evidence of an association of rs13332514 with RDS seems to be due to the subgroup of BPD infants among the RDS population. Inclusion of additional samples in the analysis further confirmed the detected associations (Table ). However, our sample sizes were still small and were not sufficient for a multiparameter analysis, especially in the analysis of BPD infants and their age‐matched controls with RDS. Thus, our data should be viewed as suggestive of an association of rs13332514 alleles with a prolonged course of RDS in the very premature infants, and the results need to be confirmed in replicate studies. This would include an adequate number of samples to be able to evaluate the role of potentially important contributing factors, such as gene‐gene interactions, the role of multiple birth, and strict (O2 requirement at 36 weeks of gestation) or broad (O2 requirement at the postnatal age of 28 days) BPD definition.
The associated SNP is located in exon 10 of the ABCA3 gene corresponding to UUC/UUU in tRNA for a phenylalanine (F) residue at amino acid position 353. By secondary structure prediction, F353F is expected to reside within the third membrane‐spanning α helix in the first transmembrane domain of the protein. According to our findings, the major allele C may be a protective factor for RDS or BPD, or the minor allele T a risk factor among very premature infants. Since frequency of the T allele in the group of very premature infants without RDS is lower compared to that in late‐preterm infants with or without RDS or in term infants (Table ), CC could be a so‐called hypernormal protective genotype, i.e. a genotype whose carriers have the lowest likelihood to develop a disease despite predisposing environmental exposure Citation25, Citation30. Since rs13332514 does not cause a change in amino acid and is not a rare polymorphism in our population (minor allele frequency 9.3%), it cannot have a drastic impact on the function of the protein. However, rs13332514 as a causal variant cannot be ruled out. Indeed, previous studies have shown that synonymous SNPs cause allele‐specific differences in mRNA stability and translation Citation31 or even substrate specificity as was recently demonstrated for a member of the ABC family, the Multidrug Resistance 1 gene (MDR1 alias ABCB1) Citation32. Alternatively, it is possible that the SNP lies in LD with a causal variant influencing the function of the protein or regulation of the gene directly. However, no apparent other causal variations could be indicated, since all the exonic polymorphisms were screened in the present study and no promoter polymorphisms are present in the putative transcription factor binding sites reported to date Citation14.
In contrast to very preterm infants, no evidence of an association between ABCA3 polymorphism and RDS was found among the late‐preterm infants born at 32–35 weeks of gestation. According to both clinical and genetic evidence, RDS in very premature and late‐preterm infants appears to represent separate disease phenotypes Citation12. In very premature infants, RDS is predominantly a direct consequence of surfactant deficiency due to the high degree of prematurity of the lung, whereas different, presumably inflammation‐related etiologic factors probably modify the course of the disease in late‐preterm infants. Accordingly, SP‐A haplotypes have different associations with RDS in very premature and late‐preterm infants Citation10, whereas the association of the haplotypes of neuropeptide S receptor‐1 (NPSR1 or GPRA alias GPR154) with RDS was restricted to late‐preterm infants Citation12. The heterogeneity of the RDS phenotype is further supported by the present finding that the genetic association was restricted to very premature infants. However, no definitive conclusions can be drawn from this data before the results have been confirmed in a larger study population. Furthermore, in our study population, the detected association among the very premature infants may be due to BPD, which is a frequent complication of very preterm infants with RDS.
The tSNP approach is a powerful means to reduce the number of assayed SNPs in association studies and therefore especially advantageous for large genes with a great number of SNPs, such as the ABCA3 gene with a genomic sequence of ∼65 kb, 33 exons, and ∼200 known SNPs. In the present study, the number of assayed polymorphisms could be reduced to nine tSNPs and a length variation which together explain >90% of the haplotypic variation. However, the current method may not have sufficient power to detect rare associating alleles. The selected tSNPs, most with a MAF >0.1, were likely to detect any associating common polymorphism but rare associating variants may have escaped our analysis. Furthermore, no promoter SNPs met the present inclusion criteria. Since the selected tSNPs cover only a 46‐kb region of the ABCA3 gene and span from intron 5 to intron 29, another limitation of this study is that the 5′and 3′ regions were not represented by our set of tSNPs. Characterization of the variation in these regions will further facilitate the analysis of ABCA3 variation in relation to the risk of RDS, BPD, and other diseases.
In addition to the SNPs identified for tagging purposes, we screened for the four nonsynonymous cSNPs indicated in dbSNP for ABCA3. Of these, two (corresponding to L101P and Q1591P) have been previously identified in term infants with fatal surfactant deficiency or chronic lung disease Citation13. In our study population, however, neither these nor the two other nonsynonymous cSNPs (corresponding to V839F and L1552P) listed in dbSNP were polymorphic, suggesting that they represent mutations rather than common polymorphisms. Additionally, we screened for the E292V mutation, which has been associated with pediatric ILD Citation22. However, E292V was not detected either in term or in preterm infants in our population.
In summary, we have performed the first haplotype tagging analysis of the ABCA3 gene and a pilot association study assessing the role of ABCA3 variation in the pathogenesis of RDS and mild BPD. We present preliminary evidence for an association between a synonymous coding SNP and a prolonged course of RDS in very premature infants, suggesting that ABCA3 plays a role in the etiology of these diseases. This is consistent with accumulating evidence for requirement of ABCA3 in the intracellular transport of surfactant lipids and lamellar body biogenesis Citation15, Citation20, Citation21, Citation33. Further studies are required to verify the detected association using more extensive sample sizes and to resolve the mechanism by which the associating SNP could predispose very premature infants to RDS or BPD. The interaction of ABCA3 gene variation with other genes, particularly those involved in the synthesis and processing of surfactant, may elucidate further the critical molecular events in respiratory adaptation.
Acknowledgements
The authors wish to thank Ms Maarit Haarala for excellent technical assistance. Riitta Marttila, MD, PhD, and Ms Eija Rautio are acknowledged for the collection of clinical data. The work was supported by Sigrid Juselius Foundation and Academy of Finland.
References
- Dean M., Hamon Y., Chimini G. The human ATP‐binding cassette (ABC) transporter superfamily. J Lipid Res 2001; 42: 1007–17
- Kaminski W. E., Piehler A., Wenzel J. J. ABC A‐subfamily transporters: structure, function and disease. Biochim Biophys Acta 2006; 1762: 510–24
- Yamano G., Funahashi H., Kawanami O., Zhao L. X., Ban N., Uchida Y., et al. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett 2001; 508: 221–5
- Serrano A. G., Perez‐Gil J. Protein‐lipid interactions and surface activity in the pulmonary surfactant system. Chem Phys Lipids 2006; 141: 105–18
- Avery M. E., Mead J. Surface properties in relation to atelectasis and hyaline membrane disease. AMA J Dis Child 1959; 97: 517–23
- Jobe A. H., Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med 2001; 163: 1723–9
- Clark H., Clark L. S. The genetics of neonatal respiratory disease. Semin Fetal Neonatal Med 2005; 10: 271–82
- Hallman M., Haataja R. Genetic basis of respiratory distress syndrome. Front Biosci 2007; 12: 2670–82
- Bhandari V., Gruen J. R. The genetics of bronchopulmonary dysplasia. Semin Perinatol 2006; 30: 185–91
- Ramet M., Haataja R., Marttila R., Floros J., Hallman M. Association between the surfactant protein A (SP‐A) gene locus and respiratory‐distress syndrome in the Finnish population. Am J Hum Genet 2000; 66: 1569–79
- Rova M., Haataja R., Marttila R., Ollikainen V., Tammela O., Hallman M. Data mining and multiparameter analysis of lung surfactant protein genes in bronchopulmonary dysplasia. Hum Mol Genet 2004; 13: 1095–104
- Pulkkinen V., Haataja R., Hannelius U., Helve O., Pitkanen O. M., Karikoski R., et al. G protein‐coupled receptor for asthma susceptibility associates with respiratory distress syndrome. Ann Med 2006; 38: 357–66
- Shulenin S., Nogee L. M., Annilo T., Wert S. E., Whitsett J. A., Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med 2004; 350: 1296–303
- Yoshida I., Ban N., Inagaki N. Expression of ABCA3, a causative gene for fatal surfactant deficiency, is up‐regulated by glucocorticoids in lung alveolar type II cells. Biochem Biophys Res Commun 2004; 323: 547–55
- Cheong N., Madesh M., Gonzales L. W., Zhao M., Yu K., Ballard P. L., et al. Functional and trafficking defects in ATP binding cassette A3 mutants associated with respiratory distress syndrome. J Biol Chem 2006; 281: 9791–800
- Brasch F., Schimanski S., Muhlfeld C., Barlage S., Langmann T., Aslanidis C., et al. Alteration of the pulmonary surfactant system in full‐term infants with hereditary ABCA3 deficiency. Am J Respir Crit Care Med 2006; 174: 571–80
- Matsumura Y., Ban N., Ueda K., Inagaki N. Characterization and classification of ATP‐binding cassette transporter ABCA3 mutants in fatal surfactant deficiency. J Biol Chem 2006; 281: 34503–14
- Garmany T. H., Moxley M. A., White F. V., Dean M., Hull W. M., Whitsett J. A., et al. Surfactant composition and function in patients with ABCA3 mutations. Pediatr Res 2006; 59: 801–05
- Stahlman M. T., Besnard V., Wert S. E., Weaver T. E., Dingle S., Xu Y., et al. Expression of ABCA3 in developing lung and other tissues. J Histochem Cytochem 2007; 55: 71–83
- Fitzgerald M. L., Xavier R., Haley K. J., Welti R., Goss J. L., Brown C. E., et al. ABCA3 inactivation in mice causes respiratory failure, loss of pulmonary surfactant, and depletion of lung phosphatidylglycerol. J Lipid Res 2007; 48: 621–32
- Ban N., Matsumura Y., Sakai H., Takanezawa Y., Sasaki M., Arai H., et al. ABCA3 as a lipid transporter in pulmonary surfactant biogenesis. J Biol Chem 2007; 282: 9628–34
- Bullard J. E., Wert S. E., Whitsett J. A., Dean M., Nogee L. M. ABCA3 mutations associated with pediatric interstitial lung disease. Am J Respir Crit Care Med 2005; 172: 1026–31
- Johnson G. C., Esposito L., Barratt B. J., Smith A. N., Heward J., Di Genova G., et al. Haplotype tagging for the identification of common disease genes. Nat Genet 2001; 29: 233–7
- Haataja R., Ramet M., Marttila R., Hallman M. Surfactant proteins A and B as interactive genetic determinants of neonatal respiratory distress syndrome. Hum Mol Genet 2000; 9: 2751–60
- Haataja R., Marttila R., Uimari P., Lofgren J., Ramet M., Hallman M. Respiratory distress syndrome: evaluation of genetic susceptibility and protection by transmission disequilibrium test. Hum Genet 2001; 109: 351–5
- Marttila R., Haataja R., Ramet M., Pokela M. L., Tammela O., Hallman M. Surfactant protein A gene locus and respiratory distress syndrome in Finnish premature twin pairs. Ann Med 2003; 35: 344–52
- Chen X., Kwok P. Y. Homogeneous genotyping assays for single nucleotide polymorphisms with fluorescence resonance energy transfer detection. Genet Anal 1999; 14: 157–63
- Carlson C. S., Eberle M. A., Rieder M. J., Yi Q., Kruglyak L., Nickerson D. A. Selecting a maximally informative set of single‐nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 2004; 74: 106–20
- Barrett J. C., Fry B., Maller J., Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–5
- Morton N. E., Collins A. Tests and estimates of allelic association in complex inheritance. Proc Natl Acad Sci U S A 1998; 95: 11389–93
- Duan J., Wainwright M. S., Comeron J. M., Saitou N., Sanders A. R., Gelernter J., et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet 2003; 12: 205–16
- Kimchi‐Sarfaty C., Oh J. M., Kim I. W., Sauna Z. E., Calcagno A. M., Ambudkar S. V., et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007; 315: 525–8
- Nagata K., Yamamoto A., Ban N., Tanaka A. R., Matsuo M., Kioka N., et al. Human ABCA3, a product of a responsible gene for abca3 for fatal surfactant deficiency in newborns, exhibits unique ATP hydrolysis activity and generates intracellular multilamellar vesicles. Biochem Biophys Res Commun 2004; 324: 262–8
- den Dunnen J. T., Antonarakis S. E. Nomenclature for the description of human sequence variations. Hum Genet 2001; 109: 121–4
Supplementary references
- den Dunnen J. T., Antonarakis S. E. Nomenclature for the description of human sequence variations. Hum Genet 2001; 109: 121–4
Electronic supplementary material
Supplementary Figure 1 The sequence surrounding intron‐15 length variation in the ABCA3 gene. The length variation is composed of four nearly identical 80‐bp repeats indicated by single (first and third repeats) or double underline (second and fourth repeats). The database SNP (single nucleotide polymorphism) rs323044 is written in lower case. Positions refer to the NCBI (National Center for Biotechnology Information) entry NT_037887.

Supplementary Table I. ABCA3 SNPs genotyped in 176 infants to determine the linkage disequilibrium structure for tagging SNP selection, and primers used in PCR and SNP extension.
Supplementary Table II. Genotypic association of rs13332514 to RDS and BPD in very premature infants.a