Abstract
Infants born before term (<37 weeks) have an increased risk of neonatal mortality as well as other health problems. The increasing rate of preterm birth in recent decades, despite improvements in health care, creates an impetus to better understand and prevent this disorder. Preterm birth likely depends on a number of interacting factors, including genetic, epigenetic, and environmental risk factors. Genetic studies may identify markers, which more accurately predict preterm birth than currently known risk factors, or novel proteins and/or pathways involved in the disorder. This review summarizes epidemiological and genetic studies to date, emphasizing the complexity of genetic influences on birth timing. While several candidate genes have been reportedly associated with the disorder, inconsistency across studies has been problematic. More systematic and unbiased genetic approaches are needed for future studies to examine the genetic etiology of human birth timing thoroughly.
Preterm birth is a major public health concern. In the United States, 12.7% of births occur before term (<37 weeks) Citation1. Infants born before term have an increased risk of neonatal mortality as well as serious health problems, such as respiratory illness, blindness, and cerebral palsy Citation2. Moreover, the severity and incidence of these problems worsen with decreasing gestational age Citation3. The impact of this disorder grows with the increasing rate of preterm birth in recent decades Citation2.
Although 20%–30% preterm births are medically indicated to avoid or minimize maternal and/or fetal complications, most preterm births result from spontaneous preterm labor or preterm premature rupture of membranes (PPROM) Citation3. While a number of risk factors have been identified, accurate prediction and prevention are difficult Citation3. For example, biomarkers of preterm birth, such as serum protein concentrations of interleukin (IL)‐6, IL‐8, tumor necrosis factor (TNF)‐α or relaxin, while strong predictors of preterm birth, may not be useful in large low‐risk populations Citation4. One problem may be that such markers vary over time or among individuals, making it difficult to determine the levels at which risk for preterm birth is increased. In contrast, genetic factors are stable over time and therefore may be better predictors of risk. As a result, genetic studies may identify markers which more accurately predict preterm birth than currently known risk factors. Genetic studies may also identify novel proteins and/or pathways involved in the disorder. This new information will augment our general understanding of parturition and provide new targets for drug therapies.
Key messages
A wealth of evidence suggests that genetics are important in birth timing.
A number of case‐control studies have identified potential candidate genes associated with preterm birth and birth timing; however, several statistical genetic concerns should be considered to improve the robustness of these findings in the future.
Systematic genome‐wide scans and other unbiased approaches may help identify novel genes involved in birth timing and preterm birth.
Evidence for influence of genetics on birth timing
Birth timing across pregnancies in the same woman
A wealth of evidence suggests that genetics are important in birth timing. For example, both preterm and postterm births tend to recur in mothers Citation5–10. Moreover, the most likely age for a recurrent preterm birth is same week as the first preterm birth Citation7, Citation11, Citation12, suggesting that factors that are stable over time, such as genetics, affect birth timing.
Birth timing trends among family members
Familial trends for birth timing also suggest that genetics influence this trait. Women who are born preterm are more likely to have had a preterm delivery themselves Citation13, indicating that mothers and their daughters share risk. Sisters of women who have had a preterm delivery also have an increased risk for preterm delivery Citation10. Due to the nature of family studies, environmental factors shared between mothers and daughters or between sisters cannot be untangled from genetic influences. As a result, it is difficult to determine the relative importance of genetic versus environmental factors from these studies alone.
Partitioning variance in birth timing into genetic versus environmental components
In contrast to family studies, twin studies measure the relative importance of genes in overall trait variance within a population. By comparing concordance rates between monozygotic and dizygotic twins, which share 100% and approximately 50% of their genes, respectively, one can model the genetic and environmental factors that influence a trait. Clausson and colleagues Citation14 estimated that genetics account for 36% and 31% of variation in preterm birth (<37 weeks) and gestational age as continuous trait, respectively. Another study estimated that genetics account for 27% of variation in preterm birth in any pregnancy and 17% in the first pregnancy Citation15. Based on these estimates, genetics play a significant role in preterm birth, in addition to environmental factors. However, the exact percentage of variation in birth timing due to genetics is not known, as both studies had small sample sizes from which estimates were made. Moreover, Treloar and colleagues Citation15 used ‘more than 2 weeks early’ as their phenotype, rather than the standard definition of preterm birth (<37 weeks). It is unclear how well this measure correlates with standard definitions and to what extent one can compare the results of Treloar et al. Citation15 to other studies.
A similar method was used to estimate the influence of maternal and fetal genetic factors by Lunde and colleagues Citation16. Comparing concordance rates among full and half siblings for gestational age, the authors estimated that 11% of variation for this trait is due to fetal genetic factors and 14% of variation is due to maternal genetic factors Citation16. Such comparisons use the degree of genetic relatedness (on average 50% for full siblings and 25% for half siblings) and trait concordance to estimate the relative importance of genetic versus environmental factors. Because siblings that are not monozygotic twins display some variability in their percent genetic identity and may differ in important dominance or interactive genetic effects, these estimates are more difficult to make using nontwin siblings. As a result, the estimates by Lunde et al. Citation16, in addition to those made by Clausson et al. Citation14 or Treloar et al. Citation15, probably do not accurately measure the amount of variance in preterm birth due to genetics. Despite the limitations in estimating the heritability, all three of these studies indicate that genetics play an important role in preterm birth.
Another approach to separating genetic and environmental factors is the coefficient of kinship. This measure depicts the degree of genetic relatedness within a population. Ward and colleagues Citation17 used this measure to examine genetic influences on preterm birth in a Utah population. The Utah population from which the families were drawn was established by 10,000 people who moved to the state to establish the Mormon religion Citation17. Because Mormons are discouraged from using alcohol or tobacco and have low rates of substance abuse and sexually transmitted diseases, this population may represent individuals with relatively few environmental risk factors for preterm birth Citation17. As a result, detecting genetic effects may be easier in this cohort of Utah preterm families. In this study, Ward and colleagues found that families with preterm deliveries had a significantly lower coefficient of kinship than controls Citation17, indicating that these families are more closely genetically related than control families. This evidence suggests that the increased rate of preterm birth in these families can be explained by genetic factors. It is important to note that the authors of this study did not report the relative abundance of any environmental risk factors for preterm birth in the two populations. It is possible that one or more important environmental risk factors differ between these groups, in addition to genetic relatedness. Hence, the results of Ward et al. Citation17 support the significance of genetics in preterm birth, but do not address their relative importance compared to known environmental risk factors.
Mendelian disorders
Certain Mendelian disorders are associated with preterm birth, further supporting genetic effects on birth timing. Ehlers‐Danlos syndrome [EDS] represents a diverse group of Mendelian disorders affecting connective tissue, primarily inherited in an autosomal dominant manner Citation18. Women with vascular EDS have an increased risk of delivering preterm, primarily due to PPROM Citation18. Since this disorder is inherited in an autosomal dominant pattern, there is a 50% chance the fetus has inherited the disorder, making it less clear whether the mother or infant genome contributes to the increase in PPROM risk.
Racial disparities in preterm birth
Ethnic differences in preterm birth rates also may be suggestive of genetic effects. Pregnancies in which either the mother Citation5, Citation8 or father Citation19 is black are at increased risk for preterm delivery. According to a study by Goldenberg and colleagues Citation20, these racial disparities are not explained entirely by measured environmental risk factors, implying that genetic differences among races may also contribute.
While the human population as a whole is genetically very similar, differing by 0.1% on average, important genetic differences exist across populations. Studies have found that approximately 10%–15% of total genetic variation in humans can be explained by differences between sub‐Saharan Africans, Northern Europeans, and East Asians Citation21. Moreover, specific polymorphisms can be used to group individuals based on ethnic groups with high accuracy Citation22. These results suggest that traditional racial designations parallel genetic differences to a certain extent, supporting the usefulness of race as an index of genetic composition.
Some investigators assert that only environmental and other nongenetic risk factors may account for the observed racial disparity in preterm birth rates Citation23. Indirect evidence for this hypothesis includes the observation that foreign‐born blacks and US‐born blacks differ in rates of preterm birth and low birth weight (often used as a proxy for gestational age). However, complex gene‐gene and gene‐environment interactions are not considered in this interpretation. For example, admixture in blacks, such that their genomes are partially African and partially European in ancestry, does not necessarily predict that Africans will have higher rates of preterm birth due to higher frequencies of predisposing alleles of African origin. Admixed blacks may have higher rates of preterm birth due to gene‐gene interactions that are only present when African‐ and European‐derived variation are combined into a single genome, or because of novel environmental risk factors that uncover important gene‐environment interactions not present in either US‐born whites or foreign‐born blacks. Other more subtle genetic effects, such as genetic variation affecting gene expression patterns, known to differ among races Citation24, may also explain observed differences in preterm birth rates. Hence, while environmental risk factors do significantly contribute to racial disparities, genetic effects on racial disparities in birth timing should not be discounted.
Additionally, studies comparing foreign‐born and US‐born blacks have rarely considered the race or birthplace of the father. Previous studies observed that preterm birth rates are higher whether the mother or father of a infant born preterm is black Citation25, suggesting that fetal race also influences birth timing. As a result, previous studies of black mothers may underestimate the role of genetics in such observed racial differences.
Modeling of genetic effects on birth timing
The complexity of genetic effects on birth timing cannot be sufficiently underscored. Genetic studies of preterm birth should not be thought of as a nature‐versus‐nurture debate, but rather an effort to characterize to how nature and nurture interact together to influence birth timing. There is increasing evidence that preterm birth can be conceptualized as a common, complex disorder. In contrast to Mendelian disorders, in which alterations of a single gene can lead to disease, complex diseases are influenced by a variety of factors, none necessary and sufficient to cause the disorder by itself. As a result, there is not a direct relationship between genotypes and phenotypes Citation26. These disorders likely depend on a number of interacting factors, including genetic, epigenetic, and environmental risk factors Citation26. Moreover, because environmental and epigenetic factors are also important, specific genes may be more influential in increasing risk in the context of particular environmental factors. For example, individuals with a particular genotype who also experienced high stress levels may be at higher risk for preterm birth than less stressed people of the same genotype. This has been demonstrated in relation to the development of major depression, whereby individuals carrying a particular version of the serotonin transporter gene showed an elevated risk of depression only in the context of stressful life events Citation27. Additionally, certain genes may be more important in increasing risk when found in combination with polymorphisms in other genes. For example, in testing several candidate genes for hypertension, Williams and colleagues Citation28 found no significant single site associations; however, a two‐locus model including polymorphisms in angiotensin I‐converting enzyme (ACE) and G protein‐coupled receptor kinase 4 (GRK4) was associated with the blood pressure phenotype studied. Hence, interactions between genes and/or the environment likely add complexity to genetic influences on birth timing.
A variety of evidence supports the complexity of preterm birth. Modeling procedures used by twin studies suggest that additive genetic factors and environmental risk factors that are not shared among siblings both influence preterm birth Citation14, Citation15. Additionally, interactions between genes Citation29, Citation30 have been associated with preterm birth risk. Several studies suggest that gene‐environment interactions, such as interactions between inflammatory gene risk alleles and bacterial infections Citation31–33, also influence the disorder. Together, these studies imply that the etiology of preterm birth likely involves genetic as well as environmental factors in complex interactions.
In addition to the complexity of genetic effects described above, several issues complicate how investigators think about the disorder. First, it is not clear whether the mother or the infant from a preterm delivery should be considered the proband. As a result, it is not clear which individual's DNA should be interrogated. Additionally, preterm delivery as a trait can be thought of in two ways. First, it can be thought of as discrete, resulting from genetic factors that lead to either disease or nondisease outcomes. Alternatively, gestational age can be thought of as a quantitative trait, with preterm ages as extremely low values. Consequently, genetic effects may be quantitative trait loci (QTLs) that influence the value of gestational age in both term and preterm deliveries. As disease models shape how one approaches identifying genes, it is important to consider the uncertainty about how to conceptualize preterm birth when evaluating various approaches taken to study this trait.
Specific human genes tested for association with preterm birth
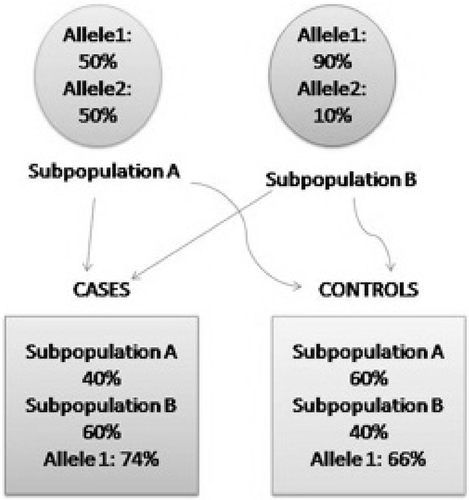
The majority of human genetic studies of birth timing to date have been case‐control comparisons. In such studies, polymorphism frequencies are compared in affected and unaffected individuals. If the polymorphism significantly differs in frequency across these groups, the polymorphism is said to influence risk of preterm birth. One advantage of using this study design is that it is relatively easy to recruit individual mothers and/or infants from a preterm delivery as cases, as opposed to the more extensive family recruitment needed for family‐based association or linkage tests. However, differences between case and control groups other than affection status, such as ethnic background, can create spurious associations. For example, case and control groups may come from two subpopulations that differ in allele frequencies for a particular locus. If the two groups do not have similar representations of the populations, an allele may be appear to be associated in the mixed sample, but does not actually contribute to disease. To illustrate, if allele 1 of a biallelic polymorphism was more common in subpopulation 1 and this subpopulation was overrepresented in the affected group, an association could be detected between allele 1 and the disease regardless of whether the gene was actually involved in the disorder ().
Figure 1 Illustration of population stratification in case‐control studies. Case and control groups come from two subpopulations, A and B, which differ in allele frequency for the polymorphism tested, 50% versus 90% for allele 1. If the case and control groups differ in the number of individuals coming from subpopulation A (40% versus 60%), this will create a difference in observed allele 1 frequencies between the groups (74% versus 66%) simply due to population substructure, rather than any relationship between allele 1 and disease status.
Candidate genes in a variety of pathways believed to be important in parturition have been assessed, such as inflammation and infection, stress response, functional loss of progesterone, remodeling of connective tissues, uterine contractility, and placental function (please see supplementary online materials for details).
Inflammation and infection
Many of the genetic studies of preterm birth to date have focused on genes involved in immunity and inflammation. Inflammation and infection are commonly associated with preterm birth. Women who deliver as a result of preterm labor have high rates of positive bacterial culture from amniotic fluid Citation34. Moreover, increases in inflammation factors may represent an early event in the transition to active labor in both term and preterm pregnancies Citation35. Less fulminant infections, such as bacterial vaginosis Citation36 and periodontal disease Citation37, all increase the risk for preterm birth. Whether these infections are due to microbes or variations in host response to identified microbes remains uncertain as prophylactic antibiotic treatment fails to alter the risk for preterm delivery Citation38–40. The parturition process at both term and preterm is associated with induction of many proinflammatory mediators, suggesting induction of these molecules as central components of the parturition cascade in humans Citation41.
Tumor necrosis factor‐α and receptors
The most frequently studied gene has been tumor necrosis factor‐α (TNF) a pro‐inflammatory cytokine. Many studies have tested the −308 polymorphism in the promoter of this gene, which affects expression levels, with mixed results. Several studies have found association between this polymorphism in mothers Citation32, Citation42, while others have not Citation30, Citation43–45. A meta‐analysis of seven studies of this polymorphism in mothers Citation46 found no association for this polymorphism, providing additional negative evidence for this gene. One study found that only mothers with PPROM showed association with −308, suggesting that heterogeneity in preterm birth phenotype may contribute to the varied results Citation47. Studies examining infants as affected also show mixed results Citation43, Citation48, Citation49.
Other polymorphisms in TNF have been tested for effects on preterm birth. While one study found association with another promoter polymorphism at −863 Citation43, another study examining multiple single nucleotide polymorphisms (SNPs) across the gene region found no association Citation45. One possible explanation for the lack of replication across studies may be that haplotypes of multiple polymorphisms may be more informative than individual polymorphisms. For example, Annells and colleagues Citation50 noted that the ‘−308, −238, +488’ AGG haplotype was associated with preterm birth less than 29 weeks, but none of the individual polymorphisms were significantly associated. Overall, it is not clear whether or not this gene plays a significant role in preterm birth without further study.
In addition to TNF, investigators have tested for its receptors for genetic effects on preterm birth. Menon and colleagues Citation45 found significant differences in allele frequencies between blacks and whites, and some suggestive association within blacks with both TNF receptors 1 (TNFR1) and 2 (TNFR2). A later study with the same population found significant association with a promoter polymorphism in TNFR1 and an intronic polymorphism in TNFR2Citation30, supporting the previous study's results. While these studies provide suggestive evidence for a role of TNF receptors in preterm birth, replication in other study populations would further solidify these findings.
Interleukins and their receptors
Interleukins form another class of inflammatory genes that have been studied extensively. To date, interleukins 1α Citation31, Citation50, Citation51, 1β Citation31, Citation42, Citation50, 2 Citation31, 8 Citation51, Citation52, 11 Citation51, 12A Citation51, and 18 Citation51 have shown no association with preterm birth. In contrast, interleukins 4, 6, and 10 have shown mixed results. Kalish and colleagues Citation53 found that homozygotes for a promoter polymorphism in interleukin 4 increased risk approximately 11‐fold over the corresponding low‐risk genotype. However, association with the opposite homozygous genotype Citation50 and no association with the gene Citation51 and preterm birth risk were reported in two other studies. Similarly, a few studies have reported main effects of interleukin 6 Citation54, Citation55 on preterm birth, but others failed to replicate this association Citation31, Citation33, Citation45, Citation50. Also, the ‘−1082, −819, −592’ ATA haplotype of interleukin 10 was associated in one study Citation50; however, this haplotype was not tested by others Citation51, Citation53. Neither of these studies Citation51, Citation53 reported association with the interleukin 10 gene for the markers that were interrogated.
Interleukin receptor genes also show mixed results. Interleukin 1 receptor 1 (IL1R1) Citation50 and interleukin 10 receptor A (IL10RA) Citation51 show no association with preterm birth. In contrast, Hao and colleagues Citation51 found association with interleukin 1 receptor 2 (IL1R2) in black mothers. Interleukin 1 receptor agonist (IL1RN) shows association with the allele 2 of an intronic variable number tandem repeat in some studies Citation56, Citation57, yet association with allele 1 Citation58, and no association Citation50, Citation51 in others. Similarly, one study of interleukin 6 receptor (IL6R) demonstrated association Citation33; however, no association was observed for this gene in an earlier study Citation30.
Other infection and inflammation‐related genes
Other infection and inflammation‐related genes have also been examined. The cytokine, CSF3, and IFNGR1, encoding the ligand‐binding chain alpha of the gamma interferon receptor, have not been associated with preterm birth Citation51. Additionally, TGFB1, which interacts with cytokine receptors Citation50, and ALOX5AP, involved in other aspects of the inflammation process Citation52, also have not been associated with the disorder. In contrast, LTA, lymphotoxin alpha, has shown mixed results. One study reported association with a promoter polymorphism Citation31, but two others did not Citation51, Citation52.
Pathogen recognition genes
Investigators have also examined genes involved in pathogen recognition. For example, MBL2 encodes a soluble mannose‐binding protein that recognizes mannose and N‐acetylglucosamine on bacterial pathogens. A promoter and two missense polymorphisms in MBL2 have been reportedly associated with preterm birth Citation50, Citation59. In addition, a member of the TNF superfamily, TNFRSF6, which encodes apoptosis (APO‐1) antigen 1 (FAS), member 6, was associated with preterm births limited to PPROM cases Citation60. An earlier study of TNFRSF6 including all spontaneous preterm births found no association Citation50. Another study Citation29 found that association with CD14, a surface protein preferentially expressed on monocytes and macrophages, was associated with PPROM cases only, in contrast to an earlier negative association with this gene Citation54. Additionally, two toll‐like receptors, TLR2 and TLR4, involved in pathogen recognition and activation of innate immunity, have been associated with preterm birth in some Citation61, Citation62 but not all Citation54, Citation63 studies analyzing their polymorphisms. Moreover, nucleotide‐binding oligomerization domain containing 2 (NOD2), which plays a role in the immune response to intracellular bacterial lipopolysaccharides, was associated with PPROM in black infants in one study Citation63, but was not associated with preterm birth in another study Citation54.
Remodeling of connective tissues
Physical remodeling of the cervix and fetal membranes are important steps in the overall normal delivery process and may represent an important control point in parturition. Indeed, preterm premature rupture of membranes is a common cause of preterm birth. Matrix metalloproteinases are responsible for degrading collagen structures and have been used as a biomarker for preterm birth. A promoter polymorphism in MMP1, matrix metalloproteinase 1, was found to be associated with PPROM in blacks Citation64. In another study of PPROM in blacks, a promoter polymorphism in MMP9, matrix metalloproteinase 9, increased risk for the disorder Citation65. SERPINH1, encoding the heat shock protein 47, is thought to be a molecular chaperone involved in the maturation of collagen molecules. As PPROM membranes have reduced collagen content, Wang and colleagues Citation66 sought to investigate this gene for association with PPROM. The authors found that a promoter polymorphism was associated with PPROM in a discovery as well as replication set Citation66. Additionally, Erichsen and colleagues Citation67 examined several genes involved in vitamin C absorption, solute carrier family 23 members 1 and 2, since this vitamin is required for collagen synthesis. One gene, SLC23A2, showed association, while another, SLC23A1, did not Citation67. PLAT, encoding plasminogen activator, tissue, also involved in tissue remodeling, was not associated with preterm birth Citation52.
Premature uterine contraction
The progression of the uterus from a quiescent tissue inhibited from contraction by a number or mediators, to a synchronously, forcefully contracting tissue characterizes the onset of active labor. For example, the β2‐adrenergic receptor (ADRB2) modulates uterine muscle contraction by promoting uterine smooth muscle relaxation. Studies to date have observed association of the Gly16Ala missense polymorphism in this gene Citation68, Citation69 with risk for preterm birth. A different missense polymorphism, Gln27Glu, in this gene showed association in one study Citation52, but not in two others Citation68, Citation69. Another neurotransmitter receptor involved in uterine contraction, dopamine receptor D2 (DRD2), was not associated with preterm birth Citation51. Prostaglandins also help promote uterine contractility. However, no associations were found for any of the prostaglandin pathway genes tested, prostaglandin E receptor 2 (PTGER2), prostaglandin E synthase (PTGES), or prostaglandin F receptor (PTGFR) Citation51. In contrast, phosphodiesterase 4D (PDE4D) degrades cAMP, a signaling molecule that promotes smooth muscle relaxation, but also showed no association with preterm birth Citation52.
Placental function
Placental functioning is critical for the maintenance of pregnancy and relies on appropriate blood flow between mother and fetus. Alteration in genes whose products are associated with formation of blood vessels or thrombosis may result in abnormal placental functioning and preterm birth. Vascular endothelial growth factor (VEGF) induces angiogenesis. One study observed an association between an intronic polymorphism in VEGF and preterm birth risk Citation70. Additionally, several hemostasis genes have been studied. One study found that factor V (F5) was significantly associated Citation51; however, two additional studies failed to replicate this result Citation52, Citation54. Factor VII (F7) was associated in both infants and mothers, and factor XIII (F13A1) was associated in infants only, in a study by Hartel and colleagues Citation71. A missense polymorphism in thrombomodulin (THBD) has been associated in infants as well Citation52. Other hemostasis genes assayed, factor II (F2) Citation52, Citation71, protein C receptor, endothelial (EPCR) Citation52, protein C (PROC) Citation71, and tissue factor pathway inhibitor (TFPI) Citation52, were not associated.
Some genes associated with blood pressure regulation have been associated with preterm birth risk. Hao and colleagues Citation51 found that one of two nitric oxide synthases tested, NOS2A, showed association with preterm birth in whites, but not the other, NOS3. In contrast, Gibson and colleagues Citation52 found association with both genes. Other genes linked to appropriate blood flow and pressure, annexin A5 (ANXA5), adducin 1 (ADD1), fibrinogen beta chain (FGB), serpin peptidase inhibitor, clade E member 1 (SERPINE1), and serpin peptidase inhibitor, clade B member 2 (SERPINB2), have not been associated with preterm birth Citation52.
Response to environmental toxins
Certain environmental toxins may play a role in birth timing by disrupting normal regulation of signaling pathways. For example, tobacco and other substance abuse by mothers increase the risk for preterm birth Citation34. Polymorphisms in genes responding to such compounds may make some mothers and/or infants more sensitive to their effects. PON1, paraoxonase 1, is a HDL‐C bound enzyme that metabolizes a variety of compounds, including drugs and pesticides Citation72. Lawlor and colleagues Citation72 tested a missense polymorphism associated with reduced activity and found a significant increase in risk that worsened with increasing number of risk alleles. Chen and colleagues Citation73 also found association with this polymorphism in PON1, as well as another missense polymorphism in paraoxonase 2, PON2, in Han Chinese infants who were born preterm. Additionally, OPRM1, opioid receptor mu 1, involved in the response to opioids, has been associated with preterm birth risk in Hispanics Citation51. In contrast, ADH1C, an enzyme involved in alcohol metabolism, did not show association Citation51. Several genes involved in the response to smoke exposure were examined by Nukui and colleagues Citation74 in both mothers and children from preterm deliveries. Of the polymorphisms tested, the glutathione S‐transferase theta 1, GSTT1, ‘null’ genotype was associated in mothers, but not infants Citation74. None of the other genes assayed, cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1), cytochrome P450, family 2, subfamily E, polypeptide 1 (CYP2E1), glutathione S‐transferase M1 (GSTM1), and N‐acetyltransferase 2 (NAT2), showed any association with preterm birth in mothers or children Citation74.
Fetal stress pathway
Stress has been thought to play an important role in parturition, based on data from animal studies. In many species, activation of the hypothalamic‐pituitary‐adrenal (HPA) axis, the endocrine stress pathway, with glucocorticoid production is a necessary prelude to progesterone withdrawal and active labor Citation75, Citation76. In humans, glucocorticoids do not precipitate labor, but corticotropin‐releasing hormone (CRH), a neuropeptide important in HPA axis control, is synthesized by the placenta Citation77. Circulating levels of CRH have been correlated with preterm and postterm pregnancy, depending upon whether they are above or below the normal range, respectively Citation78, Citation79. To date, only corticotropin‐releasing hormone‐binding protein (CRHBP) has been tested for association with preterm birth in humans and was not associated with the disorder Citation51.
Functional loss of progesterone
Progesterone withdrawal is extremely important in nonprimate parturition. Progesterone withdrawal, however, does not occur in human pregnancy. It has been postulated that functional loss of progesterone may initiate labor in primates. Altered function of PGR, encoding the progesterone receptor, may play a role of functional loss of progesterone human parturition. However, PGR was not associated with preterm birth in Hao et al. Citation51. Alterations in progesterone receptor coactivators and corepressors have been implicated as mediating progesterone resistance; however, specific genes have not been studied for variants predisposing to preterm birth.
Other
Folate is required for DNA replication and metabolism. Insufficient dietary intake of folate during pregnancy has important impacts on infant health. Functional changes in genes involved in folate metabolism may lower levels of important intermediates in DNA replication and metabolism, affecting the fetus in the same manner as reduced intake of folate. To illustrate, missense polymorphisms in two such genes, 5‐methyltetrahydrofolate‐homocysteine methyltransferase reductase (MTRR) and serine hydroxymethyltransferase 1 (SHMT1), were associated with increased risk of preterm birth in the white subset of the cohort studied by Engel et al. Citation80. Several other folate metabolism genes, 5,10‐methylenetetrahydrofolate reductase (MTHFR) Citation52, Citation80, Citation81, 5‐methyltetrahydrofolate‐homocysteine methyltransferase (MTR) Citation80 and cystathionine‐beta‐synthase (CBS) Citation80, were not associated with the disorder.
Peroxisome proliferator‐activated receptor gamma (PPARG) is a transcription factor in the nuclear hormone receptor subfamily involved in adipose tissue differentiation. This gene has been associated with type 2 diabetes and heart disease for which preterm infants are at increased risk of developing later in life Citation82. Carriers of the Ala12 allele of a Pro12Ala missense polymorphism had an increased risk for preterm birth in one study Citation82, possibly explaining the correlation observed between these disorders.
Genetic, environmental interactions, and preterm birth
Several studies suggest that gene‐gene interactions, in addition to gene main effects, influence preterm birth. A study of CD14 found both main effects for the gene and interactive effects with HSP‐70‐2 and with ILRNCitation29. In contrast, a study found interactive effects for TNFα×IL‐6×IL6R, but no main effects for any of the genes Citation30. As a result, epistasis analyses may identify genes that would not otherwise be detected.
Interactions between inflammatory gene risk alleles and bacterial infections have also been shown to influence preterm birth. Two studies of IL6 found association with the gene and disorder only in the presence of bacterial vaginosis Citation31 or microbial invasion of the intra‐amniotic cavity Citation33. Hence considering gene‐gene and gene‐environment interactions may detect genes without main effects in some instances. In contrast, Macones and colleagues Citation32 found that TNFα had both a main effect and an interactive effect with bacterial vaginosis on preterm birth risk. A gene involved in folate metabolism, SHMT1, also showed both main and interactive effects with dietary folate intake in black women Citation80. As a result, many of the genes for which main effects have been detected may also interact with environmental factors to further increase risk for preterm birth.
Some polymorphisms may only increase risk in the context of other genetic polymorphisms or certain environmental factors; as a result, considering polymorphisms in multiple genes or gene‐environment interactions may increase our power to detect genetic effects. However, large sample sizes are required in order to detect such effects with confidence. Because studies to date have been relatively small, such observed interactions are tentative. Further tests with larger sample sizes are needed to verify these observations.
Limitations of previous studies
Genes studied to date biased by current understanding of physiology
The majority of human genetic studies to date have focused on one or a few candidate genes, chosen based on our current understanding of physiology. A few studies have taken a larger‐scale approach to candidate genes; however, no genes have been identified by unbiased approaches yet. One of the many benefits of systematic genetic screens is elucidating pathways that would not otherwise have been considered important for a particular disease. For example, evidence from linkage and genome‐wide association screens identified transcription factor 7‐like 2 (TCF7L2), a transcription factor believed to be involved in blood glucose homeostasis, located on chromosome 10q, as a risk factor for type 2 diabetes Citation83, Citation84. While this gene's function has a plausible role in diabetes pathology, it probably would not have been identified as a candidate gene by physiology. The association between TCF7L2 and type 2 diabetes has been replicated across numerous studies and populations, demonstrated by a global meta‐analysis Citation85, underscoring the ability of unbiased approaches to identify important novel genes. Moreover, pleiotropic effects of TCF7L2 on birth weight in addition to diabetes have been demonstrated Citation86, further signifying the utility of such approaches. Hence, systematic genome‐wide scans may help identify novel genes involved in birth timing and preterm birth.
Range of genetic variation within a gene has not been sampled sufficiently
Moreover, the lack of sufficient coverage of genetic variation within genes tested to date has been problematic. Many of the studies described in this review have genotyped one functional SNP in a particular gene. If this particular polymorphism is itself disease‐promoting, or is tightly correlated with an unsampled variant in the gene, then an association may be detected. However, additional variants may contribute to disease that are unrepresented in these studies, creating false negatives. An ideal study design would consider patterns of linkage disequilibrium (LD), i.e. the degree of correlation among polymorphisms in a region, such that each known variant in the gene is either tested directly or represented by other polymorphisms highly correlated to it. The studies that have chosen one or a few polymorphisms to test, rather than sampling the entire LD structure of the gene, have usually focused on polymorphisms in the promoter region or nonsynonymous coding changes. While such polymorphisms are highly likely to affect protein expression and/or function, other types of changes have been tied to disease as well. Synonymous coding changes can be disease‐promoting. For example, a Gly (GGC) to Gly (GGT) mutation in the calpain 3 (LGMD2A) gene alters mRNA splicing, introducing a frameshift that is thought to lead to limb girdle muscular dystrophy Citation87. Similarly, subtle changes in introns may promote disease by altering splicing patterns Citation88. Changes in the 3′ untranslated region may also be related to disease by altering mRNA stability and/or translation efficiency, such as by altering microRNA binding sites Citation89. As a result, a range of polymorphism types may be involved in birth timing. Because the majority of studies to date have not genotyped candidate genes thoroughly, studies with negative association results may represent false negatives. Hence, genes with negative association but incomplete gene coverage cannot be excluded from involvement in preterm birth without further study.
Heterogeneity within samples studied
Genetic heterogeneity within cases of preterm birth may also contribute to the difficulty to detect genes influencing this disorder. There are likely a variety of genetic pathways that lead to preterm birth. Genetic vulnerability factors influencing the risk for the disorder might fall into several broad categories: infection and/or inflammation, response to stress, deficits in placental function, and premature uterine contractions. Additionally as preterm birth is associated with a variety of other disorders, including type 2 diabetes and heart disease Citation82, some genes may have pleiotropic effects on other traits. Factors from one or more of these categories might contribute to risk in an individual, such that a sample of preterm birth cases may represent numerous genetic pathways important in the disorder. Hence, heterogeneity within the sample may reduce the power to detect a particular gene because it is important in only a subset of affected individuals. It follows that essential for gene identification will be careful, consistent phenotyping to minimize heterogeneity of cases.
Replication issues
Replication across studies is an important validation for any detected genetic association. Few positive association findings for preterm birth have been consistent. To illustrate, for TNFα, the most extensively studied gene, seven studies report major effects of the gene Citation32, Citation42, Citation43, Citation47–50; yet, five others report no major effects of the gene Citation30, Citation31, Citation44–46, including a meta‐analysis of seven studies Citation46. Similarly, IL1RN was associated in three studies Citation56–58, but not in another two Citation50, Citation51. One possible explanation may be that some polymorphisms are significant only in the context of a particular environmental factor, such as infection. For example, IL6 has been associated in two studies without considering environmental influences Citation54, Citation55, associated in another two studies only in the context of infection Citation31, Citation33, and not associated in an additional two studies Citation30, Citation50. Also, there has been little consistency about case or control exclusion criteria across studies, such that phenotypic and genetic heterogeneity may exist across studies. For example, preterm birth cases with PPROM have been inconsistently included in samples studied to date; however, some studies have identified association only in PPROM subsets of their samples Citation47, Citation60, suggesting that these individuals may represent a distinct genetic pathway from other cases.
Associations replicated across populations are considered particularly robust. However, such replications have been difficult to assess in preterm birth studies to date because many consist of mothers and/or children from more than one ethnic group. Few studies have included analyses separately by race or attempted to correct for possible population substructure. As a result, it is difficult to determine whether any association detected in mixed samples is due to a particularly strong signal in one ethnic group or an equally strong signal in multiple groups. In fact, the majority of positive association findings to date have been in mixed samples. Because any significant deviation between case and control groups on ethnic composition could create false positive results, the need for replication across studies is greater. A mixture of ethnic backgrounds may also reduce power to detect a gene that affects the disorder in one ethnic group but not others, possibly creating false‐negative results. Thus, future studies should consider the racial compositions of their samples carefully, in light of these issues.
Sample size and power to detect small effects
Another potential problem with studies to date has been sample size. A study's ability to detect a genetic effect is intimately related to sample size. Smaller sample sizes will only be able to detect strong genetic effects, while larger samples may detect much smaller effects. For example, assuming a discrete trait model, a sample size of 100 cases and 100 controls tested at markers with minor allele frequencies between 0.15–0.35 only has ⩾80% power at α = 0.05 to detect genetic factors with relative risks ⩾3 (calculated with Citation90). Because of the complex nature of genetic influences on preterm birth, it is difficult to determine a priori whether genes of small or large effect size are important in this disorder. It may be the case that very large sample sizes are needed to detect genes of interest for this disorder.
Comment
Overall, a wealth of evidence suggests that genetics are important in birth timing. Epidemiological studies demonstrate that mothers, sisters, and daughters share risk for preterm birth. Twin and other studies separating genetic and environmental factors indicate that genetics influence birth timing and preterm birth, in addition to environmental risk factors. A number of case‐control studies have identified potential candidate genes associated with preterm birth and birth timing; however, several statistical genetic concerns should be considered to improve the robustness of these findings in the future.
A variety of alternative genetic approaches may be undertaken to identify specific genes involved in preterm birth. For example, unbiased genome‐wide screens using either linkage (e.g. affected sib pairs, variance component QTL) or association methods may be used. The ease and relatively low cost of genotypic microarray platforms make whole‐genome association scans an attractive option. Both case‐control and family‐based association methods, in which population stratification is not an issue, could be used for genome‐wide scans. Additionally, nonadditive genetic effects, such as copy number or structural variation, may be important avenues for future research. Such approaches may enable investigators to identify novel genes and pathways involved in birth timing, with important clinical applications.
References
- Hamilton B. E., Minino A. M., Martin J. A., Kochanek K. D., Strobino D. M., Guyer B. Annual summary of vital statistics: 2005. Pediatrics 2007; 119: 345–60
- Esplin M. S. Preterm birth: a review of genetic factors and future directions for genetic study. Obstet Gynecol Surv 2006; 61: 800–6
- Green N. S., Damus K., Simpson J. L., Iams J., Reece E. A., Hobel C. J., et al. Research agenda for preterm birth: recommendations from the March of Dimes. Am J Obstet Gynecol 2005; 193: 626–35
- Goldenberg R. L., Goepfert A. R., Ramsey P. S. Biochemical markers for the prediction of preterm birth. Am J Obstet Gynecol 2005; 192((5 Suppl))S36–46
- Adams M. M., Elam‐Evans L. D., Wilson H. G., Gilbertz D. A. Rates of and factors associated with recurrence of preterm delivery. JAMA 2000; 283: 1591–6
- Basso O., Olsen J., Christensen K. Study of environmental, social, and paternal factors in preterm delivery using sibs and half sibs. A population‐based study in Denmark. J Epidemiol Community Health 1999; 53: 20–3
- Kistka Z. A., Palomar L., Boslaugh S. E., DeBaun M. R., DeFranco E. A., Muglia L. J. Risk for postterm delivery after previous postterm delivery. Am J Obstet Gynecol 2007; 196: 241.e1–6
- Kistka Z. A., Palomar L., Lee K. A., Boslaugh S. E., Wangler M. F., Cole F. S., et al. Racial disparity in the frequency of recurrence of preterm birth. Am J Obstet Gynecol 2007; 196: 131.e1–6
- Mercer B. M., Goldenberg R. L., Moawad A. H., Meis P. J., Iams J. D., Das A. F., et al. The preterm prediction study: effect of gestational age and cause of preterm birth on subsequent obstetric outcome. National Institute of Child Health and Human Development Maternal‐Fetal Medicine Units Network. Am J Obstet Gynecol 1999; 181: 1216–21
- Winkvist A., Mogren I., Hogberg U. Familial patterns in birth characteristics: impact on individual and population risks. Int J Epidemiol 1998; 27: 248–54
- Bloom S. L., Yost N. P., McIntire D. D., Leveno K. J. Recurrence of preterm birth in singleton and twin pregnancies. Obstet Gynecol 2001; 98: 379–85
- Melve K. K., Skjaerven R., Gjessing H. K., Oyen N. Recurrence of gestational age in sibships: implications for perinatal mortality. Am J Epidemiol 1999; 150: 756–62
- Porter T. F., Fraser A. M., Hunter C. Y., Ward R. H., Varner M. W. The risk of preterm birth across generations. Obstet Gynecol 1997; 90: 63–7
- Clausson B., Lichtenstein P., Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG 2000; 107: 375–81
- Treloar S. A., Macones G. A., Mitchell L. E., Martin N. G. Genetic influences on premature parturition in an Australian twin sample. Twin Res 2000; 3: 80–2
- Lunde A., Melve K. K., Gjessing H. K., Skjaerven R., Irgens L. M. Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population‐based parent‐offspring data. Am J Epidemiol 2007; 165: 734–41
- Ward K., Argyle V., Meade M., Nelson L. The heritability of preterm delivery. Obstet Gynecol 2005; 106: 1235–9
- Volkov N., Nisenblat V., Ohel G., Gonen R. Ehlers‐Danlos syndrome: insights on obstetric aspects. Obstet Gynecol Surv 2007; 62: 51–7
- Palomar L., DeFranco E. A., Lee K. A., Allsworth J. E., Muglia L. J. Paternal race is a risk factor for preterm birth. Am J Obstet Gynecol 2007; 197: 152.e1–7
- Goldenberg R. L., Cliver S. P., Mulvihill F. X., Hickey C. A., Hoffman H. J., Klerman L. V., et al. Medical, psychosocial, and behavioral risk factors do not explain the increased risk for low birth weight among black women. Am J Obstet Gynecol 1996; 175: 1317–24
- Bamshad M. Genetic influences on health: does race matter?. JAMA 2005; 294: 937–46
- Allocco D. J., Song Q., Gibbons G. H., Ramoni M. F., Kohane I. S. Geography and genography: prediction of continental origin using randomly selected single nucleotide polymorphisms. BMC Genomics 2007; 8: 68
- David R., Collins J Jr. Disparities in Infant Mortality: What's Genetics Got to Do With It?. Am J Public Health 2007; 97: 1191–7
- Spielman R. S., Bastone L. A., Burdick J. T., Morley M., Ewens W. J., Cheung V. G. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 2007; 39: 226–31
- Tan H., Wen S. W., Walker M., Demissie K. Parental race, birth weight, gestational age, and fetal growth among twin infants in the United States. Early Hum Dev 2004; 80: 153–60
- Joober R., Sengupta S., Boksa P. Genetics of developmental psychiatric disorders: pathways to discovery. J Psychiatry Neurosci 2005; 30: 349–54
- Caspi A., Sugden K., Moffitt T. E., Taylor A., Craig I. W., Harrington H., et al. Influence of life stress on depression: moderation by a polymorphism in the 5‐HTT gene. Science 2003; 301: 386–9
- Williams S. M., Ritchie M. D., Phillips J. A 3rd., Dawson E., Prince M., Dzhura E., et al. Multilocus Analysis of Hypertension: A Hierarchical Approach. Hum Hered 2004; 57: 28–38
- Kalish R. B., Vardhana S., Normand N. J., Gupta M., Witkin S. S. Association of a maternal CD14‐159 gene polymorphism with preterm premature rupture of membranes and spontaneous preterm birth in multi‐fetal pregnancies. J Reprod Immunol 2006; 70: 109–17
- Menon R., Velez D. R., Simhan H., Ryckman K., Jiang L., Thorsen P., et al. Multilocus interactions at maternal tumor necrosis factor‐alpha, tumor necrosis factor receptors, interleukin‐6 and interleukin‐6 receptor genes predict spontaneous preterm labor in European‐American women. Am J Obstet Gynecol 2006; 194: 1616–24
- Engel S. A., Erichsen H. C., Savitz D. A., Thorp J., Chanock S. J., Olshan A. F. Risk of spontaneous preterm birth is associated with common proinflammatory cytokine polymorphisms. Epidemiology 2005; 16: 469–77
- Macones G. A., Parry S., Elkousy M., Clothier B., Ural S. H., Strauss J. F 3rd. A polymorphism in the promoter region of TNF and bacterial vaginosis: preliminary evidence of gene‐environment interaction in the etiology of spontaneous preterm birth. Am J Obstet Gynecol 2004; 190: 1504–8
- Velez D. R., Menon R., Thorsen P., Jiang L., Simhan H., Morgan N., et al. Ethnic differences in interleukin 6 (IL‐6) and IL6 receptor genes in spontaneous preterm birth and effects on amniotic fluid protein levels. Ann Hum Genet 2007; 71: 586–600
- DeFranco E., Teramo K., Muglia L. Genetic influences on preterm birth. Semin Reprod Med 2007; 25: 40–51
- Smith R. Parturition. N Engl J Med 2007; 356: 271–83
- Leitich H., Bodner‐Adler B., Brunbauer M., Kaider A., Egarter C., Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: a meta‐analysis. Am J Obstet Gynecol 2003; 189: 139–47
- Khader Y. S., Ta'ani Q. Periodontal diseases and the risk of preterm birth and low birth weight: a meta‐analysis. J Periodontol 2005; 76: 161–5
- Carey J. C., Klebanoff M. A., Hauth J. C., Hillier S. L., Thom E. A., Ernest J. M., et al. Metronidazole to Prevent Preterm Delivery in Pregnant Women with Asymptomatic Bacterial Vaginosis. N Engl J Med 2000; 342: 534–40
- Lamont R. F. Can antibiotics prevent preterm birth; the pro and con debate. BJOG 2005; 112(suppl 1)67–73
- McDonald H., Brocklehurst P., Parsons J. Antibiotics for treating bacterial vaginosis in pregnancy. Cochrane Database Syst Rev 2005, CD000262
- Keelan J. A., Blumenstein M., Helliwell R. J., Sato T. A., Marvin K. W., Mitchell M. D. Cytokines, prostaglandins and parturition—a review. Placenta 2003; 24: S33–46
- Moore S., Ide M., Randhawa M., Walker J. J., Reid J. G., Simpson N. A. An investigation into the association among preterm birth, cytokine gene polymorphisms and periodontal disease. BJOG 2004; 111: 125–32
- Amory J. H., Adams K. M., Lin M. T., Hansen J. A., Eschenbach D. A., Hitti J. Adverse outcomes after preterm labor are associated with tumor necrosis factor‐alpha polymorphism −863, but not −308, in mother‐infant pairs. Am J Obstet Gynecol 2004; 191: 1362–7
- Dizon‐Townson D. S., Major H., Varner M., Ward K. A promoter mutation that increases transcription of the tumor necrosis factor‐alpha gene is not associated with preterm delivery. Am J Obstet Gynecol 1997; 177: 810–3
- Menon R., Velez D. R., Thorsen P., Vogel I., Jacobsson B., Williams S. M., et al. Ethnic differences in key candidate genes for spontaneous preterm birth: TNF‐alpha and its receptors. Hum Hered 2006; 62: 107–18
- Menon R., Merialdi M., Betran A. P., Dolan S., Jiang L., Fortunato S. J., et al. Analysis of association between maternal tumor necrosis factor‐alpha promoter polymorphism (−308), tumor necrosis factor concentration, and preterm birth. Am J Obstet Gynecol 2006; 195: 1240–8
- Roberts A. K., Monzon‐Bordonaba F., Van Deerlin P. G., Holder J., Macones G. A., Morgan M. A., et al. Association of polymorphism within the promoter of the tumor necrosis factor alpha gene with increased risk of preterm premature rupture of the fetal membranes. Am J Obstet Gynecol 1999; 180: 1297–302
- Aidoo M., McElroy P. D., Kolczak M. S., Terlouw D. J., ter Kuile F. O., Nahlen B., et al. Tumor necrosis factor‐alpha promoter variant 2 (TNF2) is associated with pre‐term delivery, infant mortality, and malaria morbidity in western Kenya: Asembo Bay Cohort Project IX. Genet Epidemiol 2001; 21: 201–11
- Chen D., Hu Y., Wu B., Chen L., Fang Z., Yang F., et al. Tumor necrosis factor‐alpha gene G308A polymorphism is associated with the risk of preterm delivery. Beijing Da Xue Xue Bao 2003; 35: 377–81
- Annells M. F., Hart P. H., Mullighan C. G., Heatley S. L., Robinson J. S., Bardy P., et al. Interleukins−1, −4, −6, −10, tumor necrosis factor, transforming growth factor‐beta, FAS, and mannose‐binding protein C gene polymorphisms in Australian women: Risk of preterm birth. Am J Obstet Gynecol 2004; 191: 2056–67
- Hao K., Wang X., Niu T., Xu X., Li A., Chang W., et al. A candidate gene association study on preterm delivery: application of high‐throughput genotyping technology and advanced statistical methods. Hum Mol Genet 2004; 13: 683–91
- Gibson C. S., MacLennan A. H., Dekker G. A., Goldwater P. N., Dambrosia J. M., Munroe D. J., et al. Genetic Polymorphisms and Spontaneous Preterm Birth. Obstet Gynecol 2007; 109: 384–91
- Kalish R. B., Vardhana S., Gupta M., Perni S. C., Witkin S. S. Interleukin‐4 and ‐10 gene polymorphisms and spontaneous preterm birth in multifetal gestations. Am J Obstet Gynecol 2004; 190: 702–6
- Hartel C., Finas D., Ahrens P., Kattner E., Schaible T., Muller D., et al. Polymorphisms of genes involved in innate immunity: association with preterm delivery. Mol Hum Reprod 2004; 10: 911–5
- Simhan H. N., Krohn M. A., Roberts J. M., Zeevi A., Caritis S. N. Interleukin‐6 promoter −174 polymorphism and spontaneous preterm birth. Am J Obstet Gynecol 2003; 189: 915–8
- Murtha A. P., Nieves A., Hauser E. R., Swamy G. K., Yonish B. A., Sinclair T. R., et al. Association of maternal IL‐1 receptor antagonist intron 2 gene polymorphism and preterm birth. Am J Obstet Gynecol 2006; 195: 1249–53
- Witkin S. S., Vardhana S., Yih M., Doh K., Bongiovanni A. M., Gerber S. Polymorphism in intron 2 of the fetal interleukin‐1 receptor antagonist genotype influences midtrimester amniotic fluid concentrations of interleukin‐1beta and interleukin‐1 receptor antagonist and pregnancy outcome. Am J Obstet Gynecol 2003; 189: 1413–7
- Genc M. R., Onderdonk A. B., Vardhana S., Delaney M. L., Norwitz E. R., Tuomala R. E., et al. Polymorphism in intron 2 of the interleukin‐1 receptor antagonist gene, local midtrimester cytokine response to vaginal flora, and subsequent preterm birth. Am J Obstet Gynecol 2004; 191: 1324–30
- Bodamer O. A., Mitterer G., Maurer W., Pollak A., Mueller M. W., Schmidt W. M. Evidence for an association between mannose‐binding lectin 2 (MBL2) gene polymorphisms and pre‐term birth. Genet Med 2006; 8: 518–24
- Kalish R. B., Nguyen D. P., Vardhana S., Gupta M., Perni S. C., Witkin S. S. A single nucleotide A>G polymorphism at position −670 in the Fas gene promoter: relationship to preterm premature rupture of fetal membranes in multifetal pregnancies. Am J Obstet Gynecol 2005; 192: 208–12
- Krediet T. G., Wiertsema S. P., Vossers M. J., Hoeks S. B., Fleer A., Ruven H. J., et al. Toll‐like Receptor 2 Polymorphism Is Associated With Preterm Birth. Pediatr Res 2007; 62: 474–6
- Lorenz E., Hallman M., Marttila R., Haataja R., Schwartz D. A. Association between the Asp299Gly polymorphisms in the Toll‐like receptor 4 and premature births in the Finnish population. Pediatr Res 2002; 52: 373–6
- Ferrand P. E., Fujimoto T., Chennathukuzhi V., Parry S., Macones G. A., Sammel M., et al. The CARD15 2936insC mutation and TLR4 896 A>G polymorphism in African Americans and risk of preterm premature rupture of membranes (PPROM). Mol Hum Reprod 2002; 8: 1031–4
- Fujimoto T., Parry S., Urbanek M., Sammel M., Macones G., Kuivaniemi H., et al. A single nucleotide polymorphism in the matrix metalloproteinase‐1 (MMP‐1) promoter influences amnion cell MMP‐1 expression and risk for preterm premature rupture of the fetal membranes. J Biol Chem 2002; 277: 6296–302
- Ferrand P. E., Parry S., Sammel M., Macones G. A., Kuivaniemi H., Romero R., et al. A polymorphism in the matrix metalloproteinase‐9 promoter is associated with increased risk of preterm premature rupture of membranes in African Americans. Mol Hum Reprod 2002; 8: 494–501
- Wang H., Parry S., Macones G., Sammel M. D., Kuivaniemi H., Tromp G., et al. A functional SNP in the promoter of the SERPINH1 gene increases risk of preterm premature rupture of membranes in African Americans. Proc Natl Acad Sci U S A 2006; 103: 13463–7
- Erichsen H. C., Engel S. A., Eck P. K., Welch R., Yeager M., Levine M., et al. Genetic variation in the sodium‐dependent vitamin C transporters, SLC23A1, and SLC23A2 and risk for preterm delivery. Am J Epidemiol 2006; 163: 245–54
- Doh K., Sziller I., Vardhana S., Kovacs E., Papp Z., Witkin S. S. Beta2‐adrenergic receptor gene polymorphisms and pregnancy outcome. J Perinat Med 2004; 32: 413–7
- Landau R., Xie H. G., Dishy V., Stein C. M., Wood A. J., Emala C. W., et al. beta2‐Adrenergic receptor genotype and preterm delivery. Am J Obstet Gynecol 2002; 187: 1294–8
- Papazoglou D., Galazios G., Koukourakis M. I., Kontomanolis E. N., Maltezos E. Association of −634G/C and 936C/T polymorphisms of the vascular endothelial growth factor with spontaneous preterm delivery. Acta Obstet Gynecol Scand 2004; 83: 461–5
- Hartel C., von Otte S., Koch J., Ahrens P., Kattner E., Segerer H., et al. Polymorphisms of haemostasis genes as risk factors for preterm delivery. Thromb Haemost 2005; 94: 88–92
- Lawlor D. A., Gaunt T. R., Hinks L. J., Davey Smith G., Timpson N., Day I. N., et al. The association of the PON1 Q192R polymorphism with complications and outcomes of pregnancy: findings from the British Women's Heart and Health cohort study. Paediatr Perinat Epidemiol 2006; 20: 244–50
- Chen D., Hu Y., Chen C., Yang F., Fang Z., Wang L., et al. Polymorphisms of the paraoxonase gene and risk of preterm delivery. Epidemiology 2004; 1: 466–70
- Nukui T., Day R. D., Sims C. S., Ness R. B., Romkes M. Maternal/newborn GSTT1 null genotype contributes to risk of preterm, low birthweight infants. Pharmacogenetics 2004; 14: 569–76
- Challis J. R. G., Brooks A. N. Maturation and activation of hypothalamic‐pituitary‐adrenal function in fetal sheep. Endocr Rev 1989; 10: 182–204
- Challis J. R. G., Lye S. J. Parturition. The Physiology of Reproduction, E Knobil, J. D Neill. Raven Press, Ltd., New York 1994; 985–1031
- Frim D. F., Emanuel R. L., Robinson B. G., Smas C. F., Adler G. K., Majzoub J. A. Characterization and gestational regulation of preprocorticotropin releasing hormone messenger RNA in human placenta. J Clin Invest 1988; 82: 287–92
- Majzoub J. A., McGregor J. A., Lockwood C. J., Smith R., Taggart M. S., Schulkin J. A central theory of preterm and term labor: putative role for corticotropin‐releasing hormone. Am J Obstet Gynecol 1999; 180: S232–41
- McLean M., Bisits A., Davies J., Woods R., Lowry P., Smith R. A placental clock controlling the length of human pregnancy. Nat Med 1995; 1: 460–3
- Engel S. M., Olshan A. F., Siega‐Riz A. M., Savitz D. A., Chanock S. J. Polymorphisms in folate metabolizing genes and risk for spontaneous preterm and small‐for‐gestational age birth. Am J Obstet Gynecol 2006; 195: 1231.e1–11
- Lauszus F. F., Gron P. L., Klebe J. G. Association of polymorphism of methylene‐tetrahydro‐folate‐reductase with urinary albumin excretion rate in type 1 diabetes mellitus but not with preeclampsia, retinopathy, and preterm delivery. Acta Obstet Gynecol Scand 2001; 80: 803–6
- Meirhaeghe A., Boreham C. A., Murray L. J., Richard F., Davey Smith G., Young I. S., et al. A possible role for the PPARG Pro12Ala polymorphism in preterm birth. Diabetes 2007; 56: 494–8
- Duggirala R., Blangero J., Almasy L., Dyer T. D., Williams K. L., Leach R. J., et al. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 1999; 64: 1127–40
- Grant S. F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., et al. Variant of transcription factor 7‐like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006; 38: 320–3
- Cauchi S., El Achhab Y., Choquet H., Dina C., Krempler F., Weitgasser R., et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta‐analysis. J Mol Med 2007; 85: 777–82
- Freathy R. M., Weedon M. N., Bennett A., Hypponen E., Relton C. L., Knight B., et al. Type 2 diabetes TCF7L2 risk genotypes alter birth weight: a study of 24,053 individuals. Am J Hum Genet 2007; 80: 1150–61
- Richard I., Beckmann J. S. How neutral are synonymous codon mutations?. Nat Genet 1995; 10: 259
- Takahara K., Schwarze U., Imamura Y., Hoffman G. G., Toriello H., Smith L. T., et al. Order of intron removal influences multiple splice outcomes, including a two‐exon skip, in a COL5A1 acceptor‐site mutation that results in abnormal pro‐alpha1(V) N‐propeptides and Ehlers‐Danlos syndrome type I. Am J Hum Genet 2002; 71: 451–65
- Abelson J. F., Kwan K. Y., O'Roak B. J., Baek D. Y., Stillman A. A., Morgan T. M., et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science 2005; 310: 317–20
- Purcell S., Cherny S. S., Sham P. C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003; 19: 149–50