?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
De-identification of electronic health records (EHR) is a vital step toward advancing health informatics research and maximizing the use of available data. It is a two-step process where step one is the identification of protected health information (PHI), and step two is replacing such PHI with surrogates. Despite the recent advances in automatic de-identification of EHR, significant obstacles remain if the abundant health data available are to be used to the full potential. Accuracy in de-identification could be considered a necessary, but not sufficient condition for the use of EHR without individual patient consent. We present here a comprehensive review of the progress to date, both the impressive successes in achieving high accuracy and the significant risks and challenges that remain. To best of our knowledge, this is the first paper to present a complete picture of end-to-end automatic de-identification. We review 18 recently published automatic de-identification systems -designed to de-identify EHR in the form of free text- to show the advancements made in improving the overall accuracy of the system, and in identifying individual PHI. We argue that despite the improvements in accuracy there remain challenges in surrogate generation and replacements of identified PHIs, and the risks posed to patient protection and privacy.
Introduction
The application of machine learning research using EHR has the potential to revolutionize health care. There is an abundance of health data available and maximizing the utility of this data will result in improving health care, especially in patient care, medical outcomes, surgical outcomes, risk prediction, clinical decision support and medical diagnosis.
Use of patient data typically requires individual patient consent. For research, without individual consent, the data must be de-identified such that the patient’s identity or privacy is not breached. Obtaining individual patient consent for massive datasets is time-consuming and is a challenging task. Hence there is a great interest in automating the de-identification process such that EHR can be used in research to improve the health care and quality of patient care without compromising the identity of the patient.
There is growing interest internationally in applying big data techniques to electronic health records. However, privacy laws in many jurisdictions — including New Zealand’s Health Information Privacy Code and the United States Health Insurance Portability and Accountability Act (HIPAA) – require accurate de-identification of medical documents (such as discharge summaries and electronic health records) before they can be shared outside of their originating institutions.
The sharing of records is crucial for advancing health research. For example, the 2014 Heart Disease Risk Factors Challenge involved participating research groups attempting to predict heart disease risk factors in diabetic patients from longitudinal clinical narratives. As noted above, such a challenge would not have been possible under United States law if the narratives (1,304 medical records from 296 diabetic patients) were not de-identified first. In this case, the records were de-identified manually by multiple medical professionals. Since most institutions will not be able to afford the costs of manual de-identification, automating the process is crucial therefore for sharing data and advancing health research.
We present our findings in three main groups: achievements, challenges, and risks, associated with generating an automatic de-identification. Achievements of automatic de-identification primarily focus on identification of PHI in EHR. Challenges are associated with the surrogate generation and replacement. Risks outline the issues relating to re-identification and medical correctness and usability of de-identified data.
The rest of the paper is structured such that a brief background on de-identification is presented in Section 2, achievements of recent de-identification systems in Section 3, challenges in Section 4, risks in Section 5 and finally a discussion in Section 6.
Background on De-Identification
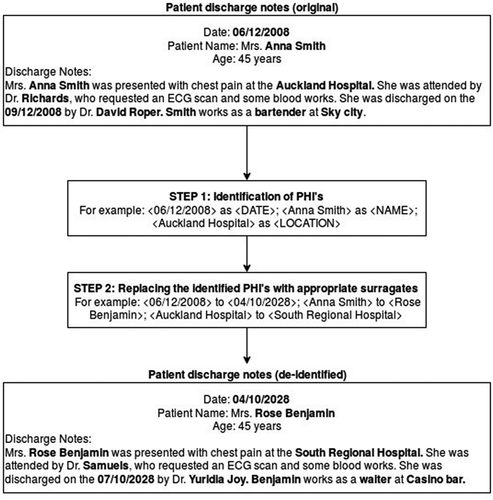
De-identification is a two-step process where PHI is identified in EHR and replaced with suitable surrogates such that patient privacy and confidentiality is not at risk. provides a detailed example of de-identification of EHR, where original patient discharge notes are de-identified.
This figure also outlines the two-step de-identification process. It is important to note that step two requires the use of appropriate surrogates to replace the original PHI and hence automating surrogate generation is a vital step in creating a longitudinal narrative end-to-end automatic de-identification system. Although EHR is in the form of tabular structures (i.e. tables), free-form narratives, and images, this study focuses on medical data in the free form longitudinal text.
De-identification should be considered a means of satisfying rather than circumventing the legal and ethical requirements created to protect patient privacy across the world. Individual countries have different requirements, for example HIPAA in the United States of America (Garfinkel (Citation2015); Stubbs et al. (Citation2015a); Yogarajan, Mayo, and Pfahringer (Citation2018b)), the European Union’s new General Data Protection Regulation (GDPR) (Brasher (Citation2018); Polonetsky, Tene, and Finch (Citation2016)), and New Zealand’s own health information privacy code (Health & Disability Commissioner (Citation2009); Office of the Privacy Commissioner (Citation2013); Yogarajan, Mayo, and Pfahringer (Citation2018a)). HIPAA is arguably the gold standard, and both HIPAA’s regulations on Expert Determination and Safe Harbor are used as the standard benchmarks for de-identification of EHR in the form of free text. We use HIPAA’s Safe Harbor guidelines as the basis of assessing the accuracy of the de-identification systems (for details on HIPAA’s Safe Harbor and the 18 categories see Yogarajan, Mayo, and Pfahringer (Citation2018b)).
A superior de-identification system will not only meet legal requirements but will also help build societal consent by assuring the public that their privacy and medical data will be protected. This consent is vital if large-scale research involving medical records is to be accepted in the same way as, for example, Statistics New Zealand’s Integrated Data Infrastructure. Acceptance of the latter is arguably in part due to measures were taken by Statistics New Zealand to de-identify data (Ragupathy and Yogarajan (Citation2018); Statistics New Zealand (Citation2016)).
Achievements
In the recent years there has been a substantial development in natural language processing tasks, including de-identification, primarily due to the development in deep learning (Dalianis (Citation2018); Goldberg (Citation2017)). Improving accuracy of de-identification of EHR – step 1 from – has been the primary focus of research in this field, and several de-identification systems have achieved remarkable success. The main reason for such development is the EHR de-identification competitions (Kumar et al. (Citation2015); Stubbs, Filannino, and Uzuner (Citation2017); Stubbs et al. (Citation2015); Stubbs and Uzuner (Citation2015c, Citation2017); Uzuner and Stubbs (Citation2015)). For a complete review of these competitions and significance see Yogarajan, Mayo, and Pfahringer (Citation2018b). It is important to note that these competitions provide open access data which allowed the research to grow rapidly. In addition to these competition datasets, the MIMIC dataset (Goldberger et al. (Citation2000); Johnson et al. (Citation2016)) is another open access dataset that has been used to develop de-identification systems.
In this section, we outline the most significant achievement of automating end-to-end de-identification system: improving accuracy. It has been argued that as far as de-identification is concerned, perfection cannot be achieved; however, 95% accuracy is considered to be the rule of thumb and universally accepted value (Stubbs, Filannino, and Uzuner (Citation2017); Stubbs, Kotfila, and Uzuner (Citation2015)). We use 18 de-identification systems, as outlined in , to show that several of these systems have achieved an overall F-measure of 0.95. Also, we outline the fact that these systems have also identified the majority of the HIPAA PHI with the F-measure of
0.95. These achievements have been a significant milestone in automating end-to-end de-identification of EHR and has been a significant breakthrough in this area of research.
Table 1. De-identification systems summary. Machine learning indicates systems that uses machine learning techniques only. Hybrid systems indicates systems that used a combination of machine learning techniques and hand crafted rules.
This section will be structured such that a brief overview of the datasets will be provided. This is followed by an outline of the systems that obtained an overall F-measure of 0.95, and also a summary of systems that recorded F-measure of
0.95 for individual PHIs. The techniques and datasets used for these results are also outlined.
Overview of Datasets
In this section, we provide a quick overview of the most commonly used datasets by the 18 de-identification systems outlined in . The most commonly used datasets were introduced by the following three competitions: the 2006 Informatics for Integrating Biology and the Bedside (i2b2) competition (Uzuner, Luo, and Szolovits (Citation2007)); the 2014 i2b2/UTHealth shared task (Stubbs et al. (Citation2015); Stubbs and Uzuner (Citation2015c)); and the 2016 Centers of Excellence in Genomic Science (CEGS) and Neuropsychiatric Genome-Scale and RDOC Individualized Domain (N-GRID) shared task (Stubbs, Filannino, and Uzuner (Citation2017); Stubbs and Uzuner (Citation2017)).
The dataset for the 2006 competition included 889 unannotated discharge summaries, also used for smoking challenges, manually broken into sentences and tokenized. The dataset for the i2b2/UTHealth shared task 2014 included 2—5 records for each patient over a fixed period and was obtained from two large academic tertiary hospitals: Massachusetts General Hospital (MGH), and Brigham and Women’s Hospital (BWH) . It includes 296 diabetics patients with 1304 longitudinal medical records and contains three cohorts based on the diagnosis of coronary artery disease (CAD) (Kumar et al. (Citation2015); Stubbs et al. (Citation2015); Stubbs and Uzuner (Citation2015a, Citation2015c))).
The 2016 CEGS N-GRID shared task used psychiatric data, making it the first ever competition to use psychiatric intake records (Lee et al. (Citation2017); Stubbs, Filannino, and Uzuner (Citation2017)). The data for the 2016 competition reflected the records “as is” (Stubbs, Filannino, and Uzuner (Citation2017); Uzuner, Stubbs, and Filannino (Citation2017)): the state at which data was received from the sources. Unlike other medical data, such as that of the 2014 challenge, psychiatric data contains an abundance of information related to the patients such as places lived, jobs held, children’s ages, hobbies, traumatic events, patients’ relatives’ relationship information, and pet names. This makes it a much more significant challenge to de-identify (Bui, Wyatt, and Cimino (Citation2017b); Stubbs, Filannino, and Uzuner (Citation2017)).
MIMIC III is one the most extensive publicly available database (Goldberger et al. (Citation2000); Johnson et al. (Citation2016)). It contains health records of approximately sixty thousand admissions of patients in critical care units. The database includes information such as demographics, laboratory test results, procedures, medications, and physician notes.
Also, there were other data used by individual systems such as Chinese data by S1 and Dutch data by Menger et al. (Citation2018). Although we do not describe these systems in this paper, it is important to note that these systems also presented with high F-measure.
Overall F-Measure of De-Identification System
presents the de-identification systems that recorded an overall F-measure of 0.95. Each entry also outlines the datasets used to obtain such results. The highest recorded overall F-measure was obtained by S3 and S9 using MIMIC III dataset. One possible reason for such high accuracy obtained using MIMIC III dataset could be due to the duplicates created by cut and paste (Gabriel et al. (Citation2018)). The i2b2 2014 is the most commonly used dataset. It is important to point out S7 as the best performing system from the actual i2b2 2014 competition. As shown in there has been a substantial improvement in F-measure since the 2014 competition. Unfortunately, this might partly be due to overfitting of the now known and freely available test set.
Table 2. De-identification systems with overall F-measure 0.95.
provides an outline of the techniques used by the de-identification systems in . Machine learning only systems favor deep learning approaches. Hybrid systems with the incorporation of handcrafted rules and dictionary-based approaches are also used by a couple of the de-identification systems to achieve high F-measure.
Table 3. De-identification systems with overall F-measure 0.95.
F-Measure of Individual PHIs
In this section, we provide an overview of the systems that recorded F-measure 0.95 for individual HIPAA PHIs. Where the F-measure was
0.95, the highest recorded score is presented. We also provide some possible issues relating to the PHIs that have lower F-measure. This section also provides an overview of the i2b2 PHIs. These are the additional PHIs introduced by the i2b2 2014 and 2016 competitions (Stubbs et al. (Citation2015); Stubbs and Uzuner (Citation2015a, Citation2015c)). Although legally, as per HIPAA rules, there are no requirements for these additional PHIs to be de-identified, the competition organizers argue that these extra PHIs provide more security over re-identification of data. Since i2b2 2014 and 2016 datasets are most commonly used in the advancement of de-identification research, we feel it is vital to also present the successes in these additional PHIs.
provides an overview of the systems that achieved high F-measures. It also outlines the datasets that were used to obtain such results. Except for Fax and Device all other PHIs have obtained an F-measure of 0.95. This is an incredible achievement and a significant improvement to the results obtained in i2b2 competitions (Yogarajan, Mayo, and Pfahringer (Citation2018b)). Although Fax and Device recorded
0.95 F-measure, it is important to note that only a very few instances (
10) were found in the datasets for both of these PHIs. This makes improving the accuracy using machine learning approaches very hard.
Table 4. F-measure 0.95 for HIPAA categories for de-identification. On occasions where F-measure was not
0.95, the highest score is presented. CONTACT: URL and IP address; ID: BioID, Healthplan, Social Security no, and Vehicle license plate no; Face photo; and Any other unique code are PHIs that were not present in any of the dataset, hence not included here.
provides an overview of the techniques used to obtain the F-measures presented in . As observed in there is a clear increase in deep learning methods. With a combination of handcrafted rules, de-identification systems have achieved high F-measure for the majority of the PHI. In several cases, hand-crafted rules only also achieve high F-measure. Good examples are License and E-Mail, where regular expressions work very well.
Table 5. Techniques used for the F-measures presented in for HIPAA PHI categories.
provides an overview of F-measures for i2b2 introduced extra PHIs. These PHIs are not part of the legal requirement as per HIPAA regulations but are additional security for ensuring that patient privacy and confidentiality are maintained. Compared to the recorded results in the i2b2 2014 and 2016 competitions, there is a substantial increase in the F-measure. Clearly, Organization, Location-others, Profession and Country are the PHIs yet to reach the 0.95 F-measure. These were also the PHIs that recorded a very low F-measure in both competitions (see Yogarajan, Mayo, and Pfahringer (Citation2018b) for details). The main issue with Country and Organization is that the data is very sparse. Location-others only occurs in thirteen instances in the dataset. The sparsity of the data and the very low frequencies of same values make achieving higher F-measures very hard. However, there is still an improvement in results compared to that recorded in the competitions.
Table 6. The best F-measure for i2b2 extra categories for de-identification. This table includes categories not included in , but were introduced by i2b2 competitions as additional categories (Stubbs et al. (Citation2015); Stubbs and Uzuner (Citation2015a)). It also provides the techniques used to achieve these F-measures.
In Summary
This section showed the achievements in automating de-identification research, with substantial improvement in F-measure of identifying PHI in the overall systems and individual PHIs (notably the HIPAA required PHIs).
Challenges
The biggest challenge in automating end-to-end de-identification is surrogate generation and surrogate replacement (step 2 in ). At first sight, this appears to be superficially simple when compared to step 1. However, when one considers it in detail, there are many complex subtleties associated with the surrogate generation and surrogate replacement. Unlike the research toward increasing accuracy in identifying PHI, as seen in section 3, this is an area where very little research progress has been made. There have been only a few papers published in the recent years regarding surrogate generation and surrogate replacement for the de-identification problem, with the schema developed in the 2014 i2b2 competitions being the prominent one to date (Stubbs and Uzuner (Citation2015b); Stubbs et al. (Citation2015b)).
PHI are categorized into explicit identifiers and quasi-identifiers. Explicit identifiers such as name, phone number and social security number are directly linked to a patient. Quasi-identifiers such as age, gender, race and zip code are not directly connected to a patient but can be linked to external data sources and consequently be used to identify a patient, hence posing the same risk to patient privacy as explicit identifiers.
In this section, we present examples of standard practices used in surrogate replacement and challenges faced. Automation in the surrogate generation is arguably still a very challenging and unsolved problem.
Examples of Surrogate Replacement of PHI
provides an outline of some PHIs and the standard practices used in a surrogate generation while creating de-identified data. Although all of these practices are based on hand-crafted rules and pre-compiled tables, there was also a need to do a manual check after the data is de-identified. This is to ensure that medical correctness, readability and consistency are maintained across the health data. also indicates where manual checking after de-identification was required. Surrogates need to maintain the same form as the original, and where possible same internal temporal and co-reference relationships. Also, as illustrated in semantic links must be maintained, for example between LOCATION and PROFESSION.
Table 7. Common practices used in surrogate generation and replacement of PHI as outlined in (Johnson et al. (Citation2016); Pantazos, Lauesen, and Lippert (Citation2017); Stubbs and Uzuner (Citation2015b); Stubbs et al. (Citation2015b)).
It is important to note that it is relatively easy to create surrogates randomly and maintain co-references for PHONE, FAX, URLs and ID (Stubbs and Uzuner (Citation2015b); Stubbs et al. (Citation2015b)). Any ambiguous words appearing as part of a name, medical term or acronym were replaced using a set of hand-crafted rules (Pantazos, Lauesen, and Lippert (Citation2017)). This is primarily because, in medicine, it is common to have diseases, signs and symptoms being named after the person first describing it. One such example is “Aaron” which can refer to a name of a person, or be part of a medical term: Aaron sign referring the pain felt in the epigastrium.
Issues and Challenges Due to Surrogate Replacement of PHI
provided an overview of techniques used in the surrogate generation and replacement. However, there are many practical issues with some of these rules and techniques which creates challenges in maintaining medical correctness and usability of de-identified data in health advancement research. Moreover, it is important to note in most cases there was a need to manually check the surrogate replaced data to ensure consistency and accuracy is maintained across patient data.
When DATE is changed to just the year or randomly changed it removes inferrable information such as the “season” which could result in missing any pandemic outbreak (Li and Qin (Citation2017)). There is a need to maintain the semantic link between the LOCATION and DATE to ensure such information is not missed. Also, for PHIs DATE and AGE, medical correctness is a major issue. Birth dates have to be transformed such that the patient age is in a similar age range. Otherwise diagnosis patterns will become inapplicable. For example, a 20-year-old de-identified to be a 60-year-old will cause issues in medical diagnosis.
When LOCATION such as zip code is replaced by random zip codes (even from a pre-compiled list), geographical information is distorted. For example, a patient living in a high socioeconomic area being moved to low decile area, or vice versa, will result in relevant information about the living conditions and life expectancy changing. This could mislead patient diagnosis, or miss vital information in patient care. In addition to socioeconomic issues relating to LOCATION, there is also ethnicity information. For example, in New Zealand, there are parts of the country, such as Northland, where there is known to be a higher population of New Zealand’s indigenous Maori people. If everyone from Northland is moved to another LOCATION or spread across several LOCATIONS, the ethnicity information is also lost in the de-identified data. It is very challenging to ensure such information is not lost without introducing systemic bias toward a sub-population, e.g. Maori people in the New Zealand example above.
With NAME, if the patient’s name, for example, “John”, is replaced by “Jack”, then there is a need to ensure all of his medical records reflect this change. For instance, in addition to the free text data that was replaced, the change must also be made consistently across all of his longitudinal data, including but not restricted to his structured data and medical images. In addition, the name change should also reflect correctly on his family’s medical records, i.e. his wife’s records and his childrens. This does allow the consistency and medical correctness to be maintained in de-identified data (Pantazos, Lauesen, and Lippert (Citation2017)). The need to maintain consistency and medical correctness makes automating de-identification a very challenging task and does require manual checks and inputs (Pantazos, Lauesen, and Lippert (Citation2017); Stubbs et al. (Citation2015b)). Also, in order to maintain readability, a patient name must be replaced by a new name that looks real and consistency should also be taken into consideration. For example, the frequency of the name in a database needs to be consistent. A rare name occurring more frequently will not look real.
One of the many challenges faced in de-identifying medical, free text data is ambiguous words. In many cases, it is challenging to differentiate between an everyday word, medical word and part of the patient name. This may result in errors with surrogate replacement where for instance a medical term is replaced by a person’s name surrogate.
In Summary
This section presented common practices used in surrogate generation and replacement, where most of the techniques rely on hand-crafted rules and pre-compiled tables. We outline some of the important challenges faced in this step of de-identification and argue that automation in surrogacy is still an open question with many obstacles to overcome.
Risks
De-identified data in addition to protecting patient privacy should also meet the following standards: medical correctness, readability and consistency across data (Pantazos, Lauesen, and Lippert (Citation2017)). Risks around de-identification of health data can be classified into two main areas: the risk of re-identification and the risk of losing usability, medical correctness and consistency across data. In this section, we provide a brief overview of these two areas.
Re-Identification
Re-identification is a process where a person’s identity is identified from the de-identified data. This does result in a serious breach of patient privacy and confidentiality. Explicit identifiers such as person’s name and address can be considered obvious identifiers. However, even quasi-identifiers can result in re-identification of a person (Johnson et al. (Citation2018); Li and Qin (Citation2017); Sweeney (Citation2002)). There have been many examples of such occurrences where quasi-identifiers have been matched with external resources to identify patients. For example, it was proven that attributes such as gender, date of birth and zip code could be matched with external sources such as voting data to identify a patient (Li and Qin (Citation2017); Sweeney (Citation2002)). Also, other examples demonstrate that a combination of a small subset of quasi-identifiers, with or without other medical data, may even be enough to identify the individual patient and pose serious threat to patient privacy (El Emam et al. (Citation2006); Mayo and Yogarajan (Citation2019)).
In addition to explicit identifiers and quasi-identifiers, there are also the sensitive attributes such as psychiatric diseases, HIV, and cancer, which patients are not willing to be associated with. Due to the specific nature of these sensitive attributes and the need for special care facility these attributes when combined with other identifiers makes re-identification of a patient much more feasible (Gkoulalas-Divanis, Loukides, and Sun (Citation2014)).
The risk of re-identification is real and can lead to serious breaches of patient privacy and confidentiality. While designing an automatic de-identification system, it is important to consider the re-identification risk and take appropriate measures to minimize such risk. Also, there needs to be transparency in acknowledging such concerns. The main questions when it comes to re-identification are:
What is the accepted level of risk with re-identification?
Who makes that decision, the de-identification system designers, the users or the patients?
There is no easy or correct answer to these questions, but they still need to be considered when designing a de-identification system. There is a need for human input in making such decisions and deciding the boundaries of acceptable risk associated with de-identification of a medical system.
Medical Correctness and Usability of De-identified Data
Maintaining medical correctness, consistency, readability and usability of data is a difficult problem and the risks associated with this are usually overlooked. Compared to de-identification of structured data, unstructured free text is very challenging. It contains medical information about a patient that needs to be preserved for medical correctness. However, it also contains personal details such as name, phone number, family members names and other personal identifying items. Although the accuracy of identifying PHI in such data has improved considerably, ensuring these PHI are replaced with appropriate surrogates, and medical correctness maintained, is an open question. This poses a great risk in using de-identified data for machine learning based health research, as the de-identified health records may compromise the accuracy and outcome of the resulting model. For example, accidentally replacing a word that resembles a name but is not a name (maybe an abbreviation for a disease, or a disease name itself) can result in readability and medical correctness errors (Pantazos, Lauesen, and Lippert (Citation2017)). The hope is that the original data and the de-identified data of a particular problem will result in the same outcome. However, there is no clear evidence that it does. In reality, the only way to check if it does or does not, is by building models for both versions of the data and comparing them.
Many of the surrogate replacements use randomized identifiers. However, in such cases, the readability and consistency are compromised (Pantazos, Lauesen, and Lippert (Citation2017)). Unless manually checked there is no guarantee these randomly replaced PHI makes much sense in the context and provide useful data outcomes.
Another significant risk is accidentally confusing patients. Lets say you have two patients in the same age range, both named Anne Smith, but one presenting with cancer and the other one with cardiovascular issues. Ensuring these two are kept separated across all of their data, especially longitudinal data, can be very hard. This will require using several PHI to match the person’s identity. However, in this case, there is an increased risk of re-identification. This poses a question around confidentiality vs verifiability, and as a result, increases the risk. This problem cannot be readily solved by using unique identifiers (such as NHI numbers, date of birth or tax numbers) to match narratives, as automated de-identification systems by design prune such de-identifiers. Furthermore, HIPPAs Safe Harbor provision mandates the removal of such unique identifiers (Garfinkel (Citation2015); Stubbs et al. (Citation2015a)). Similarly, New Zealands Privacy Act and Health Information Privacy Code set strict limits on the assignment and use of unique identifiers (Health & Disability Commissioner (Citation2009); Office of the Privacy Commissioner (Citation2013)).
Summary
This area outlined the two main risks associated with de-identification: the risk of re-identification and the risk of losing usability, medical correctness and consistency across data. Minimizing the risk posed to patient privacy and confidentiality is vital. The risk of re-identification must be considered a severe threat when designing a de-identification system. The de-identified data must also maintain medical correctness, readability and consistency. The advancement of health research using de-identified data does rely on the usability of data and medical correctness of data. There is a need for a manual check to ensure that the de-identified data resembles the original data.
Discussion
To best of our knowledge, this is the first paper to present a complete picture of end-to-end automatic de-identification of medical narratives. Noticeably, the majority of the research is done on improving the accuracy of PHI identification in the overall system and of individual PHI. We acknowledge the need for such research, and despite the recent advancements in this area, mainly due to the use of deep learning in natural language processing tasks, there is more room for improvement. At this stage the minimum requirement of 95% F-measure has been met by several systems, but this is only the minimum requirement. There is room for higher F-measures. Also, it will be nice to take these systems to the next level, where in addition to the open access data they use other sources of data. It will be interesting to see the adaptability of these systems.
One of the big downfalls to these systems is that they do not outline the surrogacy generation aspect of de-identification. However, de-identification is not just about identifying the PHI, but is also replacing the identified PHI with appropriate surrogates. As mentioned earlier there is very little research done in this area, and clearly, there are many challenges yet to be overcome. Also, most of the current practices are data specific and use hand-made rules and pre-compiled tables. This is far from achieving full automation in the de-identification problem, and there is an explicit acknowledgment of the need for manual checks after surrogate replacement. We encourage for more research in this area, where the priority is to address some of the challenges outlined in this paper and also to eliminate the need for manual checks.
The importance of automatic de-identification in advancing health research cannot be emphasized enough. However, there is still a need to be aware of the risks associated with designing such systems. This will ensure that risk to patient privacy and confidentiality is minimized while advancing the field of medicine through maximizing the potential of EHR with the use of machine learning techniques. Also, it is vital that the medical correctness, consistency, readability and usability of data are all maintained such that the resulting de-identified data provides the parallel output to that of the original data. It must be pointed out that high accuracy of de-identification is directly proportional to medical correctness. It does become harder to maintain the medical correctness and usability of data when achieving high accuracy becomes the focus. Hence, de-identification of data does become a balancing act where barriers associated with risk and benefits must both be considered. This is another area where there needs to be more research done in proving that de-identified data is providing the same outcomes as the original data.
The challenges and risks associated with de-identification have opened new avenues of research in finding alternatives. One has to ask the question: “What if proper de-identification is impossible?”. Guinney and Saez-Rodriguez (Citation2018) proposes an alternative idea for sharing confidential data called “model to data” where the flow of information between data generators and modelers is reversed. Another idea presented by Vepakomma et al. (Citation2018) proposes a deep learning model which excludes the need to share raw patient data or labels. These are merely examples of alternatives, and are just the beginning of possibly solving the problem of sharing and using EHR without the risk to patient privacy and confidentiality.
References
- Shweta, A. Kumar, A. Ekbal, S. Saha, and P. Bhattacharyya. 2016. A recurrent neural network architecture for de-identifying clinical records. Proceedings of the 13th Intl. Conference on Natural Language Processing, Varanasi, India, 188–97.
- Brasher, E. A. 2018. Addressing the failure of anonymization: Guidance from the European Union’s general data protection regulation. Columbia Business Law Review 209, 2018.
- Bui, D. D. A., M. Wyatt, and J. J. Cimino. 2017a. The UAB informatics institute and 2016 CEGS N-GRID de-identification shared task challenge. Journal of Biomedical Informatics 75:S54–S61. doi:10.1016/j.jbi.2017.05.001.
- Bui, D. D. A., M. Wyatt, and J. J. Cimino. 2017b. The UAB informatics institute and 2016 CEGS N-GRID de-identification shared task challenge. Journal of Biomedical Informatics 75:S54–S61. doi:10.1016/j.jbi.2017.05.001.
- Chen, T., R. M. Cullen, and M. Godwin. 2015. Hidden Markov model using Dirichlet process for de-identification. Journal of Biomedical Informatics 58:S60–S66. doi:10.1016/j.jbi.2015.09.004.
- Dalianis, H. 2018. Clinical text mining: Secondary use of electronic patient records. Switzerland: Springer.
- Dehghan, A., A. Kovacevic, G. Karystianis, J. A. Keane, and G. Nenadic. 2015. Combining knowledge- and data-driven methods for de-identification of clinical narratives. Journal of Biomedical Informatics 58 (Supplement):S53– S59. doi:10.1016/j.jbi.2015.06.029.
- Dernoncourt, F., J. Y. Lee, and P. Szolovits. 2017. NeuroNER: An easy-to-use program for named-entity recognition based on neural networks. CoRR abs/1705.05487.
- Dernoncourt, F., J. Y. Lee, O. Uzuner, and P. Szolovits. 2017. De-identification of patient notes with recurrent neural networks. Journal of the American Medical Informatics Association 24 (3):596–606. doi:10.1093/jamia/ocw156.
- El Emam, K., S. Jabbouri, S. Sams, Y. Drouet, and M. Power. 2006. Evaluating common de-identification heuristics for personal health information. Journal of Medical Internet Research 8 (4):e28. doi:10.2196/jmir.8.4.e28.
- Gabriel, R. A., S. Shenoy, T.-T. Kuo, J. McAuley, and C.-N. Hsu. 2018. The presence of highly similar notes within the MIMIC-III dataset. University of California, San Diego.
- Garfinkel, S. (2015). De-identification of personally identifiable information technical report. National institute of Standards and Technology (NIST), U.S. Department of Commerce.
- Gkoulalas-Divanis, A., G. Loukides, and J. Sun. 2014. Publishing data from electronic health records while preserving privacy: A survey of algorithms. Journal of Biomedical Informatics 50:4–19. doi:10.1016/j.jbi.2014.06.002.
- Goldberg, Y. 2017. Neural network methods for natural language processing. Synthesis Lectures on Human Language Technologies 10 (1):1–309. doi:10.2200/S00762ED1V01Y201703HLT037.
- Goldberger, A. L., L. A. Amaral, L. Glass, J. M. Hausdorff, P. C. Ivanov, R. G. Mark, J. E. Mietus, G. B. Moody, C.-K. Peng, and H. E. Stanley. 2000. PhysioBank, physioToolkit, and physioNet: Components of a new research resource for complex physiologic signals. Circulation 101 (23):e215–e220. doi:10.1161/01.CIR.101.23.e215.
- Guinney, J., and J. Saez-Rodriguez. 2018. Alternative models for sharing confidential biomedical data. Nature Biotechnology 36 (5):391. doi:10.1038/nbt.4128.
- He, B., Y. Guan, J. Cheng, K. Cen, and W. Hua. 2015. CRFs based de-identification of medical records. Journal of Biomedical Informatics 58 (Supplement):S39– S46. doi:10.1016/j.jbi.2015.08.012.
- Health & Disability Commissioner. 2009. The code of rights, health & disability commissioner. Accessed December 09, 2017. http://www.hdc.org.nz/the-act–code/the-code-of-rights.
- Jiang, Z., C. Zhao, B. He, Y. Guan, and J. Jiang. 2017. De-identification of medical records using conditional random fields and long short-term memory networks. Journal of Biomedical Informatics 75:S43–S53. doi:10.1016/j.jbi.2017.10.003.
- Johnson, A. E., T. J. Pollard, L. Shen, H. L. Li-wei, M. Feng, M. Ghassemi, B. Moody, P. Szolovits, L. A. Celi, and R. G. Mark. 2016. MIMIC-III, a freely accessible critical care database. Scientific Data 3:160035. doi:10.1038/sdata.2016.35.
- Johnson, K. W., J. K. De Freitas, B. S. Glicksberg, J. R. Bobe, and J. T. Dudley. 2018. Evaluation of patient re-identification using laboratory test orders and mitigation via latent space variables. Biocomputing 2019, 415-426.
- Kumar, V., A. Stubbs, S. Shaw, and Ö. Uzuner. 2015. Creation of a new longitudinal corpus of clinical narratives. Journal of Biomedical Informatics 58 (Supplement):S6– S10. doi:10.1016/j.jbi.2015.09.018.
- Lee, H.-J., Y. Wu, Y. Zhang, J. Xu, H. Xu, and K. Roberts. 2017. A hybrid approach to automatic de-identification of psychiatric notes. Journal of Biomedical Informatics 75:S19–S27. doi:10.1016/j.jbi.2017.06.006.
- Lee, J. Y., F. Dernoncourt, and P. Szolovits. 2018. Transfer learning for named-entity recognition with neural networks. In Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018), Miyazaki, Japan. European Language Resources Association (ELRA).
- Lee, J. Y., F. Dernoncourt, O. Uzuner, and P. Szolovits. 2016. Feature-augmented neural networks for patient note de-identification. Proceedings of the Clinical Natural Language Processing Workshop (ClinicalNLP), Japan, 17–22.
- Li, X.-B., and J. Qin. 2017. Anonymizing and sharing medical text records. Information Systems Research 28 (2):332–52. doi:10.1287/isre.2016.0676.
- Liu, Z., Y. Chen, B. Tang, X. Wang, Q. Chen, H. Li, J. Wang, Q. Deng, and S. Zhu. 2015. Automatic de-identification of electronic medical records using token-level and character-level conditional random fields. Journal of Biomedical Informatics 58 (Supplement):S47–S52. doi:10.1016/j.jbi.2015.06.009.
- Liu, Z., B. Tang, X. Wang, and Q. Chen. 2017. De-identification of clinical notes via recurrent neural network and conditional random field. Journal of Biomedical Informatics 75:S34–S42. doi:10.1016/j.jbi.2017.05.023.
- Mayo, M., and V. Yogarajan. 2019. A nearest neighbour-based analysis to identify patients from continuous glucose monitor data. In Asian Conference on Intelligent Information and Database Systems, Indonesia, pp. 349-360. Springer, Cham, 2019.
- Menger, V., F. Scheepers, L. M. van Wijk, and M. Spruit. 2018. DEDUCE: A pattern matching method for automatic de-identification of Dutch medical text. Telematics and Informatics 35 (4):727–36. doi:10.1016/j.tele.2017.08.002.
- Office of the Privacy Commissioner. 2013. Health information privacy code 1994. Accessed December 09, 2017. https://www.privacy.org.nz/the-privacy-act-and-codes/codes-of-practice/health-information-privacy-code-1994/.
- Pantazos, K., S. Lauesen, and S. Lippert. 2017. Preserving medical correctness, readability and consistency in de-identified health records. Health Informatics Journal 23 (4):291–303. doi:10.1177/1460458216647760.
- Phuong, N. D., and V. T. N. Chau. 2016. Automatic de-identification of medical records with a multilevel hybrid semi-supervised learning approach. 2016 IEEE RIVF International Conference on Computing & Communication Technologies, Research, Innovation, and Vision for the Future (RIVF), Vietnam, 43–48.
- Polonetsky, J., O. Tene, and K. Finch. 2016. Shades of gray: Seeing the full spectrum of practical data de-intentification. Santa Clara Law Review 56:593.
- Ragupathy, R., and V. Yogarajan. 2018. Applying the reason model to enhance health record research in the age of ‘big data’. The New Zealand Medical Journal 131 (1478):65–67.
- Statistics New Zealand. 2016. Integrated data infrastructure. In Secondary integrated data infrastructure. https://www.stats.govt.nz/integrated-data/integrated-data-infrastructure/
- Stubbs, A., M. Filannino, and Ö. Uzuner. 2017. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID shared tasks Track 1. Journal of Biomedical Informatics 75:S4–S18. doi:10.1016/j.jbi.2017.06.011.
- Stubbs, A., C. Kotfila, and Ö. Uzuner. 2015. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task track 1. Journal of Biomedical Informatics 58 (Supplement):S11– S19. doi:10.1016/j.jbi.2015.06.007.
- Stubbs, A., C. Kotfila, H. Xu, and Ö. Uzuner. 2015. Identifying risk factors for heart disease over time: Overview of 2014 i2b2/UTHealth shared task track 2. Journal of Biomedical Informatics 58 (Supplement):S67– S77. doi:10.1016/j.jbi.2015.07.001.
- Stubbs, A., and Ö. Uzuner. 2015a. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. Journal of Biomedical Informatics 58 (Supplement):S20– S29. doi:10.1016/j.jbi.2015.07.020.
- Stubbs, A., and Ö. Uzuner. 2015b. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. Journal of Biomedical Informatics 58:S20–S29. doi:10.1016/j.jbi.2015.07.020.
- Stubbs, A., and Ö. Uzuner. 2015c. Annotating risk factors for heart disease in clinical narratives for diabetic patients. Journal of Biomedical Informatics 58 (Supplement):S78– S91. doi:10.1016/j.jbi.2015.05.009.
- Stubbs, A., and Ö. Uzuner. 2017. De-identification of medical records through annotation. In Nancy Ide and James Pustejovsky (Eds.) Handbook of linguistic annotation, 1433–59. Netherlands: Springer.
- Stubbs, A., Ö. Uzuner, C. Kotfila, I. Goldstein, and P. Szolovits. 2015a. Challenges in synthesizing surrogate PHI in narrative EMRs, 717–35. Cham: Springer International Publishing.
- Stubbs, A., Ö. Uzuner, C. Kotfila, I. Goldstein, and P. Szolovits. 2015b. Challenges in synthesizing surrogate PHI in narrative EMRs. In Aris Gkoulalas-Divanis and Grigorios Loukides (Eds.) Medical data privacy handbook, 717–35. Switzerland: Springer.
- Sweeney, L. 2002. k-anonymity: A model for protecting privacy. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems 10 (05):557–70. doi:10.1142/S0218488502001648.
- Uzuner, Ö., Y. Luo, and P. Szolovits. 2007. Evaluating the state-of-the-art in automatic de-identification. Journal of the American Medical Informatics Association 14 (5):550–63. doi:10.1197/jamia.M2444.
- Uzuner, Ö., and A. Stubbs. 2015. Practical applications for natural language processing in clinical research: The 2014 i2b2/UTHealth shared tasks. Journal of Biomedical Informatics 58(Suppl):S1. doi:10.1016/j.jbi.2015.10.007.
- Uzuner, Ö., A. Stubbs, and M. Filannino. 2017. A natural language processing challenge for clinical records: Research domains criteria (RDoC) for psychiatry. Journal of Biomedical Informatics 75:S1–S3. doi:10.1016/j.jbi.2017.10.005.
- Vepakomma, P., O. Gupta, T. Swedish, and R. Raskar. 2018. Split learning for health: Distributed deep learning without sharing raw patient data. arXiv preprint arXiv:1812.00564.
- Yadav, S., A. Ekbal, S. Saha, P. S. Pathak, and P. Bhattacharyya. 2017. Patient data de-identification: A conditional random-field-based supervised approach. In Snehanshu Saha, Abhyuday Mandal, Anand Narasimhamurthy, V. Sarasvathi, Shivappa Sangam (Eds.), Handbook of research on applied cybernetics and systems science, 234–53. India: IGI Global.
- Yang, H., and J. M. Garibaldi. 2015. Automatic detection of protected health information from clinic narratives. Journal of Biomedical Informatics 58 (Supplement):S30– S38. doi:10.1016/j.jbi.2015.06.015.
- Yogarajan, V., M. Mayo, and B. Pfahringer. 2018a. Privacy protection for health information research in new zealand district health boards. The New Zealand Medical Journal 131 (1485):19–26.
- Yogarajan, V., M. Mayo, and B. Pfahringer. 2018b. A survey of automatic de-identification of longitudinal clinical narratives. arXiv preprint arXiv:1810.06765.
- Zhao, Y.-S., K.-L. Zhang, H.-C. Ma, and K. Li. 2018. Leveraging text skeleton for de-identification of electronic medical records. BMC Medical Informatics and Decision Making 18 (1):18. doi:10.1186/s12911-018-0598-6.