Abstract
Lactase non-persistence (adult-type hypolactasia) is present in more than half of the human population and is caused by the down-regulation of lactase enzyme activity during childhood. Congenital lactase deficiency (CLD) is a rare severe gastrointestinal disorder of new-borns enriched in the Finnish population. Both lactase deficiencies are autosomal recessive traits and characterized by diminished expression of lactase activity in the intestine. Genetic variants underlying both forms have been identified. Here we review the current understanding of the molecular defects of human lactase deficiencies and their phenotype-genotype correlation, the implications on clinical practice, and the understanding of their function and role in human evolution.
Introduction
Two forms of primary lactase deficiencies are known in humans. Lactase non-persistence (adult-type hypolactasia; primary lactose malabsorption; MIM#223100) is a normal, developmental phenomenon characterized by the down-regulation of lactase enzyme activity after weaning Citation[1]. Mutations have occurred in human history that have caused less than half of the human populations to retain high lactase activity throughout life (lactase persistence; LP). Lactase persistence is most common in Northern European populations and in some nomadic populations Citation[1–4]. It is a general theory that these populations have gained selected advantage for survival from the ability to digest milk lactose and thus from the increase of nutrient intake. The congenital form of lactase deficiency (CLD; OMIM#223000) is a severe gastrointestinal disorder of new-borns first described by Holzel Citation[5]. Most cases have been reported from Finland Citation[6–8]. Based on family studies, both adult-type hypolactasia and CLD are inherited in an autosomal recessive manner Citation[6–9]. Here we provide a concise review on molecular genetic findings in these two human lactase deficiencies and discuss their phenotype-genotype correlation, functional significance, and impact on diagnostics and human evolution.
Identification of the variants
The lactase gene has been localized to 2q21-22 Citation[10], and its structure has been characterized Citation[11], Citation[12]. The lactase polypeptide contains four (I–IV) conserved regions, of which regions III and IV remain in the mature lactase enzyme after posttranslational processing Citation[11]. The protein has two catalytically active sites: phlorizin hydrolase activity located in region III and lactase activity in region IV Citation[13], Citation[14].
Key messages
Congenital lactase deficiency (CLD) and adult-type hypolactasia, both with low lactase enzyme activity, are molecularly distinct conditions and represent strikingly different phenotypes.
Low lactase activity in both lactase deficiencies proposed the lactase gene (LCT) as an excellent candidate gene for molecular genetic studies. Consequently, polymorphic microsatellite markers in the LCT region were analyzed in a family-based approach using linkage, linkage disequilibrium (LD), and haplotype analysis. As a result, both traits were assigned outside the LCT gene on 2q21-22 Citation[7], Citation[15]. Sequencing of the lactase gene resulted in the identification of five distinct mutations in a total of 32 Finnish CLD patients from 24 families. Of them, a nonsense mutation Y1390X in exon 9 of the LCT gene, was present in 90% of affected chromosomes (), compatible with the founder effect . The highest carrier frequency of 1:35 was demonstrated in the late settlement region of Finland in Northern Savo Citation[8], compatible with genealogical studies Citation[6]. The rest of the mutations included a four-base-pair deletion S1666fsX1722 in exon 14, a two-nucleotide deletion S218fsX224 in exon 2, and missense mutations Q268H in exon 3 and G1363S in exon 9. Each mutation occurred in a different haplotype background, suggesting a diverse genetic origin. Interestingly, two of the mutations had occurred in the lactase persistence haplotype (T-13910 allele) and three in the non-persistence haplotype (C-13910) (discussed below). Recently, four new mutations underlying CLD have been identified Citation[16]. Two new substitutions were identified in a patient of Italian origin: c.2062T > C in exon 7 leading to a substitution of serine to proline (S688P) and c.4834G > T in exon 12 leading to glutamic acid to change to a premature stop codon (E1612X). The mutations found in Finnish patients were deletion of five bases c.1692-1696delAGTGG in exon 6 leading to a frameshift mutation V565fsX567 and a substitution c.4760G→A in exon 12 which changes arginine to histidine (R1587H). Two of the non-Finnish patients with Turkish origin were homozygous for the missense mutation G1363S in exon 9, present in a Finnish family, suggesting that this mutation has arrived Finland from the East. The mutations were located both in the pro-region and the regions that determine phlorizin hydrolase and lactase activity ().
Abbreviations
Figure 1. Schematic presentation of the locations of DNA variants of adult-type hypolactasia and the mutations underlying congenital lactase deficiency (CLD). DNA variants in bold in exon 13 of the minichromosome maintenance type 6 gene (MCM6) have been confirmed to have an effect on lactase activity by functional studies.
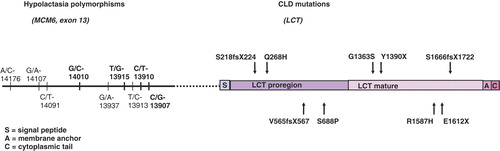
Figure 2. The approximate location of the most common DNA variants associated with lactase persistence and showing functional evidence Citation[15], Citation[18], Citation[21], Citation[22], Citation[24–26], Citation[30–32].
![Figure 2. The approximate location of the most common DNA variants associated with lactase persistence and showing functional evidence Citation[15], Citation[18], Citation[21], Citation[22], Citation[24–26], Citation[30–32].](/cms/asset/7bc9f960-578e-49a3-af4d-b874e0e1722d/iann_a_412276_f0002_b.jpg)
For the lactase persistence/non-persistence trait an analogous approach was implemented in nine Finnish families where the phenotype had been determined using a lactose tolerance test (LTT) with ethanol. Availability of the biochemical test results Citation[9] facilitated linkage analyses and the ascertainment of the haplotypes associated with lactase persistence versus non-persistence in the analyzed families with multiple generations. Although lactase persistence is very common (>80%) in the Finnish population, an enrichment of one major haplotype could be detected in 60% of the chromosomes in persistent subjects, suggesting a common variant associated with the trait. This also reflects the low recombination rate at 2q21-22. Sequence analysis of the SNPs in the core haplotype region between markers D2S3014 and D2S3015 among different phenotypic groups resulted in the identification of a single nucleotide polymorphism (SNP) C to T (rs4988235) that resided 13,910 base pairs upstream of the LCT gene in exon 13 of the minichromosome maintenance type 6 gene (MCM6) at 2q21. The linkage result could be confirmed by correlating the C/T-13910 genotypes with lactase activity and the lactase and sucrase (L/S) ratio in a case control study Citation[15] as well as in other studies Citation[17–20]. The original form, the C/C-13910 genotype, was associated with low lactase activity, whereas the genotypes containing the mutant allele (C/T-13910 and T/T-13910) were associated with high lactase activity. The frequency of the C/C-13910 genotype has been shown to be in agreement with previously published figures of adult-type hypolactasia in over 40 populations from four continents Citation[18], Citation[21–25], Citation[24–26]. Another SNP, G/A-22018, associated with a lactose tolerance test and residing 8 kb telomeric from C/T-13910, has been excluded as a functional variant based on incomplete association with lactase activity Citation[18] and on functional studies Citation[27–29].
Several single nucleotide polymorphisms (SNPs) in the immediate vicinity of the C/T-13910 in African and Arab populations have recently been identified () Citation[26], Citation[30–32]. Of them, the G/C-14010 variant residing 100 bp from the C/T-13910 variant is found in small pastoralist groups from Kenya and Tanzania and has shown significant association with lactase persistence using the indirect lactose tolerance test (LTT) () Citation[32]. The T/G-13915 variant has been found in Bedouin populations in Saudi Arabia and Jordan as well as in pastoralist and non-pastoralist groups from Cameroon, Ethiopia, Kenya, and Sudan. This variant has been shown to be the founder allele on the Arabian peninsula that perfectly correlates with lactase activity and the L/S ratio Citation[30]. The C-13907 has been found in the Afro-Asiatic Beja population in Kenya, Sudan, and Ethiopia Citation[31], Citation[32]. In addition, a rare polymorphism, T/C-13913, has been reported from some cases in Cameroon, Sudan, Ethiopia, and the Bedouin population in Saudi Arabia Citation[31]. Functional studies using a dual-luciferase reporter assay have shown that the mutant C-14010, G-13907, and G-13915 alleles increase the LCT promoter expression Citation[32]. Although many of the African variants are rare due to the small sample size, their localization near the C/T-13910 variant suggests that lactase persistence is caused by many variants in a distant enhancer region in intron 13 of the MCM6 gene.
Table I. The lactase persistence/non-persistence variants and their correlation with biochemical studies Citation[15], Citation[26], Citation[30–32].
Phenotype-genotype correlation
It is notable that lactose intolerance is not absolute in lactase non-persistence. Earlier studies have shown that an average of 250 mL whole milk is tolerated daily in lactase non-persistence Citation[33], Citation[34]. In a recent study in the Swedish population, 76% of subjects with the C/C-13910 genotype were milk drinkers compared to 90% of subjects with C/T-13910 or T/T-13910 genotypes (P < 0.01) Citation[35]. There is evidence that a natural aversion to milk is developed by subjects with lactase non-persistence Citation[36], already during childhood Citation[18]. Overall the phenotypic manifestations of lactase non-persistence are mild: flatulence, bloating, diarrhea, and abdominal pain after ingestion of dairy products containing lactose. In a recent study, 9% of adult Finnish patients from primary health care with the C/C-13910 genotype were subjectively symptomless Citation[36]. In contrast, the phenotype of CLD is strikingly different: watery diarrhea after ingestion of lactose-containing milk, leading to dehydration, acidosis, and hospital care during the first days or weeks of life Citation[6]. In CLD, a lactose-free diet alleviates the symptoms rapidly. All mutations so far identified in the LCT gene in CLD, independent of the type or location, have caused a similar phenotype Citation[8]. These data show that mutations in the coding region of LCT result in a severe disease phenotype manifested as CLD, whereas symptoms are mild or absent in the lactase non-persistence that is a normal developmental trait.
Correlation of lactase activities in lactase persistence/non-persistence genotypes shows a trimodal distribution Citation[15], Citation[17–20], Citation[30]. The mean level of lactase activity with lactase non-persistence genotypes (C/C-13910; G/G-13915) varied from 4.8–6.9 U/g protein. In CLD representing a life-threatening diarrhea, lactase activity is overlapping with the values of adult-type hypolactasia (lactase 0–7 U/g protein in CLD) Citation[6], Citation[8], Citation[16]. The weak effect of an enhancer mutation was demonstrated in a 6-year-old patient who carried the lactase-persistent genotype C/T-13910 but had exceptionally low lactase activity (6 U/g protein) Citation[19]. The reason for the low lactase activity was the founder mutation of CLD, Y1390X, in the other chromosome. The relative expression of the allele carrying the premature stop codon in LCT was at the same level as the transcript of the C-13910 allele, representing about 8% of the wild-type LCT mRNA Citation[17].
The two phenotypes with low lactase activity suggest a different regulatory mechanism of lactase activity in these conditions. Several studies have shown that lactase persistence alleles 14 kb upstream of the LCT gene regulate lactase activity at transcriptional level Citation[17], Citation[19], Citation[27], Citation[28], Citation[31], Citation[32]. In vitro studies have demonstrated an enhancer effect in lactase promoter activity by the persistent alleles compared to non-persistent alleles Citation[27], Citation[31], Citation[32]. Oct-1 transcription factor has been found to bind with higher affinity to the T-13910 variant than to the C-13910 variant site. Co-expression of Oct-1, binding the T-13910 enhancer region, with hepatic nuclear factor 1alpha (HNF1α), which has a binding site at the proximal lactase promoter, increased the effect of the T-13910 enhancer more than 133-fold over the proximal promoter activity. Using DN Aase footprint and supershift analyses binding sites for GATA transcription factor 6 (GATA-6), HNF4α, Fox/HNF3α, and Cdx-2 transcription factors, previously reported to affect lactase gene expression, were detected and characterized within the -13910 region . Mutation in any of these sites—Oct-1, GATA-6, HNF4α, or Fox/HNF3α—abolished the activity of the T-13910 enhancer, demonstrating that the presence of all of the above-mentioned binding sites is necessary for the C/T-13910 region to possess enhancer activity. It was also shown that the -13910 enhancer did not activate the MCM6 promoter in a transfection experiment Citation[29].
The mechanisms for regulation of lactase levels during normal fetal development and in infants are incompletely understood. Lactase activity is low around 26 gestational weeks but increases 3–4-fold towards term. Few data exist on the functional activity of intestinal lactase in premature babies. We have shown that lactase activity declines during childhood due to the diminishing transcription of the lactase non-persistence allele Citation[19]. However, the molecular mechanism that induces the natural down-regulation of the non-persistence alleles remains to be clarified.
Genetic testing in clinical practice
Milk containing lactose is the major nutrient and source of energy of new-born infants. Later in life, dairy products form an essential part of human diet in many cultures.
In CLD the diagnosis is based ‘at the bedside’ on clinical symptoms and Clinitest® (www.utmb.edu/Clinitest) in the acute phase, followed by determination of disaccharidase activities in an intestinal biopsy specimen at the age of 6–12 months. In diseases like CLD with sudden appearance of symptoms, genetic testing could serve as a rapid and patient-friendly diagnostic tool. However, to find all mutations, sequencing of the whole LCT gene would be required. Surprisingly, nine novel CLD cases were found in Finland in 2006, suggesting that CLD is more common than previously thought Citation[16]. Correct diagnosis and follow-up are important in CLD to ensure normal growth of the new-born Citation[37]. For these patients lactose-free formulas (e.g. Nutramigen®, Pepti-Junior®, Profylac®) are the right choice.
For the diagnosis of adult-type hypolactasia several methods have been developed to detect the C/T-13910 genotype, including minisequencing Citation[15], Citation[18], enzyme digest Citation[38], polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)-genotyping Citation[39], and pyrosequencing Citation[40], Citation[41]. Sequencing is the most reliable method to detect all variants known so far Citation[31], Citation[32] in patients originating from multi-ethnic populations. The age should be taken into account in the interpretation of the results in children Citation[18], Citation[19].
Poor correlation of abdominal symptoms with the level of lactase activity has made clinical diagnosis of adult-type hypolactasia a challenge. By definition, the diagnosis is based on the measurement of lactase, sucrase, and maltase activities and on the lactase and sucrase (L/S) ratio in intestinal biopsies Citation[42]. This technique needs expertise Citation[43], Citation[44], is tedious to the patient, expensive, and thus not suitable for primary screening of abdominal complaints. The indirect lactose tolerance test (LTT) or the breath hydrogen test (BHT) lack both specificity and sensitivity Citation[42], Citation[45], especially in children Citation[46], leading to both false positive and negative results Citation[47], Citation[48]. There is evidence that the breath hydrogen test may be an indicator of bacterial overgrowth rather than lactose malabsorption Citation[49]. Further, a H2 non-excretion is relatively common, ranging from 5%–18%, reducing the applicability of the breath hydrogen test Citation[50]. Ethanol has been used with LTT to increase the specificity of the test, but it is not suitable for testing of children and adolescents.
Evolutionary aspects
Based on the clinical features of human lactase deficiencies, CLD may have been under negative selection. This would explain the rarity of CLD to date. An enrichment of CLD in the isolated Finnish population has most likely occurred by chance due to a founder effect and genetic drift. It came to our attention in the 1960s along with the rapid development of modern pediatrics together with the invention of invasive biopsy techniques. In contrast to CLD, lactase non-persistence is not a fatal condition but a normal developmental trait. From the evolutionary point of view it is, however, understandable: down-regulation of lactase activity is a normal phenomenon that is not meant to be fatal but actually facilitate an increase in the numbers of offspring. It is still intriguing to speculate why some lactase non-persistence subjects whose lactase activity is as low as in CLD have no symptoms from milk Citation[6], Citation[8], Citation[18–20]. This difference can be explained by a reduced amount of lactose ingested per weight. However, transcription factors regulating lactase expression during childhood may also play a role, the details of the changes in the intestine remain unknown .
Positive selection for lactase persistence in both European and African populations before the DNA era has been suggested based on our ability to utilize dairy products Citation[51], Citation[52]. Extended haplotype surrounding the C/T-13910 variant has supported the model of strong positive selection Citation[21]. The selection has been calculated to be a recent event, compatible with the invention of agriculture some 7,000–10,000 years ago Citation[4], Citation[51–53].
Selection acts on the phenotype, not on the genotype Citation[54]. Based on clinical findings the phenotype alone does not unambiguously support positive selection. A significant proportion of non-persistent individuals tolerate some milk Citation[33], Citation[34] or experience only minor symptoms. Abdominal symptoms from dairy products are at least nowadays relatively common also among lactase-persistent subjects Citation[34], Citation[55–58]. Farming and agriculture were based on permanent settlement enabling an increase in family size that was the starting-point for modern civilization. It can be speculated that cultural evolution is faster than genetic evolution, and it has acted as a positive selecting factor. The study by Burger and co-authors provides some molecular evidence for this as they showed that the T-13910 allele was rare among neolithic Europeans Citation[59]. A permanent life-style has been especially important in populations living in extreme conditions where a high prevalence of lactase persistence variants has been enriched (early neolithic Europeans, nomadic populations in Arabia and Africa, as well as in some Fenno-Ugric populations in the Ural region) Citation[30], Citation[32], Citation[53]. In these conditions, the hypothesis that milk products and the ability to digest lactose have a positive effect on health has been presented, but the reasons for this have remained controversial. Several studies with methods of varying sensitivity and specificity have assessed the absorption of calcium from milk in lactase-persistent and -deficient subjects Citation[60–64], and its effect on bone health, but the results have mainly remained controversial Citation[65–69].
Conclusions
Identification of the variants in lactase persistence/non-persistence and congenital lactase deficiency provides a comprehensive example of human traits and diseases that are important in human evolution. Identification of the mutations underlying CLD and the severe diarrhea in new-borns confirm the essential role of the LCT gene in early human growth and development. The identification of polymorphisms in adult-type hypolactasia was facilitated by haplotype analysis of the region covering an obvious candidate gene, LCT, in multi-generational families and, most importantly, by demonstrating the trimodal distribution of lactase activity with the C/T-13910 genotypes and the age-related decline of lactase mRNA expression driven by the C-13910 allele. Identification of the molecule(s) that naturally reduce the expression of the LCT gene at different stages of development may give new insights into biological behavior and the evolutionary forces underlying lactase regulation in humans.
Acknowledgements
This work was funded by the Academy of Finland, the Sigrid Jusélius Foundation, Helsinki, Finland, and the Helsinki University Hospital, Research Funding (TYH6204), Helsinki, Finland. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Sahi T. Hypolactasia and lactase persistence. Historical review and the terminology. Scand J Gastroenterol Suppl. 1994; 202: 1–6
- Cook GC, al-Torki MT. High intestinal lactase concentrations in adult Arabs in Saudi Arabia. BMJ. 1975; 3: 135–6
- Flatz G, Rotthauwe HW. The human lactase polymorphism: physiology and genetics of lactose absorption and malabsorption. Prog Med Genet. 1977; 2: 205–49
- Simoons FJ. The geographic hypothesis and lactose malabsorption. A weighing of the evidence. Am J Dig Dis. 1978; 23: 963–80
- Holzel A. Sugar malabsorption due to deficiencies of disaccharidase activities and of monosaccharide transport. Arch Dis Child. 1967; 42: 341–52
- Savilahti E, Launiala K, Kuitunen P. Congenital lactase deficiency. A clinical study on 16 patients. Arch Dis Child. 1983; 58: 246–52
- Järvelä I, Enattah NS, Kokkonen J, Varilo T, Savilahti E, Peltonen L. Assignment of the locus for congenital lactase deficiency to 2q21, in the vicinity of but separate from the lactase-phlorizin hydrolase gene. Am J Hum Genet. 1998; 63: 1078–85
- Kuokkanen M, Kokkonen J, Enattah NS, Ylisaukko-Oja T, Komu H, Varilo T, et al. Mutations in the translated region of the lactase gene (LCT) underlie congenital lactase deficiency. Am J Hum Genet. 2006; 78: 339–44
- Sahi T, Isokoski M, Jussila J, Launiala K. Recessive inheritance of adult-type lactose malabsorption. Lancet. 1973; 2: 823–6
- Harvey CB, Fox MF, Jeggo PA, Mantei N, Povey S, Swallow DM. Regional localization of the lactase-phlorizin hydrolase gene, LCT, to chromosome 2q21. Ann Hum Genet. 1993; 57: 179–85
- Mantei N, Villa M, Enzler T, Wacker H, Boll W, James P, et al. Complete primary structure of human and rabbit lactase-phlorizin hydrolase: implications for biosynthesis, membrane anchoring and evolution of the enzyme. EMBO J. 1998; 7: 2705–13
- Boll W, Wagner P, Mantei N. Structure of the chromosomal gene and cDNAs coding for lactase-phlorizin hydrolase in humans with adult-type hypolactasia or persistence of lactase. Am J Hum Genet. 1991; 48: 889–902
- Zecca L, Mesonero JE, Stutz A, Poirée JC, Giudicelli J, Cursio R, et al. Intestinal lactase-phlorizin hydrolase (LPH): the two catalytic sites; the role of the pancreas in pro-LPH maturation. FEBS Lett. 1998; 435: 225–8
- Arribas JC, Herrero AG, Martin-Lomas M, Canada FJ, He S, Withers SG. Differential mechanism-based labeling and unequivocal activity assignment of the two active sites of intestinal lactase/phlorizin hydrolase. Eur J Biochem. 2000; 267: 6996–7005
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002; 30: 233–7
- Torniainen S, Freddara R, Routi T, Gijsbers C, Catassi C, Höglund P, et al. Four novel mutations in the lactase gene underlying congenital lactase deficiency (CLD). BMC Gastroenterol. 2009; 9: 8
- Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Järvelä I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003; 52: 647–52
- Rasinperä H, Savilahti E, Enattah NS, Kuokkanen M, Tötterman N, Lindahl H, et al. A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut. 2004; 53: 157–8
- Rasinperä H, Kuokkanen M, Kolho KL, Lindahl H, Enattah NS, Savilahti E, et al. Transcriptional downregulation of the lactase (LCT) gene during childhood. Gut. 2005; 54: 1660–1
- Enattah NS, Kuokkanen M, Forsblom C, Natah S, Oksanen A, Jarvela I, et al. Correlation of intestinal disaccharidase activities with the C/T(-13910) variant and age. World J Gastroenterol. 2007; 13: 3508–12
- Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004; 76: 1111–20
- Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, et al. T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet. 2004; 74: 1102–10
- Rasinperä, H, Forsblom, C, Enattah, NS, Halonen, P, Salo, K, Victorzon, M, , FinnDiane Study Group, et al. The C/C-13910 genotype of adult-type hypolactasia is associated with an increased risk of colorectal cancer in the Finnish population. Gut. 2005;54:643–7
- Myles S, Bouzekri N, Haverfield E, Cherkaoui M, Dugoujon JM, Ward R. Genetic evidence in support of a shared Eurasian-North African dairying origin. Hum Genet. 2005; 17: 34–42
- Borinskaia SA, Rebrikova DV, Nefedova VV, Kofiadi IA, Sokolova MV, Kolchina EV, et al. Molecular diagnosis and frequencies of primary hypolactasia in populations of Russia and neighboring countries. Mol Biol (Mosk) 2006; 40(6)1031–6
- Torniainen, S, Parker, MI, Holmberg, V, Lahtela, E, Dandara, C, Järvelä, I. Screening of variants for lactase persistence/non-persistence in populations from South Africa and Ghana. BMC Genetics. 2009; 10: 31
- Troelsen JT, Olsen J, Møller J, Sjöström H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003; 125: 1686–94
- Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003; 12: 2333–40
- Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, Troelsen JT. The T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet. 2005; 14: 3945–53
- Imtiaz F, Savilahti E, Sarnesto A, Trabzuni D, Al-Kahtani K, Kagevi I, et al. T/G-13915 variant upstream of the lactase gene (LCT) is the founder allele of adult-type hypolactasia in an urban Saudi population. J Med Genet. 2007; 44: e89
- Ingram CJE, Elamin FF, Mulcare CA, Weale ME, Tarekegn A, Raga TO, et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence?. Hum Genet. 2007; 120: 779–88
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007; 39: 31–40
- Newcomer AD, McGill DB. Clinical importance of lactase deficiency. N Eng J Med. 1984; 310: 42–3
- Suarez FL, Savaiano DA, Levitt MD. A comparison of symptoms after the consumption of milk or lactose-hydrolyzed milk by people with self-reported severe lactose intolerance. N Engl J Med. 1995; 333: 1–4
- Torniainen S, Hedelin M, Autio V, Rasinperä H, Bälter KA, Klint A, et al. Lactase persistence, dietary intake of milk and risk of prostate cancer in Sweden and Finland. Cancer Epidemiol Biomarkers Prev. 2007; 16: 956–61
- Anthoni SR, Rasinperä HA, Kotamies A, Pihlajamäki H, Komu HA, Kolho KL, et al. Molecularly defined adult-type hypolactasia among working age people with reference to milk consumption and gastrointestinal symptoms. World J Gastroenterol. 2007; 13: 1230–5
- Paganus A, Juntunen-Backman K, Savilahti E. Follow-up of nutritional status and dietary survey in children with cow's milk allergy. Acta Paediatr Scand. 1992; 81: 518–23
- Buning C, Ockenga J, Kruger S, Jurga J, Baier P, Dignass A, et al. The C/C (-13910) and G/G (-22018) genotypes for adult-type hypolactasia are not associated with inflammatory bowel disease. Scand J Gastroenterol. 2003; 38: 538–42
- Chao CK, Sibley E. PCR-RFLP genotyping assay for a lactase persistence polymorphism upstream of the lactase-phlorizin hydrolase gene. Genet Test. 2004; 8: 190–3
- Nilsson TK, Johansson CA. A novel method for diagnosis of adult hypolactasia by genotyping of the −13910 C/T polymorphism with Pyrosequencing technology. Scand J Gastroenterol. 2004; 39: 287–90
- Ridefelt P, Håkansson LD. Lactose intolerance: lactose tolerance test versus genotyping. Scand J Gastroenterol. 2005; 40(7)822–6
- Arola H. Diagnosis of hypolactasia and lactose malabsorption. Scand J Gastroenterol Suppl. 1994; 202: 26–35
- Langman JM, Rowland R. Activity of duodenal disaccharidases in relation to normal and abnormal mucosal morphology. J Clin Pathol. 1990; 43: 537–40
- Heitlinger LA, Rossi TM, Ping-Cheung L, Lebenthal E. Human intestinal disaccharidase activities: Correlations with age, biopsy technique, and degree of villus atrophy. J Pediatr Gastroenterol Nutr. 1991; 12: 204–8
- Waud JP, Matthews SB, Campbell AK. Measurement of breath hydrogen and methane, together with lactase genotype, defines the current best practice for investigation of lactose sensitivity. Ann Clin Bioch. 45 Pt 2008; 1: 50–8
- Krasilnikoff PA, Gudmand-Hoyer E, Moltke HH. Diagnostic value of disaccharide tolerance tests in children. Acta Paediatr Scand. 1975; 64: 693–8
- Högenauer C, Hammer HF, Mellitzer K, Renner W, Krejs GJ, Toplak H. Evaluation of a new DNA test compared with the lactose hydrogen breath test for the diagnosis of lactase non-persistence. Eur J Gastroenterol Hepatol. 2005; 17: 371–6
- Krawczyk M, Wolska M, Schwartz S, Gruenhage F, Terjung B, Portincasa P, et al. Concordance of genetic and breath tests for lactose intolerance in a tertiary referral centre. J Gastrointestin Liver Dis. 2008; 17: 135–9
- Pimentel M, Kong Y, Park S. Breath testing to evaluate lactose intolerance in irritable bowel syndrome correlates with lactulose testing and may not reflect true lactose malabsorption. Am J Gastroenterol. 2003; 98: 2700–4
- Hammer HF, Petritsch W, Pristautz H, Krejs GJ. Assessment of the influence of hydrogen nonexcretion on the usefulness of the hydrogen breath test and lactose tolerance test. Wien Klin Wochenschr. 1996; 108: 137–41
- Simoons FJ. Primary adult lactose intolerance and the milking habit: a problem in biologic and cultural interrelations. II. A culture historical hypothesis. Am J Dig Dis. 1970; 15: 695–710
- McCracken MD. Lactase deficiency: An example of dietary evolution. Current Anthropol. 1971; 12: 479–517
- Enattah NS, Trudeau A, Pimenoff V, Maiuri L, Auricchio S, Greco L, et al. Evidence for still ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet. 2007; 81: 615–25
- Hartl, DL, Clark, AG. Principles of population genetics3rd ed. Sunderland, Massachusetts, Sinauer Associates, Inc. 1997.
- Carroccio A, Montalto G, Cavera G, Notarbatolo A. Lactose intolerance and self-reported milk intolerance: relationship with lactose maldigestion and nutrient intake. J Am Coll Nutr. 1998; 17: 631–6
- Saltzman JR, Russell RM, Golner B, Barakat S, Dallal GE, Goldin BR. A randomized trial of Lactobacillus acidophilus BG2FO4 to treat lactose intolerance. Am J Clin Nutr. 1999; 69: 140–6
- Vesa TH, Korpela RA, Sahi T. Tolerance to small amounts of lactose in lactose maldigesters. Am J Clin Nutr. 1996; 64: 197–201
- de Vrese, M, Stegelmann, A, Richter, B, Fenselau, S, Laue, C, Schrezenmeir, J. Probiotics—compensation for lactase insufficiency. Am J Clin Nutr. 2001; 73(Suppl 2)421–9.
- Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase persistence allele in early Neolithic Europeans. PNAS. 2007; 104: 3736–41
- Debongnie JC, Newcomer AD, McGill DB, Phillips SF. Absorption of nutrients in lactase deficiency. Dig Dis Sci. 1979; 24: 225–31
- Cochet B, Jung A, Griessen M, Bartholdi P, Schaller P, Donath A. Effects of lactose on intestinal calcium absorption in normal and lactase-deficient subjects. Gastroenterology. 1983; 84: 935–40
- Zittermann A, Bock P, Drummer C, Scheld K, Heer M, Stehle P. Lactose does not enhance calcium bioavailability in lactose-tolerant, healthy adults. Am J Clin Nutr. 2000; 71: 931–6
- Tremaine WJ, Newcomer AD, Riggs BL, McGill DB. Calcium absorption from milk in lactase-deficient and lactase-sufficient adults. Dig Dis Sci. 1986; 31: 376–8
- Kocian J, Skala I, Bakos K. Calcium absorption from milk and lactose-free milk in healthy subjects and patients with lactose intolerance. Digestion. 1973; 9: 317–24
- Black RE, Williams SM, Jones IE, Goulding A. Children who avoid drinking cow milk have low dietary calcium intakes and poor bone health. Am J Clin Nutr. 2002; 76: 675–80
- Honkanen R, Kroger H, Alhava E, Turpeinen P, Tuppurainen M, Saarikoski S. Lactose intolerance associated with fractures of weight-bearing bones in Finnish women aged 38–57 years. Bone. 1997; 21: 473–7
- Goulding A, Taylor RW, Keil D, Gold E, Lewis-Barned NJ, Williams SM. Lactose malabsorption and rate of bone loss in older women. Age Ageing. 1999; 28: 175–80
- Kalkwarf HJ, Khoury JC, Lanphear BP. Milk intake during childhood and adolescence, adult bone density, and osteoporotic fractures in US women. Am J Clin Nutr. 2003; 77: 257–65
- Enattah NS, Sulkava R, Halonen P, Kontula K, Jarvela I. Genetic variant of lactase-persistent C/T-13910 is associated with bone fractures in very old age. J Am Geriatr Soc. 2005; 53: 79–82