Abstract
Knowledge about specific diseases is often evolving but is embedded in medical texts. We propose a technique that employs term proximity information to improve the extraction of the disease factors, which are concept terms related to specific diseases in the medical texts. The disease factors are a core knowledge base for many information systems for healthcare decision support and education. In two case studies on a broad range of diseases, the proposed technique significantly further enhances a good extraction technique to rank the diagnosis factors.
INTRODUCTION
Knowledge about specific diseases is often categorized into several aspects such as etiology, diagnosis, treatment, and prognosis. The aspects of disease knowledge are the fundamental basis of healthcare activities and medical education. Although there have been many thesauri (e.g., Medical Subject Headings, MeSHFootnote1) and ontologies (e.g., Disease Ontology; Schriml et al. Citation2012) developed for defining various diseases, the evolving knowledge concerning the disease aspects is not well curated. Therefore, the knowledge about the disease aspects is routinely created, edited, and updated in medical texts by many healthcare information providers (e.g., MedlinePlusFootnote2 and MedicineNetFootnote3). Information technology is also being developed to retrieve and classify the medical texts of various disease aspects (e.g., Lin and Demner-Fushman Citation2006; Liu Citation2007).
Given a set of medical texts about an aspect a of a disease c, automatic extraction of the disease factors (concept terms in the texts) related to a of c is essential. The disease factors are a core knowledge base for many healthcare information systems. For example, with the disease factors, an expert system can be developed to support the diagnosis and treatment of diseases (e.g., a symptom-based disease classification system; Li, 2007). An online navigation system may also be developed to support the learning of differential diagnosis skills by exploring the diseases that are correlated by certain factors. Correlations among diseases and related factors are also the fundamental basis to build problem-based learning (PBL) systems for medical education (Suebnukarn and Haddawy Citation2006).
Problem Definition and Motivation
In this article, we explore how existing text mining techniques can be enhanced to extract the disease factors from medical texts. The research is motivated by the difficulty and cost of manually constructing and maintaining the discriminating factors for a broad range of diseases. In clinical practice, there are a large number of complicated many-to-many relationships among the diseases. However, manual construction of the disease factors for even a single disease may require a great deal of time and effort, and the complexity dramatically increases when a broad range of diseases are considered and the factors may evolve as new medical findings are produced. Therefore, by extracting candidate factors from up-to-date medical texts, a text mining system could offer support to reduce the difficulty and cost.
We present and evaluate a novel technique, an Enhancer for the Extractor of Disease Factor Information (EDFI) that acts to improve discriminative factors extractors. For a candidate factor u, EDFI employs term proximity information to improve the underlying extractors. It measures how other candidate factors appear in the areas near to u in the medical texts and then associates the term proximity information with the discriminating capability of u measured by the underlying factors extractors. The idea is based on the observation that in a medical text, d talking about an aspect a of a disease c, the key factors related to a of c often appear in a nearby area of d, and hence, by associating term proximity information with the discriminating capabilities of the factors, the factor extractors might be improved.
Organization and Contribution of the Article
In the next section, we identify the main challenges of the research problem and accordingly describe the weaknesses of previous studies in tackling the challenges. “Extracting Disease Factors by Term Proximity” then presents EDFI, and to empirically evaluate EDFI, “Two Case Studies” reports two case studies in which two sources of real-world medical texts for a broad range of diseases are tested. Given a good extractor, EDFI can significantly further improve it in ranking the disease factors from both sources of medical texts. The contribution is of practical significance because EDFI enhances existing factor extractors to extract the disease factors from medical texts, which are a fundamental basis of intelligent systems for healthcare decision making and education.
TABLE 1 Main Challenges of Extracting Disease Factors from Medical Texts
MAIN CHALLENGES AND RELATED WORK
summarizes the main tasks and the challenges of EDFI. The first challenge is to extract (from the medical texts) those factors that are capable of discriminating a broad range of diseases (ref., Challenge 1 in ). It is also a task of finding the correlations among diseases and their key factors. There have been natural language processing tools (e.g., MetaMapFootnote4 and cTAKESFootnote5) that extract and recognize biomedical entities in texts. However the tools do not aim at extracting disease-factor correlations in medical texts. There are also previous studies focusing on finding the correlations between biomedical objects (e.g., proteins, genes, and diseases). They often employed sentence parsing (Özgür et al. Citation2008; Temkin and Gilder Citation2003), template matching (Domedel-Puig and Wernisch Citation2005; Ono et al. Citation2001), or integration of machine learning and parsing (Kim, Yoon, and Yang Citation2008) to extract the relationships between biomedical objects. The previous techniques often worked on single sentences within which all objects of interest (e.g., the disease and its related factors) need to be mentioned. They would have difficulties in extracting disease factors, because in a medical text about a specific disease, the disease factors may scatter in multiple sentences within which the disease is not repeatedly mentioned.
Another approach to extracting disease factors is text classification. The problem of extracting key factors for a disease may be treated as a feature selection problem in which we score and select those features (terms of factors) that are capable of discriminating the categories (diseases). The selected features may also serve as the basis on which a text classifier could be built to classify medical descriptions into related disease categories (e.g., classifying symptom descriptions into related diseases; Liu, Citation2007). Feature selection in text classification does not rely on processing individual sentences, relieving it from the weaknesses of the above parsing and template matching approaches. It employs the texts labeled with category names to measure how a feature may discriminate the categories, making it a plausible way to extract disease factors. Previous studies have developed and tested many feature selection techniques (e.g., Mladeniá et al. Citation2004; Yang and Pedersen Citation1997). However, they do not consider the proximity among the factor terms (features).
Therefore, our second challenge (ref., Challenge 2 in ) is to recognize the proximity of terms so that factor terms may be extracted more properly. The challenge is based on the observation that authors of medical texts for a disease tend to describe the factors of the disease in a nearby area in the texts, making term proximity information quite helpful to extract the factors. Term proximity was routinely noted as important information by many previous studies as well, however, previous studies mainly employed term proximity to improve text retrieval and text classification, rather than the extraction of disease factors. In text retrieval, proximity-based techniques often considered adjacent terms (Wang et al. Citation2007) and nearby terms (Liu and Huang Citation2011; Svore, Kanani, and Khan Citation2010; Cummins and O’Riordan Citation2009; Gerani, Carman, and Crestani Citation2010; Zhao and Yun Citation2009; Lv and Zhai Citation2009; Tao and Zhai Citation2007; Rasolofo and Savoy Citation2003), whereas in text classification, proximity-based techniques often considered multiple consecutive terms (Peng and Schuurmans Citation2003), and nearby terms (Liu Citation2010; Cohen and Singer Citation1996) as well. No previous techniques employed term proximity to extract disease factors, which tend to appear in nearby areas in medical texts.
Therefore, the final strength of each disease factor should be based on both the term proximity and the discriminating capability of the factor. The third challenge of the research (ref., Challenge 3 in ) is thus to integrate the term proximity information with the discriminating capability. A proper integration technique may make EDFI able to collaborate with factors extractors (i.e., the feature selection techniques as noted above). EDFI may thus act as a supplement to the extractors, with the goal of enhancing the underlying extractors in employing term proximity to improve the precision of disease factor extraction. To our knowledge, no previous studies tackled the challenge.
EXTRACTING DISEASE FACTORS BY TERM PROXIMITY
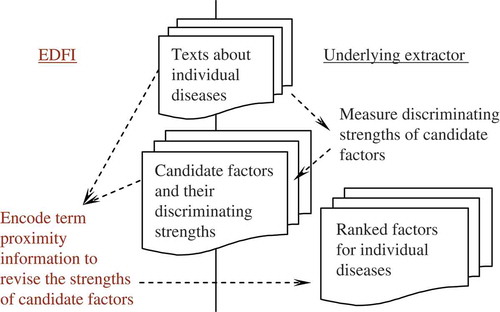
illustrates an overview of EDFI, which collaborates with the underyling factor extractor by integrating the output of the extractor (i.e., discriminating strengths of candidate factors) with term proximity information, and the resulting strengths of the factors are used to rank the factors for medical professionals to validate. The strength estimation is thus essential in reducing the load incurred to the professionals. Note that EDFI receives as input (1) the medical data employed by the underlying extractor (i.e., medical texts labeled with corresponding disease names) and (2) the output of the extractor (i.e., the candidate factors and their capabilities in discriminating the diseases). The former is used to measure term proximity among candidate factors, and the result of the proximity measurement is integrated with the latter to produce the final strengths of the factors. Therefore, EDFI may collaborate with discriminative factor extractors without needing to change the development and behaviors of the extractors.
TABLE 2 Proximity-Based Revision of the Strength of a Candidate Factor
Algorithm of EDFI
presents the algorithm of EDFI in estimating the strength of a candidate factor. Given a feature u (candidate factor term) and its discriminating strength x for a disease c (x is produced by the underlying discriminative factors extractor), EDFI measures a term proximity score for u (ProximityScore, ref. Step 4 and Step 5). The proximity score is measured based on the shortest distances (in the given medical texts) between u and other candidate features of c (ref. Step 4.1 and Step 4.2). EDFI employs a sigmoid function to transform the shortest distances into the proximity score–the shortest distance between u and another candidate feature n is transformed into a weight between 0 and 1, and a smaller distance leads to a larger weight (ref. Step 4.3). The sigmoid weighting function has a parameter α that governs the distance for which a weight of 0.5 is produced (when the distance is equal to α, the weight is 0.5). Moreover, it is interesting to note that to measure the proximity score of u, the sum of the sigmoid weights with respect to all the other candidate features is computed (i.e., RawScore, ref. Step 4.3) and normalized by the maximum possible sum of the weights (i.e., |N|, ref. Step 5). The normalization aims at measuring the contextual completeness of the other candidate features appearing near to u, and hence the proximity score of u is in the range of [0, 1] as well.
To properly integrate the proximity score with the discriminating strength of u with respect to c, EDFI computes a ranking score based on the strength rank of u among all candidate features of c (i.e. RankScore, ref., Step 6).Footnote6 The final strength of u is the sum of the ranking score and the proximity score (ref., Step 7), and hence, it may indicate the possibility of u serving as a factor for disease c–if u may discriminate c from other diseases and has individually appeared near to other candidate factors at some places in the medical texts, u may be a good factor for c.
Behavior Analysis for EDFI
With the following critical behaviors, EDFI is able to tackle all the challenges noted in “Main Challenges and Related Work”:
Tackling the weakness of sentence parsing and template matching (Challenge 1 in ): To extract key factors, EDFI collaborates with a feature (factor) selection technique, which estimates the capabilities of the disease factors in discriminating the diseases, making the system able to consider those disease factors that cannot be extracted by sentence parsing or template matching.
Employing term proximity information to enhance the feature (factor) selection technique (Challenge 2 in ):
Considering the contextual completeness of disease factors in medical texts: The proximity score of a candidate factor u for a disease c is normalized by the number of other candidate factors for c (Step 5 in ). Therefore, EDFI is concerned with the completeness of the other factors that ever appeared around u in some medical text, making the proximity score more adaptive to the characteristics of different diseases. Moreover, EDFI has a parameter (α) in the proximity weighting function (i.e, the sigmoid function, ref. Step 4.3 in ) to govern the distance for which a proximity weight of 0.5 is produced. As the factors for a disease are often collectively described in one to two sentences in a medical text, it is reasonable to hypothesize that a proper value of α is about the number of terms in one sentence (i.e., about 30 terms). Therefore, we expect that properly setting α should not be a difficult task in the domain of disease factor extraction from medical texts. We will verify the hypothesis in the case studies to be reported in “Two Case Studies.”
Consolidating proximity information from multiple texts: To estimate the proximity score of a candidate factor u, EDFI consolidates the information from other candidate factors by measuring how the other candidate factors ever appeared around u in some medical text (Step 4.2 in ). Therefore, EDFI considers term proximity in multiple texts, which is not considered by previous techniques. As noted in “Main Challenges and Related Work,” previous proximity-based techniques mainly aimed at improving text retrieval and classification (rather than the extraction of disease factors), and hence, often-collected term proximity information from individual documents (rather than viewing multiple documents as a whole to collect proximity information for a factor term). In the case studies reported in “Two Case Studies,” we will show that the consolidation of term proximity information from multiple texts is helpful for the disease factor extraction problem, because key factors for a disease often cannot be completely described in one single medical text.
Collaborating with the feature (factor) selection technique (Challenge 3 in ): To associate EDFI with feature selection techniques (i.e., factor extractors), the ranks of the candidate factors (produced by the underlying factor extractor) are integrated with the proximity scores produced by EDFI (Step 7 in ), and hence EDFI can seamlessly collaborate with the extractors, without needing to deal with the different scales of discriminating strengths derived by different extractors.
TWO CASE STUDIES
We report two case studies to empirically evaluate the contribution of EDFI. Among the common aspects of diseases noted in “Introduction,” the case studies focus on the diagnosis aspect of diseases. We measure how EDFI further enhances a good discriminative factor extractor in ranking the diagnosis factors described in two real-world sources of medical texts for a broad range of diseases. summarizes the main setup of the case studies, which is to be described in the following subsections.
Data and Resources
Recognition of biomedical terms is a basic step of factor extraction, because a factor is actually expressed with a valid biomedical term. It is also known as the problem of named entity recognition, and several previous studies focused on the recognition of new terms not included in a dictionary (e.g., Takeuchi and Collier Citation2005; Zhou et al. Citation2004). In the case studies, because we do not aim at dealing with new terms, we employ a popular and up-to-date dictionary of medical terms MeSH (medical subject headings)Footnote7 to recognize medical terms in medical texts. By including all terms and their retrieval equivalence terms in MeSH,Footnote8 we have a dictionary of 164,354 medical terms, which should be able to cover most diagnosis factor terms. A term may be a candidate factor term only if it is listed in the dictionary, making the systems able to focus on valid medical terms.Footnote9
TABLE 3 Setup of Two Case Studies to Evaluate EDFI in Ranking Diagnosis Factors
To evaluate EDFI, we need experimental data that includes a set of medical texts in which diagnosis factors for individual diseases have been manually extracted as the target factors. With such data, we can evaluate whether EDFI can help to rank the target factors higher. Because there is no such data built as a benchmark, we collect medical texts from MedlinePlus and MedicineNet, which aim at providing reliable and up-to-date information about diseases. Each medical text is for a specific disease. We recruit a university graduate, who majors in and teaches English language, to carefully read the content of each medical text and manually extract (from the text) the target diagnosis factors. The manual extraction aims at extracting those factors that are believed (by the authors of the text) to be related to the disease (rather then judging whether the factors are really related to the disease). Another person is recruited to further cross-check the result of the manual extraction. Similarly, on MedlinePlus, we examine all the diseases listed, and download the texts that MedlinePlus tags as diagnosis/symptoms texts for the diseases.Footnote10
On MedicineNet, we identify 74 diseases that are listed in MedlinePlus, and manually extract medical texts for the diseases.
A few steps are conducted to preprocess the medical terms, including transforming all characters into lower case, replacing non-alphanumeric characters and “s” with a space character, removing stop words,Footnote11 and performing word stemming.Footnote12 Moreover, the dictionary noted above is used to recognize medical terms. We then remove those texts in which no diagnosis factors terms are extracted as targets and those diseases that do not have any texts with target diagnosis factors terms. Therefore, in MedlinePlus data, we have 420 medical texts for 133 diseases,Footnote13 with 3,335 target diagnosis factor terms for the diseases, and in MedicineNet data, we have 74 medical texts for 74 diseases, with 1,482 target diagnosis factor terms for the diseases. The data may thus be used to comprehensively evaluate EDFI on a broad range of diseases using different sources of medical texts.
The Underlying Discriminative Factors Extractor
We implement χ2 (chi-square) as the underlying factor extraction technique. It is a technique that was routinely employed (e.g., Himmel, Reincke, and Michelmann Citation2009; Liu Citation2007; Yang and Pedersen Citation1997) and shown to be one of the best feature (factor) scoring techniques (Yang and Pedersen Citation1997). For a term t and a category (disease) c, χ2(t, c) = [N × (A × D - B × C)2]/[(A + B) × (A + C) × (B + D) × (C + D)], where N is the total number of documents, A is the number of documents that are in c and contain t, B is the number of documents that are not in c but contain t, C is the number of documents that are in c but do not contain t, and D is the number of documents that are not in c and do not contain t. Therefore, χ2(t, c) indicates the strength of correlation between t and c. We say that t is positively correlated to c if A × D > B × C; otherwise t is negatively correlated to c. A term may be treated as a candidate factor term for c only if it is positively correlated to c. By observing how the candidate factors are ranked before and after EDFI is applied (named χ2 and χ2 + EDFI, respectively), we can measure the contribution of EDFI to χ2.
Evaluation Criterion
Both the underlying discriminative factor extractor (i.e., χ2) and the enhanced version of the extractor (i.e., χ2 + EDFI) produce strengths for candidate factor terms, and hence produce two lists of ranked terms. Therefore, by measuring how target diagnosis factor terms appear in the two ranked lists, we can evaluate the quality of the strength estimations by χ2 and χ2 + EDFI, and accordingly measure the contribution of EDFI to the underlying discriminative factor extractor. Therefore, Mean Average Precision (MAP) is employed as the evaluation criterion:
Note that when computing P(i) for a disease, we employ the number of target terms (i.e., k) as the denominator (rather than m, which is the number of target terms in the ranked list), making it able to consider the percentage of target terms in the ranked list (i.e., a sense similar to recall). Therefore, if the ith disease has many target terms but only very few of them are in the ranked list, P(i) will be quite low no matter how the few terms are ranked in the list.
Moreover, to validate whether χ2 + EDFI performs significantly better than χ2, we conduct a two-tailed and paired t-test with a 95% confidence level. The significance test is conducted on the average precision (i.e., P(i) or AP) values on all diseases (i.e., 133 diseases on MedlinePlus and 74 diseases on MedicineNet).
Results
and show the MAP performance of the factor extractors (χ2 and χ2 + EDFI) on MedlinePlus and MedicineNet, respectively. The results show that EDFI can contribute statistically significant improvements to χ2 under all settings of the parameter α on both MedlinePlus and MedicineNet. Even when α is not well set for EDFI (e.g., α is set to 0), χ2 + EDFI significantly performs better than χ2 as well. Given that χ2 is one of the best factor scoring techniques, the enhancement provided by EDFI is of both practical and technical significance: existing feature scoring techniques may be further enhanced to extract disease factors in the medical domain.
FIGURE 2 Results on MedlinePlus: EDFI successfully further enhances the baseline extractor (χ2) to achieve significantly better performance under all settings of α (‘![]()
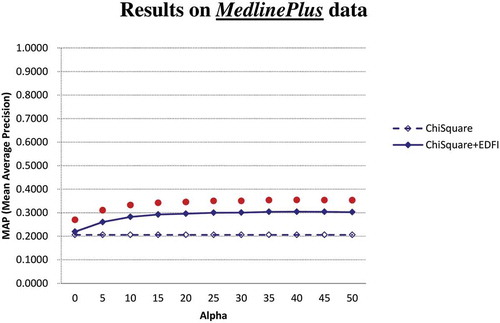
FIGURE 3 Results on MedicineNet: EDFI successfully further enhances the baseline extractor (χ2) to achieve significantly better performance under all settings of α (‘![]()
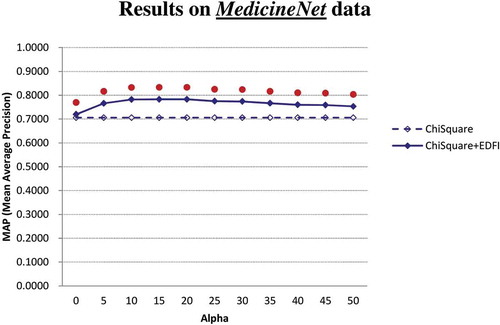
On MedlinePlus, the MAP achieved by χ2 is 0.2060, and with the parameter α set to 30, the MAP achieved by χ2 + EDFI is 0.3005, indicating that EDFI contributes 45.9% MAP improvement to χ2 (0.3005 vs. 0.2060) on MedlinePlus. Moreover, on MedicineNet both χ2 and χ2 + EDFI achieve much higher MAP than themselves on MedlinePlus. A detailed analysis shows that the large MAP difference is due to the fact that there are more candidate factors in the medical texts from MedlinePlus than from MedicineNet. Therefore, fewer candidate factors are ranked in MedicineNet, leading to higher MAP on MedicineNet. The results also show that, even when the underlying extractor has been able to rank the candidate factors well (i.e., achieving MAP of 0.7058 by χ2 on MedicineNet), EDFI may significantly further enhance the extractor under all settings of the parameter α. When α is set to 30, MAP achieved by χ2 + EDFI is 0.7739, indicating that EDFI contributes 9.6% MAP improvement to χ2 (0.7739 vs. 0.7058) on MedicineNet.
To illustrate the contribution of EDFI, consider the disease “parasitic diseases” that is of the disorder type of infections, and there are two texts for it on MedlinePlus. Average precision (i.e., P(i) in Equation (1) for the disease achieved by χ2 and χ2+EDFI are 0.2535 and 0.3933, respectively. When compared with χ2, χ2 + EDFI successfully promotes the ranks of several target diagnosis factors such as “parasite,” “diarrhea,” and “MRI scan,” as they appear at some place(s), whereas more other candidate terms occur in a nearby area. The proximity information employed by EDFI is thus helpful. However, χ2 + EDFI also fails to promote the ranks of a few target diagnosis factors such as “serology.” A detailed analysis shows that serology appears at only one place where the author of the text used several terms to explain serology with very few candidate terms appearing in the nearby area. Therefore, although EDFI has helped χ2 to achieve higher MAP on parasitic diseases, proximity information employed by EDFI cannot always be helpful for some target factors.
As another example, consider the disease “cervical cancer” that is of the disorder type of cancers, and there are two texts for it on MedlinePlus. Average precision for the disease achieved by χ2 and χ2 + EDFI are 0.0879 and 0.3448, respectively. The ranks of several target factors such as “colposcopy” and “vagina” Are promoted by χ2 + EDFI. The results verify the contribution of the strategy of EDFI in collaborating and enhancing the underlying factor extractor.
We are also concerned with the proper setting of the parameter α employed by EDFI. lists the MAP achieved by χ2 + EDFI with α set in the range of [0, 50]. We also conduct a significance test to check whether the performance of χ2 + EDFI with α = 30 is significantly different from χ2 + EDFI with α ≠ 30. The result shows that (1) on both MedlinePlus and MedicineNet, χ2 + EDFI with α = 30 does not perform significantly different from χ2 + EDFI with α = 20 and χ2 + EDFI with α = 25, (2) on MedlinePlus, χ2 + EDFI with α ≤ 15 performs significantly worse than χ2 + EDFI with α = 30, and (3) on MedicineNet, χ2 + EDFI with α ≥ 35 performs significantly worse than χ2 + EDFI with α = 30. The result thus justifies the hypothesis noted in “Behavior Analysis for EDFI”: As the factors for a disease are often collectively described in one to two sentences in a medical text, a proper value of α is about the number of terms in one sentence (i.e., about 30 terms). The proper setting of α for EDFI is thus not a difficult task in the domain of disease factor extraction from medical texts.
TABLE 4 MAP Performances of χ2 + EDFI under Different α Settings with Significance Tests with Respect to the One with α = 30: A Proper Range of α Should Be [20, 30] as There Is no Statistically Significant Differences in the Performance of the One with α = 30 and the Ones with α = 20 or 25 on Both MedlinePlus and MedicineNet, Justifying the Hypothesis That a Proper Value of α Is about the Number of Terms in a Sentence (ref. “Behavior Analysis for EDFI”) and Making the Setting of α Easier in Practice
CONCLUSION AND FUTURE WORK
Factors of a broad range of diseases are a fundamental basis for those knowledge-based systems that support healthcare decision making (e.g., an expert system to support the diagnosis and treatment of diseases) and education (e.g., an exploration system to support the learning of differential diagnosis knowledge). The identification of the disease factors requires the support provided by a text mining technique because (1) the disease factors should be discriminative among the diseases, (2) the disease factors may evolve as medical findings evolve, and (3) medical findings are often recorded in texts. We thus develop a technique, EDFI, that enhances factor extraction techniques by term proximity information, without needing to change the development and behaviors of the techniques. Two case studies on real-world medical texts verifies the contribution of EDFI, which is based on the observation that authors of a medical text for a disease tend to describe the factors of the disease in a nearby area of the text.
Based on the contribution, we are exploring several interesting issues, including automatic collection of medical texts for disease factor extraction and visualization of the disease factors for online interactive exploration. Moreover, because the disease factors extracted by EDFI are actually a simplified form of the knowledge base for disease differentiation, they should be further annotated with additional information, including (1) the preconditions of each disease factor (e.g., time and duration of a symptom, as well as the age and the gender of the patients with the symptom), and (2) the intrinsic properties of the disease-factor relationships (e.g., the negative relationship such as “symptom A is never seen in disease B, even though symptom A can be used for differential diagnosis of disease B”). We are thus interested in computer-supported annotation and validation of the disease factors (extracted by EDFI) so that more informative relationships among diseases can be constructed and shared among users and systems in various medical applications.
FUNDING
This research was supported by the National Science Council of the Republic of China under the grant NSC 99-2511-S-320-002, and by Tzu Chi University under the grant TCRPP99003.
Notes
1 MeSH is available at http://www.nlm.nih.gov/mesh/
2 Available at http://www.nlm.nih.gov/medlineplus/healthtopics.html.
3 Available at http://www.medicinenet.com.
4 MetaMap is available at http://metamap.nlm.nih.gov/.
5 cTAKES is available at http://ctakes.apache.org/.
6 EDFI does not directly employ the discriminating strength of u, because different discriminative factors extractors might produce different scales of strengths, making their integration with the proximity score more difficult.
7 We employ 2011 MeSH as the dictionary, which is available at http://www.nlm.nih.gov/mesh/filelist.html.
8 Retrieval equivalence terms of a term are from the field of “entry” for the term, because they are equivalent to the term for the purposes of indexing and retrieval, although they are not always strictly synonymous with the term (ref. http://www.nlm.nih.gov/mesh/intro_entry.html).
9 Actually, MeSH provides a tree (http://www.nlm.nih.gov/mesh/trees.html) containing various semantic types that are related to diagnosis factors, including “Phenomena and Processes” (tree node G), “diagnosis” (tree node E01), and “disease” (tree node C). However, diagnosis factor terms could still be scattered around the MeSH tree. For example, the symptom term “delusions” is under the MeSH category of Behavior and Behavior Mechanisms (tree node F01) χ Behavior χ Behavioral Symptoms χ Delusions. Therefore, we include all MeSH terms into the dictionary used in the case study.
10 The texts are collected by following the sequence: MedlinePlus → “Health Topics” → Items in “Disorders and Conditions” → Specific diseases → Texts in “diagnosis/symptoms.” We skip those diseases that have no “diagnosis/symptoms” texts tagged by MedlinePlus.
11 A stop word list by PubMed is employed: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helppubmed&part=pubmedhelp&rendertype=table&id=pubmedhelp.T43
12 We employ the Porter stemming algorithm, which is available at http://tartarus.org/˜martin/PorterStemmer/.
13 The 133 diseases fall into 7 types of disorders and conditions: cancers; inflections; mental health and behaviors; metabolic problems; poisoning, toxicology, environmental health; pregnancy and reproduction; and substance abuse problems.
REFERENCES
- Cohen, W. W., and Y. Singer. 1996. Context-sensitive mining methods for text categorization. In Proceedings of the 19th annual international ACM SIGIR conference on research and development in information retrieval, 307–315. New York, NY: ACM Press.
- Cummins, R., and C. O’Riordan. 2009. Learning in a pairwise term-term proximity framework for information retrieval. In Proceedings of the 32nd annual international ACM SIGIR conference on research and development in information retrieval, 251–258. Boston, MA, USA: ACM Press.
- Domedel-Puig, N., and L. Wernisch. 2005. Applying GIFT, a gene interactions finder in text, to fly literature. Bioinformatics 21: 3582–3583.
- Gerani S., M. J. Carman, and F. Crestani. 2010. Proximity-based opinion retrieval. In Proceedings of the 33rd annual international ACM SIGIR conference on research and development in information retrieval, 403–410. Geneva, Switzerland, July 19–23.
- Himmel, W., U. Reincke, and H. W. Michelmann 2009. Text mining and natural language processing approaches for automatic categorization of lay requests to web-based expert forums. Journal of Medical Internet Research 11(3):e25.
- Kim, S., J. Yoon, and J. Yang. 2008. Kernel approaches for genic interaction extraction. Bioinformatics 24: 118–126.
- Lin, J., and D. Demner-Fushman. 2006. The role of knowledge in conceptual retrieval: A study in the domain of clinical medicine. In Proceedings of the 29th annual international ACM SIGIR conference on research and development in information retrieval, 99–106. New York, NY: ACM.
- Liu, R.-L., and Y.-C. Huang 2011. Ranker enhancement for proximity-based ranking of biomedical texts. Journal of the American Society for Information Science and Technology 62(12):2479–2495.
- Liu, R.-L. 2007. Text classification for healthcare information support. In Proceedings of the 20th international conference on industrial, engineering & other applications of applied intelligent systems, 44–53. Kyoto University, Kyoto, Japan, June 26–29.
- Liu, R.-L. 2010. Context-based term frequency assessment for text classification. Journal of the American Society for Information Science and Technology 61(2):300–309.
- Lv, Y., and C. Zhai. 2009. Positional language models for information retrieval. In Proceedings of the 32nd annual international ACM SIGIR conference on research and development in information retrieval, 299–306. New York, NY: ACM.
- Mladeniá D., J. Brank, M. Grobelnik, and N. Milic-Frayling. 2004. Feature selection using linear classifier weights: Interaction with classification models. In Proceedings of the 27th annual international ACM sigir conference on research and development in information retrieval, 234–241. New York, NY: ACM.
- Ono, T., H. Hishigaki, A. Tanigami, and T. Takagi. 2001. Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics 17:155–161.
- Özgür, A., T. Vu, G. Erkan, and D. R. Radev. 2008. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics 24:i277–i285.
- Peng, F., and D. Schuurmans. 2003. Combining naive Bayes and n-Gram language models for text classification. In Advances in Information Retrieval: Proceedings of the 25th European Conference on IR Research (ECIR 2003), LNCS 2633: 335–350. Berlin, Heidelberg, Springer.
- Rasolofo, Y., and J. Savoy. 2003. Term proximity scoring for keyword-based retrieval systems. In Advances in Information Retrieval: Proceedings of the 25th European Conference on IR Research (ECIR 2003), LNCS 2633: 207–218. Berlin, Heidelberg, Springer.
- Schriml, L. M., C. Arze, S. Nadendla, T.-W. W. Chang, M. Mazaitis, V. Felix, G. Feng, and W. A. Kibbe. 2012. Disease ontology: A backbone for disease semantic integration. Nucleic Acids Research 40(D1): D940–D946.
- Suebnukarn, S., and P. Haddawy. 2006. Modeling individual and collaborative problem-solving in medical problem-based learning. User Modeling and User-Adapted Interaction 16:211–248.
- Svore, K. M., P. H. Kanani, and N. Khan. 2010. How good is a span of terms? Exploiting proximity to improve web retrieval. In Proceedings of the 33rd annual international ACM SIGIR conference on research and development in information retrieval, 154–161. New York, NY: ACM.
- Takeuchi, K., and N. Collier. 2005. Bio-medical entity extraction using support vector machines. Artificial Intelligence in Medicine 33:125–137.
- Tao, T., and C. Zhai. 2007. An exploration of proximity measures in information retrieval. In Proceedings of the 30th annual international ACM SIGIR conference on research and development in information retrieval, 23–27. New York, NY: ACM.
- Temkin, J. M., and M. R. Gilder. 2003. Extraction of protein interaction information from unstructured text using a context-free grammar. Bioinformatics 19:2046–2053.
- Wang, X., A. McCallum, and X. Wei. 2007. Topical n-grams: Phrase and topic discovery, with an application to information retrieval. In Proceedings of the IEEE 7th International Conference on Data Mining, 697–702. IEEE Conference Publications.
- Yang, Y., and J. O. Pedersen 1997. A comparative study on feature selection in text categorization. In Proceedings of the 14th International Conference on Machine Learning 1997), 412–420. San Mateo, CA: Morgan Kaufmann.
- Zhao, J., and Y. Yun. 2009. A proximity language model for information retrieval. In Proceedings of the 32nd annual international ACM SIGIR conference on research and development in information retrieval, 291–298. Boston, MA USA: ACM.
- Zhou, G., J. Zhang, J. Su, D. Shen, and C. Tan. 2004. Recognizing names in biomedical texts: A machine learning approach. Bioinformatics 20:1178–1190.