ABSTRACT
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of cells with potent immunosuppressive functions, which can inhibit the activation of immune responses under a steady-state condition and pathological conditions. We performed transcriptomic profiling of circulating CD33+HLA-DR+ myeloid antigen-presenting cells (APCs) and CD33+HLA-DR– myeloid cells (potentially MDSCs) in healthy individuals. We sorted both subpopulations from peripheral blood mononuclear cells (PBMCs) of 10 healthy donors and performed RNA sequencing (RNA-Seq). We found that several signaling pathways associated with the positive regulation of immune responses, such as antigen presentation/processing, FcγR-mediated phagocytosis and immune cell trafficking, phosphoinositide 3-kinase (PI3K)/Akt signaling, DC maturation, triggering receptor expressed on myeloid cells 1 (TREM1) signaling, nuclear factor of activated T cells (NFAT) and IL-8 signaling were downregulated in CD33+HLA-DR– myeloid cells. In contrast, pathways implicated in tumor suppression and anti-inflammation, including peroxisome proliferator-activated receptor (PPAR) and phosphatase and tensin homolog (PTEN), were upregulated in CD33+HLA-DR– myeloid cells. These data indicate that PPAR/PTEN axis could be upregulated in myeloid cells to keep the immune system in check in normal physiological conditions. Our data reveal some of the molecular and functional differences between CD33+HLA-DR+ APCs and CD33+HLA-DR– myeloid cells in a steady-state condition, reflecting the potential suppressive function of CD33+HLA-DR– myeloid cells to maintain immune tolerance. For future studies, the same methodological approach could be applied to perform transcriptomic profiling of myeloid subsets in pathological conditions.
Introduction
Immunosuppressive myeloid cells have been implicated in many pathological conditions, such as age-associated inflammation, autoimmune diseases, infections, organ transplantation, trauma, and cancer (Gabrilovich and Nagaraj Citation2009; Kirkwood et al. Citation2018). Myelopoiesis is disrupted during these pathological conditions, leading to increased levels of a heterogeneous myeloid cell population, which comprises cells halted at various stages of maturation/differentiation with a potent immunosuppressive activity, referred to as myeloid-derived suppressor cells (MDSCs) (Awad et al. Citation2018; Gabrilovich and Nagaraj Citation2009). MDSCs have a role in maintaining immune tolerance and circulate at very low levels in healthy individuals (Khaled et al. Citation2013). In contrast, MDSCs circulate at higher levels in disease contexts, for instance, cancer, and their numbers increase by 10-folds (Khaled et al. Citation2013).
There are multiple mechanisms by which MDSCs contribute to immunosuppression; they express immunosuppressive enzymes such as indoleamine 2,3-dioxygenase (IDO) and arginase-1 (ARG1); produce transforming growth factor-β (TGF-β), and express inhibitory immune checkpoint ligands including programmed death-ligand 1 (PD-L1) (Gabrilovich and Nagaraj Citation2009; Khaled et al. Citation2013; Saleh and Elkord Citation2019). These mechanisms in turn cause dysfunction of antigen-presenting cells (APCs) and reduction in T effector cell (Teff) activation/proliferation, and enhance T regulatory cell (Treg) recruitment, survival and function (Bernal-Estevez et al. Citation2018; Gabrilovich and Nagaraj Citation2009; Lindau et al. Citation2013; Schlecker et al. Citation2012). On the other hand, expression of major histocompatibility complex class II (MHC II, encoded by human leukocyte antigen gene complex) is preferentially limited to APCs, including monocytes, macrophages, dendritic cells (DCs) and B cells (Saraiva et al. Citation2018). T cell differentiation into mature lineages (Th1, Th2, Th17, and Treg) is dependent upon cytokine production by APCs (Zhu and Paul Citation2010). Dynamic immune responses rely on effective antigen presentation and co-stimulatory signals by APCs.
In this study, we performed transcriptomic profiling of circulating myeloid cells (CD33+HLA-DR– vs. CD33+HLA-DR+ APCs) in healthy individuals using RNA sequencing (RNA-Seq). We isolated peripheral blood mononuclear cells (PBMCs) from 10 healthy individuals, and sorted CD33+HLA-DR– myeloid cells (potentially comprise MDSCs) and CD33+HLA-DR+ cells, which represent myeloid APCs. RNA-Seq data showed that several signaling pathways associated with the regulation of immune responses were downregulated, while tumor suppressing/anti-inflammatory signaling pathways were upregulated in CD33+HLA-DR– myeloid cells.
Materials and Methods
PBMC isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples of 10 healthy donors, as we have previously described (Saleh et al. Citation2020). This study was performed under ethical approval from Qatar Biomedical Research Institute, Qatar (protocol no. 2017–006).
Fluorescence-activated cell sorting
PBMCs were washed with phosphate-buffered saline then re-suspended in 100 µl flow cytometry staining buffer and processed, as previously described (Saleh et al. Citation2020). Fc receptor (FcR) Blocking Reagent (Miltenyi Biotech, Bergisch Gladbach, Germany) was used to block FcR. Cells were stained with 7-AAD viability dye (eBioscience, San Diego, USA) to exclude dead cells and gate live cells, and also with antibodies against myeloid cell markers for sorting, as previously described (Saleh et al. Citation2020). We sorted CD33+HLA-DR+ APCs and CD33+HLA-DR– myeloid cells using a stringent gating strategy (Saleh et al. Citation2020). Purity of sorted cells was always checked and maintained at high levels with minimal sorter-induced cell stress (SICS). FlowJo V10 software (FlowJo, Ashland, USA) was used for data analyses.
RNA isolation
Total RNA was extracted from the two sorted, pure myeloid cell subpopulations of 10 healthy donors, as previously described (Saleh et al. Citation2020).
Library preparation
cDNA libraries were generated using Exome TruSeq Stranded mRNA Library Prep Kit (illumina, San Diego, USA) following the manufacturer’s protocol, and as previously described (Vishnubalaji et al. Citation2019). Quality-passed libraries were subjected to clustering using TruSeq PE Cluster Kit v3-cBot-HS (illumina). The clustered samples were sequenced on an illumina HiSeq 4000 instrument using HiSeq 3000/4000 SBS kit (illumina).
RNA sequencing data and functional annotation analyses
Quality-trimmed pair end reads were aligned to the hg19 human reference genome in CLC Genomics Workbench 12 (Qiagen, Hilden, Germany) with default settings, as previously described (Sasidharan Nair et al. Citation2020b; Vishnubalaji et al. Citation2019). The abundance of the transcript expression was determined as the score of Transcripts Per Million (TPM). Differential gene expression analysis, hierarchical clustering, and Principal component analysis (PCA) were performed on expression data, as we have previously described (Shaath et al. Citation2019), using 2.0-fold change and P value cutoff <0.05. Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems; www.ingenuity.com) was used to obtain canonical pathways and functional regulatory networks of differentially expressed genes, as previously described (Vishnubalaji et al. Citation2019). Additionally, functional gene annotation analyses were performed on Database for Annotation, visualization, and Integrated Discovery (DAVID) platform, as previously described (Sasidharan Nair et al. Citation2020a). “KEGG pathway” and “BioCarta” were obtained from significantly up/downregulated genes using DAVID platform. For heatmaps, the Z-score was calculated, as previously described (Malone et al. Citation2011), to show the fold change of each gene in CD33+HLA-DR– myeloid cells, compared to CD33+HLA-DR+ APCs. Protein–protein interaction (PPI) networks among the significantly up/downregulated genes were determined by web-based online tool, STRING V11.0 (http://string-db.org) (Szklarczyk et al. Citation2015).
Results
Clustering analyses of differentially expressed genes in circulating CD33+HLA-DR+ and CD33+HLA-DR– myeloid subsets
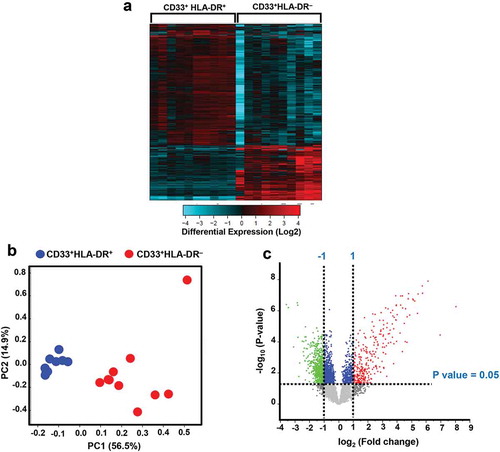
APCs are responsible for antigen processing and presentation, and activation of the adaptive immune responses, while MDSCs are known to exert suppressive activities (Kondratova et al. Citation2019). The hierarchical clustering of differentially regulated genes in circulating CD33+HLA-DR–, compared to CD33+HLA-DR+ myeloid subset in healthy controls showed distinct clusters (). We found that a total of 594 upregulated genes in CD33+HLA-DR– and 1421 downregulated genes, compared with CD33+ HLA-DR+ myeloid cells. Principal component analysis (PCA) showed that dataset in CD33+HLA-DR– myeloid cells was clustered distinctly from the control CD33+HLA-DR+ APCs, implicating the differences of the functional characteristics (). CD33+HLA-DR– myeloid cells clustered distinctly from APCs within the first two principal components, where PC1 accounting for approximately 56.5% of the observed variation (). Volcano plot shows the genes that were significantly upregulated (shown in red), significantly downregulated (shown in green) or remained unchanged (shown in grey) when comparing CD33+HLA-DR+ vs. CD33+HLA-DR– myeloid cells (). Only genes that were significantly affected, with a fold change >2 and P value cutoff <0.05, were subjected for further analyses.
Figure 1. Hierarchical clustering for differentially expressed genes in circulating CD33+HLA-DR+ and CD33+HLA-DR– myeloid cell subsets from healthy donors. Hierarchical clustering of CD33+HLA-DR+ and CD33+HLA-DR– myeloid cells based on differentially expressed genes. Each column represents a sample and each row represents a gene. Expression level of each transcript is depicted according to color scale (a). Principal component analysis (PCA) based on differentially expressed genes in each myeloid subpopulation (b). A Volcano plot shows genes that were upregulated (red), downregulated (green) or remained unchanged (grey) when comparing CD33+HLA-DR+ vs. CD33+HLA-DR– myeloid cell subsets (c). Volcano plot shows differentially expressed genes with a P value cutoff of <0.05 and Log2 fold change >1 and <-1, as indicated by the horizontal line and vertical lines, respectively
Up/downregulated signaling pathways and biological mechanisms in CD33+HLA-DR− myeloid cells
Next, we analyzed genes that were significantly up/downregulated in CD33+HLA-DR– versus CD33+ HLA-DR+ myeloid cell subsets using functional annotation analyses (). We found that genes related to antigen presentation/processing, such as HLA-DMB, HLA-DPA1, HLA-DQA1, and HLA-DQB2, were downregulated in CD33+HLA-DR–. In addition, genes related to cell-mediated immune response (such as CD1D, IRAK1, CD86, EMP1, and TLR), and genes related to IL-6 (such as JAK2, NRAS, and RAP1A), and GM-CSF and IL-3 (such as IL3A, HCK, MAP2K1, and PRKCB) signaling pathways were downregulated in CD33+HLA-DR– myeloid cells (). Additionally, genes related to cell migration and immune cell trafficking (such as HMOX1, AREG1, DDX5, and ANXA2), leukocyte extravasation (such as ITGA4, ITGAB, BTK, and GNAI3) and IL-8 (such as CHUK, NRAS, RRAS, GNB4, and IRAK1) signaling pathways were downregulated in CD33+HLA-DR– myeloid cells (). Genes related to apoptosis and cell cytotoxicity (such as KLRK1, BEX2, CLU, PRF1, KLRC1, and KLRF1) were upregulated in CD33+HLA-DR– myeloid cells (). In contrast, genes related to PI3K/Akt, TREM1, mTOR, and pattern recognition receptor (PRR) signalling pathways were downregulated in CD33+HLA-DR– myeloid cells (). These results implicate that major immune regulatory signalling pathways are downregulated in CD33+HLA-DR– myeloid cells in the circulation of healthy individuals.
Figure 2. Signaling pathway analyses of myeloid cell subsets. Functional categorization of both upregulated and downregulated genes (with P value <0.05) from CLC analysis were analyzed for CD33+HLA-DR– myeloid cells, compared with CD33+HLA-DR+ APCs using IPA. Heatmaps show the Z-score representing the fold change of each gene in CD33+HLA-DR– myeloid cells, compared to CD33+HLA-DR+ APCs from 10 healthy donors. Functional gene annotations showed that genes related to several cellular processes and signaling pathways were differentially expressed in both myeloid cell subsets (a-d). Functional categorization of top significantly affected transcripts (with P value <0.05) from CLC analysis were analyzed through IPA. The horizontal bars denote the different pathways based on the Z-scores (e).
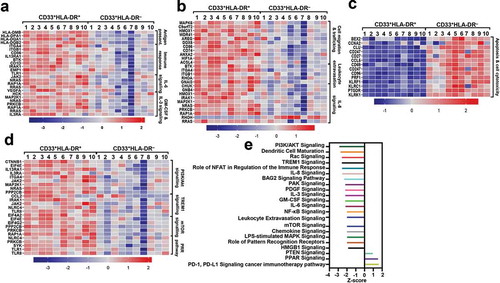
Moreover, functional annotation analyses showed that PI3K/Akt, Rac, TREM1, NFAT-mediated immune response regulation, IL-8, BAG2, PAK, PDGF, IL-3, GM-CSF, IL-6, NF-κB, mTOR, chemokine, LPS-stimulated MAPK, and HMGB1 signaling pathways were downregulated in CD33+HLA-DR– myeloid cells (−3.0 < Z-score > 2.0, ). Biological mechanisms, such as DC maturation, senescence, leukocyte extravasation, and PRR in recognition of bacteria and viruses, were all downregulated in CD33+HLA-DR– myeloid cells (). In contrast, PPAR and PTEN signaling pathways, and pathways potentially related to resistance to PD-1/PD-L1 cancer immunotherapy pathway were upregulated in CD33+HLA-DR– myeloid cells (0.9 < Z-score > 1.7, ).
Furthermore, we found that antigen presentation/processing (), FcγR-mediated phagocytosis, and chemokine/immune cell trafficking-related pathways were downregulated in CD33+HLA-DR– myeloid cells (Supplementary Figures 1 and 2). Altogether, our data show the distinct characteristics of APCs and CD33+HLA-DR– myeloid cells in physiological conditions.
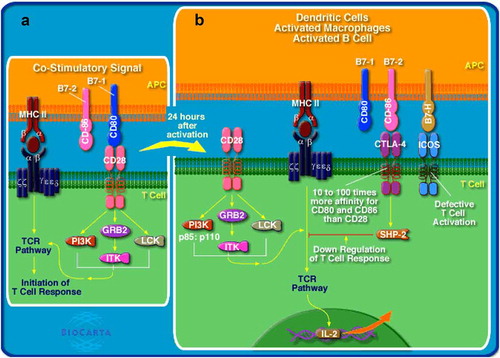
Figure 3. Antigen presentation/processing and T cell activation pathways. APCs with MHC class II-bound antigen bind to T cell receptor (TCR) on T cells and co-stimulatory molecule CD28 (on T cells) binds to CD80 (on APCs), resulting in T cell activation (a). Antigen presentation/processing and T cell activation pathways were downregulated in circulating CD33+HLA-DR– cells. Following activation, T cell response is regulated, perhaps by a negative-feedback mechanism to prevent the development of autoimmunity and shutdown unnecessary activation of T cells. CTLA-4 and ICOS on activated T cells binds to CD86 and B7-H on activated APCs, respectively, resulting in the inhibition of T cell activation (b). Figure obtained from BioCarta/DAVID functional annotation analyses
Next, we performed protein–protein interaction (PPI) and enrichment network analysis, using STRING web-based tool, to show the interactions of proteins within the significantly affected pathways between the two myeloid subsets. For this analysis, we selected two downregulated pathways (GM-CSF/IL-3 and PI3K/Akt signaling pathways) and one upregulated pathway (apoptosis and cell cytotoxicity) in CD33+HLA-DR– myeloid cells. For GM-CSF/IL-3 signaling, STRING database identified 6 nodes and 4 edges with PPI enrichment P value 0.006, average clustering coefficient of 0.5 and average node degree of 1.3 (Supplementary Figure 3a). For PI3K/Akt signaling pathway, STRING database identified 8 nodes and 10 edges with PPI enrichment P value 0.0003, average clustering coefficient of 0.696 and average node degree of 2.5 (Supplementary Figure 3b). For apoptosis and cell cytotoxicity, STRING database identified 13 nodes and 19 edges with PPI enrichment P value 2.09E-13, average clustering coefficient of 0.494 and average node degree of 2.9 (Supplementary Figure 3c).
Discussion
Transcriptomic profiling of circulating CD33+HLA-DR– myeloid cells, compared to CD33+HLA-DR+ myeloid APCs showed various signaling pathways that are differentially regulated in these cellular populations. Our RNA-Seq data revealed the distinct molecular and functional mechanisms between APCs and CD33+HLA-DR– myeloid cells, indicating the potential suppressive activity of the sorted CD33+HLA-DR– myeloid cells from healthy individuals. Our data potentially improve the current understandings of the functional characteristics of CD33+HLA-DR– myeloid cells in normal physiological conditions, and could be used for further comparisons with various pathological conditions, including cancer.
In normal physiological conditions, we found that several signaling pathways associated with the positive regulation of immune responses were downregulated in circulating CD33+HLA-DR– myeloid cells, perhaps to suppress unnecessary activation of immune response and to maintain immune tolerance. These pathways include PI3K/Akt and mTOR signaling, PAK and Rac signaling, DC maturation, Toll-like (TLR) receptor signaling, TREM1 signaling, NFAT-mediated regulation of the immune response, and IL-3, GM-CSF, IL-6 and IL-8 signaling. TLR signaling is important for the activation of macrophages in response to pathogens/stimulus (Sanaei et al. Citation2019). PI3K/Akt/mTOR pathway controls the phenotype of macrophages by regulating their response to multiple signals associated with inflammation (B. Li et al. Citation2018; Vergadi et al. Citation2017). In addition, PI3K through Rac and PAK signaling pathways can positively regulate macrophage migration in response to chemokines (Weiss-Haljiti et al. Citation2004). DC maturation is very important for the activation of adaptive immunity (Fu and Jiang Citation2018), and TREM1 signaling pathway plays a significant role in regulating innate and adaptive immunity (Arts et al. Citation2013; Roe et al. Citation2014). NFAT-mediated pathway has been shown to be important for the activation of innate immune responses post infections or inflammation, including phagocytosis and PRR response (Vandewalle et al. Citation2014). In addition, NFAT is a crucial regulator for several key pathways, including IL-3 production, which are responsible for both lymphoid and myeloid differentiation (Hawwari et al. Citation2002). GM-CSF and IL-6 are also important cytokines associated with maturation and activation of myeloid cells and T cells (Dougan et al. Citation2019; Tanaka et al. Citation2014). IL-8 signaling is known to recruit leukocytes, such as neutrophils to inflammatory or infection sites, where an inflammatory reaction is initiated (David et al. Citation2016; Krupa et al. Citation2015).
In contrast to normal physiological conditions, some of the above-mentioned pathways could be differentially regulated in CD33+HLA-DR– myeloid cells in chronic viral infections or cancer-settings. For instance, hepatitis C virus (HCV) activates PI3K signaling pathway in myeloid cells to induce the production of monocytic MDSCs, which express increased levels of the suppressive enzyme IDO and lead to T cell apoptosis (Pang et al. Citation2016). Additionally, GM-CSF and IL-6 signaling in MDSCs support tumor progression and immunosuppression via the recruitment of MDSCs to the TME, and perhaps enhancing their immunosuppressive functions (Gargett et al. Citation2016; Sasidharan Nair et al. Citation2020a). In addition, genes related to cell apoptosis and cell cytotoxicity were upregulated in CD33+HLA-DR– myeloid cells. This suggests the immunosuppressive potential for CD33+HLA-DR– myeloid cells to induce cell death of other immune cells including T cells (Sinha et al. Citation2011). Reports showed that the transcriptional upregulation of PTEN is mediated through PPARγ/PTEN axis, which is indispensable to keep host immune response in check (Patel et al. Citation2001; Song et al. Citation2012). Moreover, PTEN was considered as a pivotal tumor suppressive gene and its downregulation could lead to cancer and metabolic abnormalities (Chen et al., Citation2018; Patel et al. Citation2001; Teresi and Waite Citation2008). Interestingly, we found that PPAR/PTEN pathway was upregulated in CD33+HLA-DR– myeloid cells. These data reveal the influence of myeloid cells in the regulation of immunohomeostais in steady-state conditions, while in disease states, for example, cancer, the function of PPAR signaling pathway becomes pathological; it induces MDSC proliferation and expansion and contributes to tumor progression (Hegde et al. Citation2015; H. Li et al. Citation2011; Zhao et al. Citation2016).
We also found that antigen presentation/processing and FcγR-mediated phagocytosis genes and pathways were downregulated in CD33+HLA-DR– myeloid cells and upregulated in APCs. These data validate our sorting strategy and RNA-Seq data, and further confirm their distinct functions. APCs can activate immune cells, such as T cells, via cell contact-dependent mechanisms and through secreting molecules such as cytokines (Ennaciri and Girard Citation2009). FcγR-mediated phagocytosis in APCs is triggered upon the recognition of an antigen, which could be self-antigen in the context of autoimmunity or derived from bacteria, resulting in the generation of inflammatory responses; this involves the secretion of proteases, cytokines and chemokines, which in turn causes the induction of antibody-dependent cellular cytotoxicity, and leads to phagocytosis (Fitzer-Attas et al. Citation2000; Huang et al. Citation2011) (Supplementary Figure 1).
During inflammation and upon infections, chemokine signaling in APCs is crucial to recruit immune cells, such as macrophages, neutrophils, T cells, and B cells from the peripheral blood or lymphoid tissues to the inflammatory or infection sites, where immune responses are initiated (Esche et al. Citation2005; Sokol and Luster Citation2015) (Supplementary Figure 2). Pathways which result in leukocyte recruitment, immune cell recruitment/trafficking via chemokine signaling were also downregulated in CD33+HLA-DR– myeloid cells, indicating their potential interference with the immune response during physiological conditions to maintain immune tolerance. However, the transcriptomic profile of CD33+HLA-DR– myeloid cells in tumor microenvironment (TME) could be different. We have recently reported that colorectal tumor-infiltrating granulocytic-derived myeloid cells (PMN-MDSCs) upregulated the NF-κB/IL-1β-mediated leukocyte recruitment pathway, perhaps to recruit more MDSCs to the TME (Sasidharan Nair et al. Citation2020a).
Conclusion
Our transcriptomic data reflect the differences in molecular and functional characteristics between CD33+HLA-DR+ myeloid APCs and CD33+HLA-DR– myeloid cells in physiological conditions. However, functional studies are required to validate the biological activities of some of the significantly affected pathways in CD33+HLA-DR– myeloid cells in health. In addition, the molecular pathways regulated by CD33+HLA-DR– myeloid cells might differ in disease states and could be explored in future studies.
Supplemental Material
Download PDF (562.6 KB)Acknowledgments
We thank the staff of the Genomics Core at QBRI for performing RNA-Seq. Open Access funding provided by the Qatar National Library.
Disclosure statement
The authors have no conflicts of interest.
Data availability statement
The datasets used and/or analyzed in this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Arts RJ, Joosten LA, van der Meer JW, Netea MG. 2013. TREM-1: intracellular signaling pathways and interaction with pattern recognition receptors. J Leukoc Biol. 93(2):209–15.
- Awad RM, De Vlaeminck Y, Maebe J, Goyvaerts C, Breckpot K. 2018. Turn Back the TIMe: targeting Tumor Infiltrating Myeloid Cells to Revert Cancer Progression. Front Immunol. 9:1977.
- Bernal-Estevez DA, Garcia O, Sanchez R, Parra-Lopez CA. 2018. Monitoring the responsiveness of T and antigen presenting cell compartments in breast cancer patients is useful to predict clinical tumor response to neoadjuvant chemotherapy. BMC Cancer. 18(1):77.
- Chen CY, Chen J, He L, Stiles BL. 2018. PTEN: tumor Suppressor and Metabolic Regulator. Front Endocrinol (Lausanne). 9. doi:https://doi.org/10.3389/fendo.2018.00338
- David JM, Dominguez C, Hamilton DH, Palena C. 2016. The IL-8/IL-8R Axis: A Double Agent in Tumor Immune Resistance. Vaccines (Basel). 4. doi:https://doi.org/10.3390/vaccines4030022
- Dougan M, Dranoff G, Dougan SK. 2019. GM-CSF, IL-3, and IL-5 Family of Cytokines: regulators of Inflammation. Immunity. 50(4):796–811.
- Ennaciri J, Girard D. 2009. Immune system: maturation of myeloid cells. Methods Mol Biol. 550:195–203.
- Esche C, Stellato C, Beck LA. 2005. Chemokines: key players in innate and adaptive immunity. J Invest Dermatol. 125(4):615–28.
- Fitzer-Attas CJ, Lowry M, Crowley MT, Finn AJ, Meng F, DeFranco AL, Lowell CA. 2000. Fcgamma receptor-mediated phagocytosis in macrophages lacking the Src family tyrosine kinases Hck, Fgr, and Lyn. J Exp Med. 191(4):669–82.
- Fu C, Jiang A. 2018. Dendritic Cells and CD8 T Cell Immunity in Tumor Microenvironment. Front Immunol. 9:3059.
- Gabrilovich DI, Nagaraj S. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 9(3):162–74.
- Gargett T, Christo SN, Hercus TR, Abbas N, Singhal N, Lopez AF, Brown MP. 2016. GM-CSF signalling blockade and chemotherapeutic agents act in concert to inhibit the function of myeloid-derived suppressor cells in vitro. Clin Transl Immunology. 5(12):e119.
- Hawwari A, Burrows J, Vadas MA, Cockerill PN. 2002. The human IL-3 locus is regulated cooperatively by two NFAT-dependent enhancers that have distinct tissue-specific activities. J Immunol. 169(4):1876–86.
- Hegde VL, Singh UP, Nagarkatti PS, Nagarkatti M. 2015. Critical Role of Mast Cells and Peroxisome Proliferator-Activated Receptor gamma in the Induction of Myeloid-Derived Suppressor Cells by Marijuana Cannabidiol In Vivo. J Immunol. 5(11):5211–22.
- Huang ZY, Hunter S, Chien P, Kim MK, Han-Kim TH, Indik ZK, Schreiber AD. 2011. Interaction of two phagocytic host defense systems: fcgamma receptors and complement receptor 3. J Biol Chem. 286(1):160–68.
- Khaled YS, Ammori BJ, Elkord E. 2013. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. 91(8):493–502.
- Kirkwood KL, Zhang L, Thiyagarajan R, Seldeen KL, Troen BR. 2018. Myeloid-Derived Suppressor Cells at the Intersection of Inflammaging and Bone Fragility. Immunological Investigations. 47(8):844–54.
- Kondratova M, Czerwinska U, Sompairac N, Amigorena SD, Soumelis V, Barillot E, Zinovyev A, Kuperstein I. 2019. A multiscale signalling network map of innate immune response in cancer reveals cell heterogeneity signatures. Nat Commun. 10(1):4808.
- Krupa A, Fol M, Dziadek BR, Kepka E, Wojciechowska D, Brzostek A, Torzewska A, Dziadek J, Baughman RP, Griffith D, et al. 2015. Binding of CXCL8/IL-8 to Mycobacterium tuberculosis Modulates the Innate Immune Response. Mediators Inflamm. 2015:1–11.
- Li B, Xi P, Wang Z, Han X, Xu Y, Zhang Y, Miao J. 2018. PI3K/Akt/mTOR signaling pathway participates in Streptococcus uberis-induced inflammation in mammary epithelial cells in concert with the classical TLRs/NF-kB pathway. Vet Microbiol. 227:103–11.
- Li H, Sorenson AL, Poczobutt J, Amin J, Joyal T, Sullivan T, Crossno JT Jr., Weiser-Evans MC, Nemenoff RA. 2011. Activation of PPARgamma in myeloid cells promotes lung cancer progression and metastasis. PLoS One. 6(12):e28133.
- Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. 2013. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 138(2):105–15.
- Malone BM, Tan F, Bridges SM, Peng Z. 2011. Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS One. 6(9):e25260.
- Pang X, Song H, Zhang Q, Tu Z, Niu J. 2016. Hepatitis C virus regulates the production of monocytic myeloid-derived suppressor cells from peripheral blood mononuclear cells through PI3K pathway and autocrine signaling. Clin Immunol. 164:57–64.
- Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH. 2001. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr Biol. 11(10):764–68.
- Roe K, Gibot S, Verma S. 2014. Triggering receptor expressed on myeloid cells-1 (TREM-1): a new player in antiviral immunity? Front Microbiol. 5. doi:https://doi.org/10.3389/fmicb.2014.00627
- Saleh R, Elkord E. 2019. Acquired resistance to cancer immunotherapy: role of tumor-mediated immunosuppression. Semin Cancer Biol. doi:https://doi.org/10.1016/j.semcancer.2019.07.017
- Saleh R, Toor SM, Taha RZ, Al-Ali D, Sasidharan Nair V, Elkord E. 2020. DNA methylation in the promoters of PD-L1, MMP9, ARG1, galectin-9, TIM-3, VISTA and TGF-β genes in HLA-DR(-) myeloid cells, compared with HLA-DR(+) antigen-presenting cells. Epigenetics. 1–14. doi:https://doi.org/10.1080/15592294.2020.1767373
- Sanaei R, Rezaei N, Aghamohammadi A, Delbandi AA, Tavasolian P, Tajik N. 2019. Disturbed Transcription of TLRs’ Negative Regulators and Cytokines Secretion among TLR4- and 9-Activated PBMCs of Agammaglobulinemic Patients. Immunological Investigations. 48(8):860–74.
- Saraiva DP, Jacinto A, Borralho P, Braga S, Cabral MG. 2018. HLA-DR in Cytotoxic T Lymphocytes Predicts Breast Cancer Patients’ Response to Neoadjuvant Chemotherapy. Front Immunol. 9. doi:https://doi.org/10.3389/fimmu.2018.02605
- Sasidharan Nair V, Saleh R, Toor SM, Taha RZ, Ahmed AA, Kurer MA, Murshed K, Alajez NM, Abu Nada M, Elkord E 2020a. Transcriptomic profiling disclosed the role of DNA methylation and histone modifications in tumor-infiltrating myeloid-derived suppressor cell subsets in colorectal cancer. Clin Epigenetics. 12(1). doi:https://doi.org/10.1186/s13148-13020-10808-13149
- Sasidharan Nair V, Toor SM, Taouk G, Pfister G, Ouararhni K, Alajez NM, Elkord E. 2020b. Pembrolizumab Interferes with the Differentiation of Human FOXP3(+)-Induced T Regulatory Cells, but Not with FOXP3 Stability, through Activation of mTOR. J Immunol. 204:199–211.
- Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, Cerwenka A. 2012. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J Immunol. 189(12):5602–11.
- Shaath H, Toor SM, Nair VS, Elkord E, Alajez NM. 2019. Transcriptomic Analyses Revealed Systemic Alterations in Gene Expression in Circulation and Tumor Microenvironment of Colorectal Cancer Patients. Cancers (Basel). 11(12):1994.
- Sinha P, Chornoguz O, Clements VK, Artemenko KA, Zubarev RA, Ostrand-Rosenberg S. 2011. Myeloid-derived suppressor cells express the death receptor Fas and apoptose in response to T cell-expressed FasL. Blood. 117:5381–90.
- Sokol CL, Luster AD. 2015. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 7(5):a016303.
- Song MS, Salmena L, Pandolfi PP. 2012. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 13(5):283–96.
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43(D1):D447–452.
- Tanaka T, Narazaki M, Kishimoto T. 2014. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 6(10):a016295–a016295.
- Teresi RE, Waite KA. 2008. PPARgamma, PTEN, and the Fight against Cancer. PPAR Res. 2008:1–6.
- Vandewalle A, Tourneur E, Bens M, Chassin C, Werts C. 2014. Calcineurin/NFAT signaling and innate host defence: a role for NOD1-mediated phagocytic functions. Cell Commun Signal. 12(1):8.
- Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. 2017. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J Immunol. 198(3):1006–14.
- Vishnubalaji R, Sasidharan Nair V, Ouararhni K, Elkord E, Alajez NM. 2019. Integrated Transcriptome and Pathway Analyses Revealed Multiple Activated Pathways in Breast Cancer. Front Oncol. 9:910.
- Weiss-Haljiti C, Pasquali C, Ji H, Gillieron C, Chabert C, Curchod M-L, Hirsch E, Ridley AJ, Hooft van Huijsduijnen R, Camps M, et al. 2004. Involvement of Phosphoinositide 3-Kinase γ, Rac, and PAK Signaling in Chemokine-induced Macrophage Migration. J Biol Chem. 279(41):43273–84.
- Zhao T, Du H, Blum JS, Yan C. 2016. Critical role of PPARgamma in myeloid-derived suppressor cell-stimulated cancer cell proliferation and metastasis. Oncotarget. 7(2):1529–43.
- Zhu J, Paul WE. 2010. Heterogeneity and plasticity of T helper cells. Cell Res. 20(1):4–12.
