?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Many of the datasets in real-world applications contain incompleteness. In this paper, we approach the effects and possible solutions to incomplete databases in regression, aiming to bridge a gap between theoretically effective algorithms. We investigated the actual effects of missing data for regression by analyzing its impact in several publicly available databases implementing popular algorithms like Decision Tree, Random Forests, Adaboost, K-Nearest Neighbors, Support Vector Machines, and Neural Networks. Our goal is to offer a systematic view of how missing data may affect regression results. After exhaustive simulation analyzing eight public datasets from UCI and KEEL (Abalone, Arfoil, Bike, California, Compactiv, Mortage, Wankara and Wine), we concluded that the effect of missing data may be significant. The results obtained showed that K-Nearest Neighbors works better than others in the regression of data that has missing data.
Introduction
A considerable number of efficient algorithms for regression have been proposed in the literature (Bishop Citation2016; Deng and Yu Citation2013). Most of them intrinsically assume that the lack of data does not interfere in the analysis of the data sets. In this paper, we approach the effects and possible solutions to incomplete databases in regression, trying to contribute in bridging a gap between algorithms and their real-world applications. Missing data has been the object of study of many contributions that provided different methods, especially in the areas such as database and data integrity (Du, Liu, and Wang Citation2017; Ludtke, Robitzsch, and Grund Citation2017; Peng and Lei Citation2005; Zhang and Wang Citation2017).
In recent years some works have addressed the problem of missing data, such as: proposing a simple sequential method to attempt to identify the form of missingness (Madden et al. Citation2018). A methodology is proposed in Perez-Ruiz and Escarela (Citation2018) for the regression analysis of generalized linear models (GLMs). The results show that the methodology is rather robust and flexible, representing a competitive alternative to traditional techniques; To verify trend analysis of temperature data for the Narayani River basin (Nepal), the Multiple regression and empirical mode decomposition (EMD) methods were applied to fill in missing data and to detect trends (Chand et al. Citation2021); According to Li, Weng, Liao, Keel, & Brown (Citation2021), missing data can impair the analysis of distribution grid impedance and topology estimation in smart grid systems. The authors used the Factorized Ordinary Least Square method to mitigate the error of the missing data. According to Jadhav, Pramod, and Ramanathan (Citation2019) missing data is a common problem faced by researchers and data scientists. The authors compared seven data imputation methods and concluded that the kNN imputation method presented robust results in front of the other techniques, and; a machine learning approach for supporting clinical diagnosis of attention deficit hyperactivity disorder in adults is proposed in Tachmazidis et al. (Citation2021). Several methods such as support vector machine, Logistic regression, Decision Tree, K-Nearest neighbor, Random forest and, Naive Bayes were applied with out treatment of missing data. Therefore, this approach can skew the results. A new approach for handling missing data has been proposed by Ngueilbaye et al. (Citation2021). The new framework called “Module 9” proved to be efficient compared to the others in three known databases that have less than 1500 observations. The authors claim that, as a disadvantage, computational time is high due to the use of machine learning algorithms require a great deal of time for imputations and acquiring a total dataset because of the number of parameters streamlined during training.
Nevertheless, the focus of the majority of these contributions is on techniques to input the missing values (or apply them to databases without any prior treatment) and not enough effort has been dedicated to analyzing the actual effects of missing data for regression. Here, we approached this problem by assigning meager importance on how close the inputted value is from the original one since our major aim is to observe how distinct missing data treatments influence the estimation outcome in different scenarios. With the goal of observing how incomplete data may influence regression tasks, a number of scenarios were investigated in situations with varying degrees of data loss in several publicly available databases. Six popular algorithms were used, namely: Decision Tree (CART), Random Forests, Adaboost and K-Nearest Neighbors (KNN), Multi-Layer Perceptron (MLP) and Linear Support Vector Machine (SVM) (Altman Citation1992; Breiman Citation2001; Breiman et al. Citation1984; Freund and Schapire Citation1995; Haykin Citation1998; Steinwart and Christmann Citation2008), respectively. Of course, the performance of estimation depends, among other factors, on the choice of estimators, on how keen is tuning of the involved parameters and on feature selection.
Although all of these topics were approached in this contribution, our main objective was to carry out an exhaustive study of the effects of missing data, using a reasonable set of datasets using UCI and KEEL databases, which is a not much explored topic in the state-of-the-art. The main contributions of this paper are the introduction of a novel experimental framework to test the effect of missing data; a comparative performance analysis of the main regression models, namely Decision Tree, Random Forests, Adaboost, K-Nearest Neighbors (KNN), Support Vector Machines, and Neural Networks; simulations showing that the effect of missing data may vary significantly depending on a number of factors; the conclusion that KNN tends to provide better results for regression on datasets with missing data. As already discussed analyzing the related works, in general the methods when published suppose an environment, in the unusual real world, of complete data. Here, we are mitigating how baseline methods react differently when the (larger or smaller) amounts of missing data are present in the studied dataset.
As a novelty, this work addresses the different results obtained via chosen missing data imputation methods: (1) with imputation by k nearest neighbors, (2) naive imputation, (3) removal of lines with missing data, (4) removal of columns with missing data, (5) multivariate imputation by chained equations (MICE), and (6) SoftImpute. This characteristic differentiates our framework methodology from the other techniques previously discussed. More specifically, this paper presents the following contributions:
• A new experimental framework to test the effect of missing data was proposed;
• A performance analysis of the main regression models, namely Decision Tree, Random Forests, Adaboost, K-Nearest Neighbors, Support Vector Machines, and Neural Networks was performed;
• The simulation showed that the effect of missing data may vary significantly depending on a number of factors, and;
• The results concluded that KNN works better than others in the regression of data that has missing data.
The work is organized as follows: Section 2 describes the proposed methodology to analyze the effects of missing data. Section 3 shows experiments results obtained. Finally, Section 4 discusses the Conclusions.
Methodology
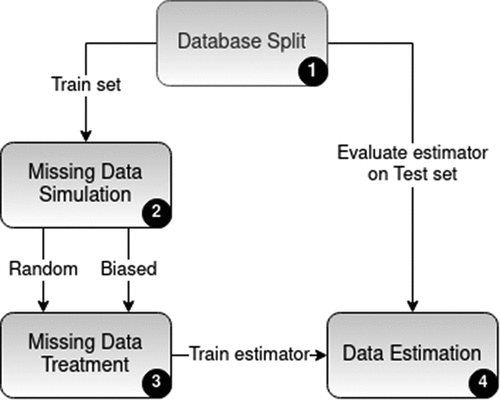
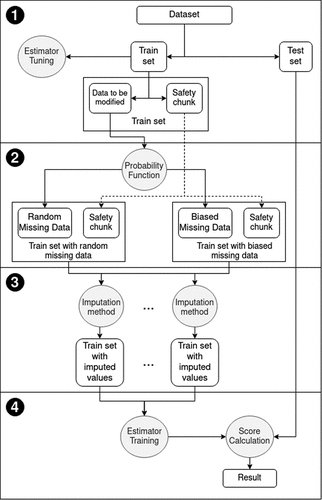
The general experimental framework (please find a flowchart on ) is divided in four main steps, namely: database split, missing data simulation, missing data treatment and data estimation. In the first of those steps, the database is bisected in test and training folds. The test fold is, as usual, reserved for estimations in the unknown population, for inferring the out-of-sample performance, and thus it is not used in any steps in model construction. In order to mimic the real world process of data regression, tuning of the hyper parameters of the regressors was necessary. Therefore, for each regressor-database pair, the tuning of these parameters was performed. The procedure consisted of a random search for the parameters’ values in an uniform distribution. The best values were selected using a 5-fold cross-validation on the training set. The second step consists in simulating missing data occurrence at the training folds by removing part of the data. This was done in two different ways, in a completely random mode and in a biased one. These folds, with missing values, are then used in a third step. As to guarantee an accurate comparison, all original databases had initially no missing data, i.e., all absences are due to artificial removal from step two.
Next steps, three and four, mimic the actions that would be taken when actually training an estimator with an incomplete database (in our experiments, created in step two). The third step consists in fulfilling the databases to produce a filled out dataset that will be later used in step four. For each dataset (with missing values) six different treatments were implemented in step three resulting in six different completed datasets that are followed to step four. Besides that, different sources for missing values generators, fully random and biased, were emulated. In the fourth step, six different estimators were trained using the datasets constructed in the three previous steps. They were also trained with the original complete training fold (without any missing data or treatment) for the sake of comparison and evaluation. The detailed flowchart is presented in . All experiments were run with scikit-learn python library version 0.18.2 Pedregosa et al. (Citation2011).
For each of the different simulated scenarios, steps two, three and four were repeated 100 times and the means and its 95% confidence interval were reported. It is important to highlight that datasets test (out-of-sample) folds were fixed at step one for all the simulations, in order to keep performance results comparable. We follow by describing the different aspects and a thorough explanation of each of these just defined steps.
Basic Definitions and Parameters Values
In order to settle the nomenclature, we define “feature” as being a measurable characteristic of an observation. Observations are entities (persons, things, etc.) of interest and a sample is a set of observations. Samples are subsets of a population to which one has never full access. Samples (data) are used to infer about the population.
We define two variables to simulate missingness in data, %A and %P. The former controls the percentage of features that will have missing information. It is calculated as the closest integer of the percentage of features indicated by %A. For instance, a database with 9 features and %A = would have three of its features containing missing data (
rounded up to 3). This variable allows comparisons among datasets with a different number of features. The latter variable controls the percentage of data removal for a specific feature. Hence, for a specific feature, in a database of
observations(rows) and %P =
, there will be
rows with missing information on this feature. As stated for %A, if the amount of rows indicated by %P is a float, it was rounded to the next integer.
For all experiments in this study we treated features similarly as presented in Santos et al. (Citation2019). Each feature is considered as being independent of each other, i.e., in a univariate manner. For a multivariate yet similar analysis, please refer to Schouten, Lugtig, and Vink (Citation2018). Furthermore, the values of %A and %P were pre-established in simulations. The possible values for %A were: ,
,
,
. We also used four possible values for %P:
,
,
,
. It is important to note that, if the value selected for %A leads to more than one feature having missing information, the drawn %P value will be the same for each feature. Finally, we assumed a general missing pattern in simulations Buuren (Citation2018), which means that the set of rows with missing for each feature is sampled independently per feature.
We used four possible values for %P: 5%, 10%, 20%, and 50%. Six algorithms were used to perform regression, namely: Decision Tree (CART), K Nearest Neighbors, Random Forest, Adaboost, Linear Support Vector Machine (SVM) and Multi Layer Perceptron (MLP) Altman (Citation1992); Breiman (Citation2001); Breiman et al. (Citation1984); Freund and Schapire (Citation1995); Haykin (Citation1998); Steinwart and Christmann (Citation2008). These were used with the implementation of scikit-learn classes AdaBoostRegressor, DecisionTreeRegressor, KNeighborsRegressor, RandomForestRegressor, LinearSVR and MLPRegressor. Six usual methods were used to treat missing data (please find details about these methods in the sequence): Nearest Neighbor Imputation, Naïve Imputation, Multivariate Imputation by Chained Equations (MICE), Soft-Imputation, Discard Rows and Discard Columns. We considered two possibilities concerning the nature of the absence of data: completely at random or in a biased way. When missing data is completely at random, all the observations (rows) have an equal chance of having missing data. In the biased missing, each row of the feature will have a different probability of being missing, depending on its value.
Databases
To perform the analysis, both synthetic and publicly available datasets were used. Synthetic databases were produced to analyze regressions performances after each missing data treatment in a controlled environment. The construction of the synthetic databases has followed the ensuing general procedure: define the number of features in the database () and the number of instances (
) and generate a matrix
where
is a random number in the interval
. Analogously a matrix
, representing the noise of these values is generated with random numbers in the interval
.
Next, a vector of weights is also generated with random values in the interval
. The target value for estimation is defined as:
The matrix , which represents the synthetic database is defined as:
To ensure covering a wide variety of scenarios, synthetic databases with
features and
instances were generated varying the Importance Ratios. To implement this variation,
was set to
and
was varied from
to
in steps of
. The higher the value of
the more dependent the target result is on the first features and the greater the Importance Ratio.
After collecting results in synthetic data, we selected eight real-world datasets to test different characteristics. These characteristics comprised: a number of observations and features, final goal, types of data (discrete, continuous). Real-world databases were taken from UCI and KEEL repositories (Alcala-Fdez et al. Citation2011; Bache and Lichman Citation2013). An overview of each database information can be found in .
Table 1. Detailed database information. Missing analysis in regression. Feature (Feat.), Integer (Int. and Continuous (Cont.).
Six usual methods were used to treat missing data (please find details about these methods in the sequence): Nearest Neighbor Imputation, Naive Imputation, Multivariate Imputation by Chained Equations (MICE), SoftImputation, Discard Rows and Discard Columns. We considered two possibilities concerning the nature of the absence of data: completely at random or in a biased way. When missing is completely at random, all the observations (rows) have equal chance of having a missing data. In the biased missing, each row of the feature will have different probability of being missing, depending on its value.
Database Split and Estimator Tuning
The first step of the experiment was to split the database, as usual, in two folds: training (in-sample) and test (out-of-sample). The training fold was submitted to a number of interventions to simulate the absence of data, while the test fold remained intact and was only used for the final out-of-sample evaluation. Training and Test folds were randomly separated following a proportion of 60% for training and 40% for test.
Concerning parameters tuning, for each regressor-database pair, the user-defined parameters of the regressor were tuned, selecting the configuration with lowest scoring Mean Absolute Percentage Error (MAPE). The choice of using MAPE in regression over Mean Squared Error (MSE) relies on: (1) MAPE provides values that are more significant for decision-makers; (2) MAPE has a more intuitive explanation regarding relative error; (3) MAPE is better at evaluating robustness. Moreover, finding the best regression model using MAPE is equivalent to using a weighted version of Mean Absolute Error (MAE) (de Myttenaere et al. Citation2016; Shokouhyar, Ahmadi, and Ashrafzadeh Citation2021). This tuning was performed based on a 5-fold cross-validation on the training set. As it will be further explained below, the Nearest Neighbors imputation method needs that at least some of the rows to be with no absent data. In order to guarantee this requirement the training fold is randomly split one more time in 10% and 90% folds. The 10%, namely “Safety Chunk”, was not used in the missing data simulation so that all its rows remained intact to fulfill the requirement. The simulation was done only with the 90% fold, namely Data To be Modified (DTBM). After the missing data simulation both 10% and 90% folds are rejoined for the remaining stages.
Missing Data Simulation
In this work, we detail how the data was removed from an originally complete dataset in order to generate a simulated dataset with missing values. The first step to emulate the absence of data was to define which features have some of its values artificially removed. Of course, the relevance of the features plays a key role. To choose the features randomly is an inadequate strategy due to the large variations from one sample to another that would be introduced.
On the other hand, to remove the less important ones is also not a good choice, since their impact on estimation would be meager and the differences in having missing data or not would be almost null. Accordingly, we focused on the more relevant features. It is important to highlight that our purpose in feature selecting is directed to simulating missing values and not in trying to improve the performance of the estimators. There are a number of methods in the literature for feature selection, broad families are Wrappers and Filters (Kohavi and John Citation1997).
Here, we create an importance rank, an ordered list from the most to the least important. This rank is built by sequentially removing one of the features, and assuming that the worse the estimation without a given feature is, the more important is this feature. The results are sorted in descending order. The first features being the more important ones, since their absence produced the worst results. Given a database/estimator rank it is possible to calculate its Importance Ratio (IR), which is defined as: .
Where is the performance score of the data estimation without the most important feature and
is the performance score for the data estimation without the least important feature.
gives a simple and fast way to acknowledge databases that are strongly dependent on a small set of features. The higher the value of
, the more dependent on key features the database is.
Missing data may be classified in three categories according to the assumptions made: Missing Completely at Random (MCAR), Missing at Random (MAR), Missing not at Random (MNAR)Buuren (Citation2018); Gelman and Hill (Citation2007); Howell (Citation2007); Little and Rubin (Citation2002); Rubin (Citation1976); Saar-Tsechansky and Provost (Citation2007); Schafer and Graham (Citation2002). To illustrate each characterization, consider a toy example of a poll in which participants were asked their age and the number of cigarettes they smoke per day. Also consider that answering age is mandatory. The resulting database contains 2 features, ’Age’ and ’Cigarettes’, in which ’Cigarettes’ may be missing and ’Age’ is completely observed, i.e., there are no missing values. The missingness characterizations are explained in what follows.
In MCAR assumption, missing data is completely unrelated to observed information. In the presented toy example, under MCAR the feature ’Cigarettes’ might be missing due to unexpected events such as participants’ typos, the person conducting the poll made a mistake when collecting data, to name a few. In other words, MCAR assumes that the probability of missing values in a certain feature is completely random.
On the other hand, MAR assumes that missing data may be related to some observed information. In the presented scenario, teenagers may be less likely to say how many cigarettes they smoke per day because they might be afraid of their parents. In this assumption, missingness of the number of cigarettes is not related to the number of cigarettes itself. The missingness probability increases as the age of the participant decreases. In other words, the probability of having missing values may depend on one or more observed information in the database.
MNAR characterization assumes that there is no randomness in missing data. Looking back at the toy scenario, higher values for ’Cigarettes’ may be missing since teenagers refuse to inform their number of cigarettes per day because they smoked a large quantity. Namely, missingness under MNAR may be dependent on both observed and missing information.
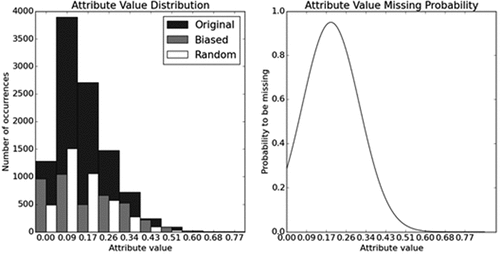
In order to analyze the effects of missing data in both purely random and biased scenarios, two probability functions were used to introduce missing values. In the first situation, each row had equal probability of being selected. Accordingly, the second scenario used a Gaussian function with mean and standard deviation equal to the feature’s mean and standard deviation of the original database, respectively. The bias is introduced by the fact that, the further the value is from mean, the lower is its probability. Moreover, the probability function was normalized so that the maximum probability of a feature to be missing is , i.e., a row of one feature will have a maximum probability of
of being missing when its value is equal to the mean of the feature.
Due to the purely random intrinsic nature of MCAR assumption, we chose to use it for the scenario in which missing data are randomly introduced. For the biased scenario, MNAR assumption was used. It is also important to note that, the percentage of missing data in each simulation is dictated by the parameter %P and it is calculated using the 90% fold previously explained.
shows how distribution of the feature was transformed by data removal, as well as the probability function used for the biased missing. One can see that random missing keeps the same distribution shape whilst biased missing severely alters it, introducing bias. After the simulation was completed the 90% fold is reunited with the 10% one (the Safety Chunk that did not participate on the simulation) and are headed to the next step.
Missing Data Treatment
At this point, after systematically erasing some entries in order to simulate missing data, we follow by dealing with the datasets, with incomplete data, in the same manner one would have to do in real situations. Two main options were used to handle this problem: simply discard the records with missing values, or to somehow fulfill (input) the absent data. Two techniques were employed for the former option and four for the latter, totaling six techniques. Concerning the first option, to keep the same dimensionality through all rows, missing values were discarded in two different ways: (1) by removing all the observations (rows) with missing values – we named “Discard Rows” –, (2) by removing any feature (column) containing missing values – we named “Discard Columns.” To illustrate the discard methods, let be a dataset with
observations and
features, %A =
and %P =
. After ”Discard Rows” and ”Discard Columns” methods, the resulting datasets have
observations with
features and
observations with
features, respectively.
Concerning imputation, the four selected approaches were: the Naive, the Nearest Neighbors, Multivariate Imputation by Chained Equations (MICE) and SoftImpute (Mazumder, Hastie, and Tibshirani Citation2010; van Buuren and Groothuis-Oudshoorn Citation2011). The Naive Imputation fulfills every missing data related to a feature with a central tendency measure, usually the mode for discrete feature or the mean for continuous one.
The Nearest Neighbors Imputation consists in inputting each value based on its similarities with other observations recorded in the database. Let us refer to an observation (row) which contains missing values as , and let
be a set of vectors representing the rows on the database which have no missing values. Denominate
as a subset of
with the
nearest vectors to
. We define as backbones, the subset of the features without any missing values in the whole training database and restrict to those the calculation of the distance to find the nearest vector. For instance, if a database with 5 features (numbered 1 to 5) has missing values in features 3 and 5, features 1, 2 and 4 would be used as backbones. For each feature a on
with missing data, its value was inputted using the mean or the mode (for discrete values) of a in the vectors in
. The number of neighbors,
, was fixed at 20 in all runs of the experiment. The “Safety Chunk,” earlier explained, was necessary in order to guarantee that
in all cases.
The MICE is performed by inputting a missing value several times and setting its value as the mean of all the imputations. Detailing this procedure, start by addressing every missing value and input it with the mean of the correspondent feature. Next, for each feature that has incomplete data, a regression is performed considering
as being dependent of the other features. The addressed missing values are then inputted with the result of this regression. This procedure is repeated for each feature with missing data and this regression cycle is repeated
times. Finally, the addressed missing values are replaced with the mean of the results of this n regressions. In the our experiments,
was set to 100 and the regression method utilized was Bayesian Interpolation MacKay (Citation1992). SoftImpute Mazumder, Hastie, and Tibshirani (Citation2010) is based on the premise that matrix
representing the database, can be generated by a matrix of parameters
where the rank of
is significantly lower than
or
.
Therefore, a low rank approximation (Markovsky Citation2009) of is performed. This is done by first constructing a matrix
in which every missing value on
is replaced by
. Sequentially, a Single Value Decomposition (SVD) is performed on
. A matrix Dr is reconstructed based on this SVD, but using only the single values that exceeds a defined threshold
.
is produced by replacing
missing values with the values found in
. An error
is calculated as:
Where is the set of indexes where
has missing values. While
,
is set to
and a new
is recalculated. The process is repeated until
converges to
. In our experiments
is set to 0.001 and the single values are selected in a way that every single value is at least 1/50 of the maximum single value. Hence
where
is the set of single values of
. These methods were chosen based on their different complexities and characteristics, aiming to analyze a broad scenario. Other methods include, but not limited to, the Hot Deck Replacement and the Maximum Likelihood (Andridge and Little Citation2010; Schafer and Graham Citation2002).
Data Estimation
Once the missing data problem is treated, the final step is estimation. Our procedure was firstly, to train the estimator using the original complete training database (with all values) and keep this for benchmarking. Then, the estimator was trained with databases originally containing simulated missing values that were fulfilled or removed as just described. For all cases, the calculation begins by training of the estimator with a training database and estimating the out-of-sample performance in a test database. The full procedure is addressed in Algorithm 2.5. Formerly the predictions are compared to the real values of the test database using the MAPE for the performance metric. In order to normalize the results and allow comparison between different databases the Non-improvement Ratio () was calculated as follows:
.
Where and
are respectively the performance metrics for the estimator trained with the original training database and the database that had missing values artificially introduced. Note that higher MAPE reflects a poor performance, hence, concerning
, the higher the value of
the worst was the performance of
compared to
. Non-improvement Ratio provides an easy way to compare different databases performances efficiently reveling good and bad results.
1 begin
2 Set test size, training size, %A values, %P values, ,
;
3 Split data into and
according to training and test size, respectively;
4 foreach regressor do
5 Apply random search of parameters using an uniform distribution and store the best parameters;
6 Select of
randomly and store in
;
7 ;
8 foreach pair of values (%a %A, %p
%P) do
9 ;
10 ;
11 Select the most important columns according to the Importance Rank calculated;
12 randomly erase values in the
most important columns independently using Algorithm 2;
13 use bias to erase values in the
most important columns independently using Algorithm 3;
14 ;
15 ;
16 foreach missing data treatment do
17 Apply missing data treatment to and
;
18 Train regressor on and evaluate on
;
19 Train regressor on and evaluate on
;
20 end
21 end
22 end
23 end
Algorithm 1: Pseudo-code of the experimental procedure.
1 begin
2 Set as the
most important columns in
according to Importance Rank;
3 foreach column in do
4 Erase values sampled according to an uniform random distribution;
5 end
6 end
Algorithm 2: Pseudo-code of random missing data generation.
1 begin
2 Set as the
most important columns in
according to Importance Rank;
3 foreach column in do
4 Calculate mean and standard deviation
;
5 Erase values sampled according to an Gaussian distribution with mean
and standard deviation
;
6 end
7 end
Algorithm 3: Pseudo-code of biased missing data generation.
Measuring Performance
To measure regression performance, we used Mean Average Percentage Error (MAPE):
Here n is the total number of data predicted, is the true value of the data on the ith test instance and
is the predicted value of the data for such instance. The greater the value of MAPE the worse the prediction. Next, we detail each of the four steps of the main experiment.
Results and Discussions
In this section, we present and discuss the main results of this contribution. Three key aspects were focused aiming at evaluating the effect of missing values: How data was missing (biased or completely at random); How the results were affected by the way databases are fulfilled and; How missing data impact in different regression algorithms.
The Effects of How Data Was Missing
Concerning the manner data was missing, the goal was to evaluate how the MCAR and the MNAR reacted under the same environment (same proportion of features with missing data (); same proportions of rows with missing data (
); same treatment method and finally the same regression algorithm). In order to make this comparison, for each combination of these four aspects, the result of the data estimation performance was recorded for the database with biased missing data and for the database with random missing data. The performances were signaled with
for the random database and with
for the biased database. The percentage difference (
) of
in respect to
was then calculated as:
. The PD was calculated for every of the four just described aspects. The results are shown on . The mean and 95% confidence interval was calculated for each group of results (e.g. when grouped by a database and a treatment method the interval was calculated using the results of such database and treatment performed under the different values of
,
and distinct estimators).
Table 2. Percentage difference means for the regression datasets (according referenced in ). 5% and 95% confidence intervals in parenthesis.
shows that the mean for the PD was below 3.61% for all the treatment methods. Naive Imputation revealed to be the treatment method with higher PD values and percentiles. It becomes clear that in the majority of cases the difference in the quality of the regression is negligible, suggesting that not knowing how data is missing might not greatly effect the regression outcome. This looks good since oftentimes acknowledging such information is not possible. Which method handles imputation more uniformly is clearly a database-dependent information. Discard Rows performed similar in Compactiv database while the other imputation methods were more diverged. The opposite was observed with Airfoil Wankara regressions differed uniquely among each treatment method, while Abalone and Wine Quality results behaved similar throughout the different methods and missing types.
Nevertheless it becomes clear that the relative behavior of the difference on the estimations results is database specific, since their relative values considered a fixed method is almost constant. Compactiv, Wankara and Mortgage have the highest difference means in every method, while Wine Quality and Abalone have the lowest. The Discard Columns method is not shown in because it does not utilize any feature with missing data and the bias is completely removed from the training dataset which the estimators are fed. Consequently PD is always 0% for such method showcasing absolutely no difference. We next focus on the impact of the way the datasets are fulfilled. Our results have shown that the best option on this matter significantly vary depending on: which estimator is used, what is the proportion of missing data, how many features have missing data.
Discard Rows Vs. Imputation Methods and the Amount of Features and Rows with Missing Data
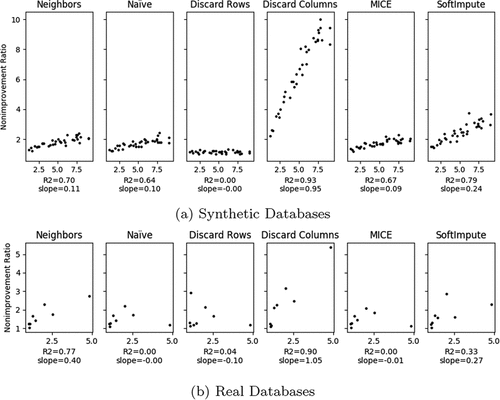
Discarding rows has shown to be a good method of choice albeit its simplicity. Nevertheless, as the number of features with missing data increases, the performance shows a decline when compared with the imputation treatment methods. This behavior can be observed in the results of the experiment with synthetic databases showcased in (left). In this experiment the number of features with missing data was increased, however the proportion of missing data was maintained in every database generated. This was achieved by increasing the number of total features in the same proportion as the number of features with missing data. Moreover was held at a constant value of 20% in every experiment.
Figure 4. As the number of features with missing data increases, Discard rows become a worse choice than the imputation methods (left). As the proportion of features with missing data () increases the outcome in data regression gets worse (right).
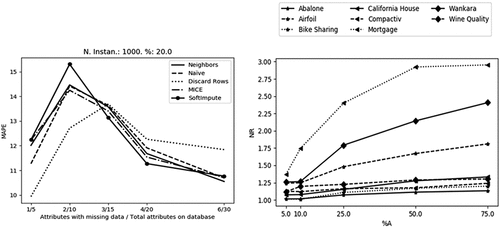
In the real-world databases it is not possible to increase the number of features in a database in order to keep the proportion of missing data and emulate the exposed synthetic experiment. Nevertheless, it is clear by (right) that every database treated with Discard Rows method has its performance seriously jeopardized as the proportion of features with missing data () increases. The Discard-Rows method shows a higher deterioration of the quality of its results when the number of features with missing increases. This deterioration is not so steep for non-discarding methods like imputation ones. Moreover as shown in left) this observation holds true even if the proportion of missing data is kept the same, with only the number of features being changed.
Relation between Treatments and Importance Ratio
Clearly, the most relevant features play a key role on the estimation outcome, however, it was not clear how the performance of the estimators were affected by data missing on these features. Therefore, an experiment was made in order to observe this behavior in not only real-world datasets but also in synthetic data with different Importance Ratios.
As it can be seen in , except from the Discard Rows method all other methods’ performances showed some degree of dependency with the Importance Ratio, Discard Columns clearly being the one most affected by it. This is expected since the more important
a feature is for regression the worst the performance will be if we discard it. By projecting the results on the real databases it can be seen that Nearest Neighbors, Discard Rows and Discard Columns mirror previous conclusions almost exactly (with Nearest Neighbors showing an even higher dependency).
SoftImpute, which showed the second-highest correlation on synthetic databases did not translated this results on the real dataset, mainly because of one outlier result caused by database Bike Sharing. Naive Imputation and MICE showed no relationship whatsoever, which is unsurprising since their synthetic correlation were among the weakest. Although a higher number of databases would be necessary to draw further conclusions it is clear how many features of the behavior found on the synthetic experiments are also seen on real databases.
Treatment-method Winners
In each experiment the winning treatment method for missing data is defined as the method which does the smallest damage in the estimation quality of the database when compared to the estimation of the complete and original database. Therefore a snapshot of the winners in each scenario was taken in order to observe how each treatment method is induced by the different variables studied. This snapshot was split in four, focusing on the number of times a treatment was a winner for each ,
, estimator and database as it can be seen in .
Figure 6. Winner methods separated by ,
, databases and regressors (AdaBoost, DecisionTree, KNeighbors, MLP, RandonForest, SVR).
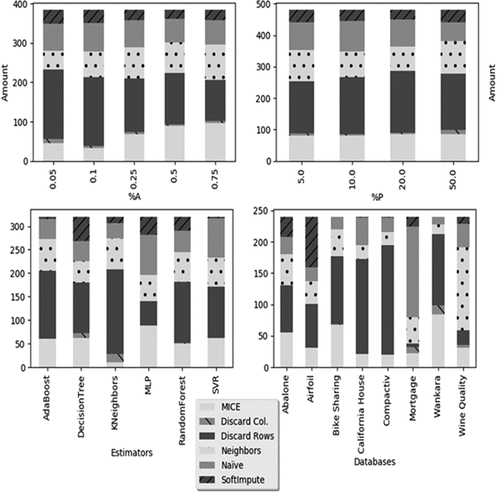
Considering the experiment analyzed by the segregation there is little difference to be noted in the amount of winners, independently of how many rows have missing data. It becomes clearer that besides its simplicity, Discard Rows is the better choice in the majority of cases, independently of
. MICE, Naive and Nearest Neighbors Imputation follows with similar amounts. SoftImpute wins considerably less and Discard Columns winnings are almost skippable except when the number of rows with missing data is high.Shifting focus to the amount of features with missing data (
) it is clear that discard methods are more preferable in situations where the magnitude of
is low. When higher number of features have missing data MICE and Nearest Neighbors imputation are preferable, nevertheless Discard Rows still win a considerable amount of times in every value of
analyzed. When the snapshot is split by estimators more drastic behaviors are observed. Clearly to Discard Rows it not preferred when using Multi-Layer Perceptron for regression. Moreover Discard Columns only won on experiments ran with Nearest Neighbors and Decision Tree regressors.
Support Vector Machine and AdaBoost Regressors had almost none winners when treated with the SoftImpute method also. However, the most distinct scenarios are seen when the results are separated by database. Although being the preferred method in the majority of the cases, Discard Rows winning results were almost nonexistent for the Mortgage and Wine Quality databases. The former had an unusual amount winning for the nearest neighbors method. Similarly SoftImpute only showcased expressive results with Abalone and Airfoil databases, while Discard Columns only won with Mortgage and Wankara, these are among the databases with fewer observations but with 10 or more features, making them more horizontal and therefore more prone to feature removal. Therefore it is possible to observe that simply discarding observations with missing data can possibly be the better choice when it comes to regression performance, even with a high number of missing data. However this situation is mitigated when using a Neural Network and the behavior has clearly a database influence more than anything else.
Nearest Neighbors Vs. Naive Imputation on Tree Based Algorithms
In the Bike Sharing regression database something curious happened. The results for nearest neighbors imputation were way worse than for Naive Imputation and usually these two imputation methods perform in similar fashion. For this specific database, the most important features are discrete, hence its missing value is inputted by mode calculation.
The reason for the drastically different outcomes is because the CART algorithm splits the database based on one feature’s values. As Naive Imputation inputs all missing data in one feature with the same value (the mode), CART can easily split out this value, preserving an intact set of values that will lead to a better database split. With Nearest Neighbors imputation this is not possible, since each time a different value can be inputted, and therefore, in the split phase, the algorithm cannot rule-out these many values. To confirm this observation this behavior was reproduced using artificially generated databases as it can be seen in .
Table 3. Percentage diference means for the regression databases. 5% and 95% confidence intervals in parenthesis.
On every database of this kind the error for Naive imputation was way lower than for Nearest Neighbors imputation. This behavior was not found and could not be reproduced with other kinds of estimators or with decision tree based estimators where the affected feature was continuous. To produce the synthetic databases for this experiment some changes were made in the generation of synthetic databases from Section 2.7. We used: In order to make the first feature discrete, after
is generated
is set to [
].
Conclusions
We showed that the effect of missing data may significantly vary depending on a number of factors and consequently, the choice of an appropriate police concerning missing data has a key impact on the results. It should be noticed however that the performance of the six tested approaches were linked with with the amount of features with missing data, but mainly with which estimator and which database the experiment was performed.
In many cases more popular and complex imputation solutions performed worse than simple methods (SoftImpute rarely was the better option and MICE frequently lost in performance to Discard Rows). Moreover it was shown that under specific, but not uncommon, conditions the complexity of Nearest Neighbors method can possibly not payoff against the simplest of methods like Naive imputation or Discard Columns.
The data removal methods showed to behave very differently between them, as expected. Discard Rows was more successful, being the method which performed better in most overall cases. It is noticeable, however, that the way features were selected in the missing data simulation (from the most to the least important) is a clear disadvantage for the Discard Columns method. Hence, in real world situations where the feature with missing data may not be important, Discard Columns can be a good and simple alternative. Concerning how data is missing, it was shown that although it induces serious bias in the database the fact that the data is missing is more hurtful than a possible introduced bias. Therefore, although the damage had great magnitude in some cases, the difference between data Missing Completely at Random (MCAR) to Missing not at Random (MNAR) was not significant in most cases.
Possibly the best general policy is to first infer the importance of the features with missing data, if they are not significant at all the Discard Columns method is the one that will allow no observation loss in the training process and will perform fast. Nevertheless, if features which are missing are indeed important, Discard Rows may be a feasible choice depending on the amount of missing values. However if data generalization is poor and an imputation is required MICE or Nearest Neighbor will provide a greater gamut of imputation techniques, notwithstanding that results here showed that in various cases a similar performance can be achieved with the simpler Naive Imputation.
Acknowledgments
The authors would like to acknowledge financial support from the following Brazilian research agencies: CAPES, CNPq and FAPERJ. We would like to say thank you to Susan Hussey ([email protected]) for the native English review.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
References
- Alcala-Fdez, J., A. Fernandez, J. Luengo, J. Derrac, S. Garcia, L. Sanchez, and F. Herrera. 2011. Keel data-mining software tool: Data set repository, integration of algorithms and experimental analysis framework. Journal of Multiple-Valued Logic and Soft Computing 17 (2–3):255–2182.
- Altman, N. 1992. An introduction to kernel and nearest-neighbor nonparametric regression. The American Statistician 46 (3):175–85.
- Andridge, R., and R. Little. 2010. A review of hot deck imputation for survey non-response. International Statistical Review 78 (1):40–64. doi:10.1111/j.17515823.2010.00103.x.
- Bache, K., and Lichman. (2013). Uci machine learning repository. Available in http://archive.ics.uci.edu/ml.
- Bishop, C. (2016). Pattern recognition and machine learning. DOI 10.1117/1.2819119.
- Breiman, L. 2001. Random forests. 1–33. doi: 10.1017/CBO9781107415324.004.
- Breiman, L., J. Friedman, R. Olshen, and C. Stone (1984). Classification and regression trees. CRC Press.
- Buuren, S. 2018. Flexible imputation of missing data. doi:10.1201/b11826.
- Chand, M., B. Bhattarai, N. Pradhananga, and P. Baral. 2021. Trend analysis of temperature5252 data for the Narayani river basin, Nepal. Science 1:38. doi:10.3390/sci1020038.
- de Myttenaere, A., B. Golden, B. Le Grand, and F. Rossi. 2016. Mean absolute percentage error for regression models. Neurocomputing 192:38–48. doi:10.1016/j.neucom.2015.12.114.
- Deng, L., and D. Yu. 2013. Deep learning: Methods and applications. Foundations and Trends R in Signal Processing 7 (3–4):197–387. doi:10.1561/2000000039.
- Du, H., F. Liu, and L. Wang. 2017. A Bayesian fill-in method for correcting for publication bias in meta-analysis. Psychological Methods 22 (4):799–817. doi:10.1037/met0000164.
- Freund, Y., and R. Schapire. 1995. A decision-theoretic generalization of on-line learning and an application to boosting. Computational Learning Theory 55:119–39.
- Gelman, A., and J. Hill. 2007. Data analysis using regression and multilevel/hierarchical models. doi:10.2277/0521867061.
- Haykin, S. 1998. Neural networks: A comprehensive foundation. 2nd ed. Upper Saddle River, NJ, USA: Prentice Hall PTR.
- Howell, D. C. 2007. The treatment of missing data. The Sage Handbook of Social Science Methodology 1: 208–24.
- Jadhav, A., D. Pramod, and K. Ramanathan. 2019. Comparison of performance of data imputation methods for numeric dataset. Applied Artificial Intelligence 33 (10):913–33. doi:10.1080/08839514.2019.1637138.
- Kohavi, R., and G. John. 1997. Wrappers for feature subset selection. Artificial Intelligence 97 (1–2):273–324. doi:10.1016/S0004-3702(97)00043-X.
- Li, H., Y. Weng, Y. Liao, B. Keel, Brown, and K. E. Brown. 2021. Distribution grid impedance and topology estimation with limited or no micro-pmus. International Journal of Electrical Power and Energy Systems 129:106794. doi:10.1016/j.ijepes.2021.106794.
- Little, R., and D. Rubin. 2002. Statistical analysis with missing data. doi:10.2307/1533221.
- Ludtke, O., A. Robitzsch, and S. Grund. 2017. Multiple imputation of missing data in multilevel designs: A comparison of different strategies. Psychological Methods 22 (1):141–65. doi:10.1037/met0000096.
- MacKay, D. 1992. Bayesian interpolation. Neural Computation 4 (3):415–47. doi:10.1162/neco.1992.4.3.415.
- Madden, G., N. Apergis, P. Pappoport, and A. Banerjee. 2018. An application of nonparametric regression to missing data in large market surveys. Journal of Applied Statistics 45 (7):1292–302. doi:10.1080/02664763.2017.1369498.
- Markovsky, I. 2009. Applications of structured low-rank approximation. IFAC Proceedings Volumes . 42(10): 1121–1126 https://doi.org/10.3182/20090706-3-FR-2004.00186.
- Mazumder, R., T. Hastie, and R. Tibshirani. 2010. Spectral regularization algorithms for learning large incomplete matrices. Journal of Machine Learning Research 11:2287–322.
- Ngueilbaye, A., H. Wanga, D. Ahmat Mahamat, and S. Junaidu. 2021. Modulo 9 model-based learning for missing data imputation. Applied Soft Computing Journal 103:107167. doi:10.1016/j.asoc.2021.107167.
- Pedregosa, F., G. Varoquaux, A. Gramfort, and V. Michel. 2011. Scikit-learn: Machine learning in python. Journal of Machine Learning Research 12:2825–30.
- Peng, L., and L. Lei. 2005. A review of missing data treatment methods. Intelligent Information Management Systems and Technology 1 (3):412–19.
- Perez-Ruiz, L., and G. Escarela. 2018. Joint regression modeling for missing categorical covariates in generalized linear models. Journal of Applied Statistics 45:2741–59. doi:10.1080/02664763.2018.1438376.
- Rubin, D. B. 1976. Inference and missing data. Biometrika 63 (3):581–92. doi:10.1093/biomet/63.3.581.
- Saar-Tsechansky, M., and F. Provost. 2007. Handling missing values when applying classification models. Journal of Machine Learning Research 8:1625–57.
- Santos, M. S., R. C. Pereira, A. F. Costa, J. P. Soares, J. Santos, and P. H. Abreu. 2019. Generating synthetic missing data: A review by missing mechanism. IEEE Access 7:11651–67. doi:10.1109/ACCESS.2019.2891360.
- Schafer, J., and J. Graham. 2002. Missing data: Our view of the state of the art. Psychological Methods 7 (2):147–77. doi:10.1037/1082-989X.7.2.147.
- Schouten, R. M., P. Lugtig, and G. Vink. 2018. Generating missing values for simulation purposes: A multivariate amputation procedure. Journal of Statistical Computation and Simulation 88 (15):2909–30. doi:10.1080/00949655.2018.1491577.
- Shokouhyar, S., S. Ahmadi, and M. Ashrafzadeh. 2021. Promoting a novel method for warranty claim prediction based on social network data. Reliability Engineering & System Safety 216:108010. doi:10.1016/j.ress.2021.108010.
- Steinwart, I., and A. Christmann. 2008. Support vector machines. 1st ed. Berlin: Springer Publishing Company, Incorporated.
- Tachmazidis, I., T. Chen, M. Adamou, and G. Antoniou. 2021. A hybrid ai approach for supporting clinical diagnosis of attention deficit hyperactivity disorder (ADHD) in adults. Health Information Science and Systems 9 (1):1–8. doi:10.1007/s13755-020-00123-7.
- van Buuren, S., and K. Groothuis-Oudshoorn. 2011. mice: Multivariate imputation by chained equations in r. Journal of Statistical Software 45 (3): 1–67. https://doi.org/10.18637/jss.v045.i03.
- Zhang, Q., and L. Wang. 2017. Moderation analysis with missing data in the predictors. Psychological Methods 22 (4):649666. doi:10.1037/met0000104.