Abstract
Background. Cytokine transforming growth factor (TGF) is involved in regulation of tissue repair after injury. More recently, TGF–β1 codon 10 gene polymorphism has been shown to be associated with circulating TGF-β levels. We tested whether TGF-β1 genotype polymorphism was predictive of renal allograft function decline. Patients and Methods. The study population consisted of 129 consecutive cadaveric or living related renal transplant recipients at our center between 1985 and 2001. The recipient TGF-β1 genotype polymorphism was determined from peripheral blood leucocytes DNA. The primary endpoint was rate of glomerular filtration rate decline between the first year and the third year of transplant. Results. Baseline glomerular filtration rate as estimated by MDRD study equation at 1 year measured 50 ± 17 mL/min/1.73 m2. At the end of the 3-year follow-up period, 52 patients (40%) experienced biopsy-confirmed acute rejections. Frequency and severity of allograft rejection did not differ with TGF-β genotypes. However, the decline in glomerular filtration rate was significantly greater in Leu/Leu (TT) than Leu/Pro (CT) recipients, 6.3 ± 16.9 mL/min/1.73 m2 versus 0.1±10.2 mL/min/1.73 m2, p = 0.04. Conclusion. Our results demonstrate that recipient TGF-β1 codon 10 Leu/Leu homozygosity is a potential risk factor of kidney allograft function decline.
Introduction
Transforming growth factor-beta (TGF-ß) is a cytokine with potent stimulation of extracellular matrix production and deposition, with known consequence of kidney fibrosis from its overexpression.Citation[1-3] However, it has been recently recognized that TGF-β exerts antiinflammatory effects, thus playing an important role in regulation of tissue repair and regeneration after tissue injury.Citation[4-6] The beneficial role of TGF-β was further supported by a more recent molecular analysis of 52 renal biopsies, which demonstrated significantly higher tubulointerstitial TGF-β mRNA levels among patients with stable or improving glomerular filtration rate function between baseline and 1 month.Citation[7]
Significant associations have been demonstrated between polymorphisms of TGF-β gene and production of the cytokine. Among them, a leucine-proline substitution T869C (Leu 10Pro) at codon 10 was implicated; the molecule containing the proline substitution is said to have a longer biological half-life. Stated another way, the number of proline allele correlates positively with circulating levels of TGF.Citation[8&9] It would be therefore important to examine the role of genetic variations on human kidney disease process such as those characterized by fibrosis and wound healing after renal transplantation. Herein, we hypothesize that the fibrogenic potential and healing effect on renal graft would therefore vary according to such genetic variation in the TGF-β system. To address these issues, we investigated and characterized the predictive value of the TGF-β gene polymorphism on transplant outcomes, including renal function decline and urinary protein excretion in a prospective cohort study of transplant population.
Patients and methods
Subjects
The ethics committee of the Chinese University of Hong Kong approved the study. Informed consents were obtained from all subjects. Between 1985 and 2001, 134 consecutive patients underwent cadaveric or living related renal transplants at our center. Four transplant recipients, whose renal allograft failure occurred within 3 months after transplantation, were excluded from the study because we wanted to avoid possible confounding influences of any complications of surgery. One patient was excluded because of incomplete data.
Genotyping, Transplant Outcome, and Covariates
Genomic DNA was isolated from peripheral blood leucocytes of transplant recipients by standard technique. DNA was amplified using polymerase chain reaction (PCR) with sequence-specific primers for codon 10 of TGF-β1, as had been reported elsewhere.Citation[10] All the reagents for PCR were purchased from Roche Diagnostics (Roche, Switzerland). The primer sequences for codon 10 amplification were 5′-TTC AAG ACC ACC CAC CTT CT-3′ (sense) and 5′-TCG CGG GTG CTG TTG TAC A-3′ (antisense). The amplification protocol consisted of an initial denaturing at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 30 s, and one final elongation step of 72°C for 5 min.
Samples were then electrophoresed through a 2% agarose gel and visualized by ethidium bromide staining. The genotypes for + 869 (codon 10) restriction site polymorphism were TT (Leu/Leu) (285 bp), TC (Pro/Pro) (273 bp), and CC (Leu/Pro) (285 bp and 273 bp).
Data including the acute rejection episodes and severity, demographic, and clinical variables of transplantation were collected. All cases of renal allograft dysfunction were assessed by core biopsy in our center to secure the diagnosis, after exclusion of nonimmune causes. Diagnosis of acute rejection was based on serum creatinine level, plus either renal scintigram or Doppler ultrasound, and core biopsy confirmation before the initiation of antirejection therapy or within 24 hours of treatment. All biopsies were scored according to the Banff 97 classificationCitation[11] by a single pathologist, who was blinded to the patients' genotype. Acute cellular rejection was treated with three consecutive daily doses of 0.5 to 1.0 g methylprednisolone given intravenously. OKT3 and plasmapheresis were instituted for steroid-resistant rejection, depending on its severity. We also examined covariates including human histocompatibility leukocyte antigen (HLA) matching and the doses of immunosuppressants used.
Primary endpoint was defined a priori as glomerular filtration rate (in milliliters per minute per 1.73 m2 of body surface are, as estimated by the MDRD study equation)Citation[12] decline between the first year and the third year. The MDRD study equation was chosen because it is more accurate and precise than the Cockcroft-Gault equation for persons with a glomerular filtration rate less than 90 mL/min/1.73 m2. This equation has also been validated in kidney transplant recipients.Citation[13] Other variables investigated were the urinary protein excretion and graft survival after transplantation.
Statistical Analysis
The Kolmogorov-Smirnov test was used to determine if continuous variables are normally distributed. Our results were expressed as mean ± standard deviation (SD) for normally distributed data and median or range for skewed data. The Statistical Package for the Social Sciences (Windows version 11.5; SPSS, Inc., Chicago, IL) program was employed. Categorical variables were analyzed using chi-square test. Continuous variables were compared using Student's t test and Mann-Whitney test where appropriate. A two-tailed p value of less than 0.05 is defined as significant.
Results
We enrolled 129 consecutive renal transplant patients, who were followed up for at least the first 3 years after transplantation. Observed genotype frequency, in agreement with the Hardy-Weinberg equilibrium and previously published data,Citation[10] was 16.2%, 50.0%, and 33.8% for Leu/Leu (TT), Leu/Pro (CT), and Pro/Pro (CC), respectively. Baseline characteristics did not differ significantly between the groups () for kidney-donor and recipient age, gender, body weight, degree of HLA mismatching, length of ischemic exposure, treatment used, blood pressure, symptomatic cytomegalovirus infection, and prevalence of diabetes mellitus.
Table 1. Baseline demographic and clinical characteristics
Baseline glomerular filtration rate at one year measured 50 ± 17 mL/min/1.73 m2 and was comparable among patients with different TGF-β genotypes (). Follow-up data were available for all 129 patients. Within the first 3 years of transplant, a total of 52 patients (40%) had history of either biopsy-proven acute cellular or vascular rejection. There was no discernible influence of TGF-β polymorphism on the frequency and severity of rejection episodes ().
Table 2. Incidence and histopathologic findings of acute rejection
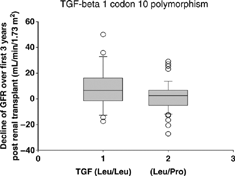
The decline of glomerular filtration rate from the first year to the third year was 6.3 ± 16.9 mL/min/1.73 m2 among recipients with Leu/Leu (TT), as compared with 1.5 ± 12.0 mL/min/1.73 m2 among those with Leu/Pro (CT) or Pro/Pro (CC) (P = 0.12). Decline in glomerular filtration rate () was significantly greater in Leu/Leu (TT) than Leu/Pro (CT) group, 6.3 ± 16.9 mL/min/1.73 m2 versus 0.1 ± 10.2 mL/min/1.73 m2, p = 0.04. Difference in glomerular filtration rate decline between Leu/Leu (TT) and Pro/Pro (CC) groups was not statistically significant, 6.3 ± 16.9 mL/min/1.73 m2 versus 3.9 ± 14.1 mL/min/1.73 m2, respectively, p = 0.55.
Figure 1. Leu/Leu (TT) genotype associated with greater decline in glomerular filtration rate. Boxes show the median values, the first and third quartiles in each group.
Urinary protein excretion rate at the third year did not differ between recipients with TGF-β Leu/Leu (TT) and other two genotypes, being 0.2 ± 0.1 g/day and 0.3 ± 0.7 g/day respectively, p = 0.42. This finding was not altered when the subjects were analyzed according to three genotypes (data not shown). Five patients had kidney allograft failure within the initial 3 years of follow-up; one of them with Leu/Leu (TT) genotype, two subjects with Leu/Pro (CT), and another two with Pro/Pro (CC), p = 0.90.
Discussion
TGF-β is believed to play an important role in various progressive kidney diseases,Citation[3] the latter being mediated by a common final pathway of tissue scar formation, regardless of the original etiology. This process results from alteration in extracellular matrix synthesis, which is in turn mediated by TGF-β. For this reason, we chose to correlate the renal function with transplant recipients possessing specific TGF-β cytokine alleles, which are influential in TGF-β synthesis. As an outcome indicator of kidney transplant, we measured the rate of change in glomerular filtration rate, which is often used in clinical studies.Citation[7], Citation[14&15]
We found a greater glomerular filtration rate decline within the first 3 years of transplant among recipients with TGF-β Leu/Leu (TT) genotypes. This effect should be interpreted under the context of association between TGF codon 10 gene polymorphism and TGF-β levels. Although TGF-β has the ability to directly stimulate the extracellular matrix synthesis, various beneficial actions have been recently ascribed to this multifunctional cytokine, such as its role in tissue repair and immunoregulatory effects. Data suggesting that TGF-β may in fact have an important positive role in renal function come from a more recent renal biopsy study confirming that high tubulointerstitial TGF-β mRNA levels were associated with favorable renal outcome as reflected by less deterioration of glomerular filtration rate 12 months after biopsy.Citation[7] This finding is in concert with observations from another study, in which higher renal cortical mRNA levels of TGF-β early after human kidney transplantation were associated with a stable graft function at later time points and absence of chronic rejection.Citation[16] Moreover, it has been suggested that progressive renal graft dysfunction from chronic rejection may well be a result of impaired repair from renal injury,Citation[17] whereas TGF-β was shown to accelerate wound healing in animal models.Citation[6] Interestingly, worse progression of renal insufficiency had been also documented among heart transplant recipients with TGF-β Leu/Leu (TT) genotypes.Citation[18] This leaves open the possibility that TGF-β gene polymorphism is linked with susceptibility to cyclosporine nephrotoxicity.
Our study has several limitations. First, it was not powered to examine allograft failure as an outcome. Moreover, neither renal TGF-β mRNA expression nor urinary TGF-β excretion were included in our assessment. Patients with higher urinary TGF-β excretion, for instance, had been shown to have significantly higher rates of chronic allograft dysfunction.Citation[19] Interestingly, intrarenal cytokine expression according to TGF-β codon 10 genotype could also be influenced by the population age and presence of chronic allograft nephropathy.Citation[20] Last but not least, we did not examine the donor TGF-β1 genomics that might have influenced the transplant outcome, such as acute rejection.Citation[21]
Although this study was not large enough to ensure the prognostic value of TGF-β codon 10 polymorphism in renal allograft function decline, it adds to the growing body of evidence that TGF-β high-producer genotype may ameliorate progressive glomerular filtration rate deterioration. Our findings may offer important clues to understanding the possible roles of TGF-β and recipients' genotypes in determining allograft outcome; further prospective study of renal TGF-β mRNA expression is warranted.
Acknowledgments
This work was supported by the Hong Kong Society of Nephrology Research Grant 2003 and the Chinese University of Hong Kong Research Grant Account 6900573.
References
- Border W A, Noble N A. Transforming growth factor β in tissue fibrosis. N Engl J Med 1994;331:1286–1292. [PUBMED], [INFOTRIEVE], [CSA]
- Border W A, Ruoslahti E. Transforming growth factor-β in disease: the dark side of tissue repair. J Clin Invest 1992;90:1–7. [PUBMED], [INFOTRIEVE], [CSA]
- Blobe G C, Schiemann W P, Lodish H F. Role of transforming growth factor β in human disease. N Engl J Med 2000;342:1350–1358. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Kitamura M, Fine L G. Evidence for TGF-β-mediated ‘defense’ of the glomerulus: a blackguard molecule rehabilitated?. Exp Nephrol 1998;6:1–6., [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Eikmans M, Baelde H J, de Heer E, Bruijn J A. RNA expression profiling as prognostic tool in renal patients: toward nephrogenomics. Kidney Int 2002;62:1125–1135. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Mustoe T A, Pierce G F, Thomason A, Gramates P, Sporn M B, Deuel T F. Accelerated healing of incisional wounds in rats induced by transforming growth factor-beta. Science 1987;237:1333–1336. [PUBMED], [INFOTRIEVE], [CSA]
- Eikmans M, Baelde H J, Hagen E C, Paul L C, Eilers P H, de Heer E, Bruijn J A. Renal mRNA levels as prognostic tools in kidney diseases. J Am Soc Nephrol 2003;14:899–907. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Yokota M, Ichihara S, Lin T L, Nakashima N, Yamada Y. Association of a T29→(C polymorphism of transforming growth factor-β1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation 2000;101:2783–2787. [PUBMED], [INFOTRIEVE], [CSA]
- Guo Z, Binswanger U, Knoflach A. Role of codon 10 and codon 25 polymorphisms on TGF-β1 gene expression and protein synthesis in stable renal allograft recipients. Transplant Proc 2002;34:2904–2906. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Wong T Y, Poon P, Chow K M, Szeto C C, Cheung M K, Li P K. Association of transforming growth factor-beta (TGF-beta) T869C (Leu 10Pro) gene polymorphisms with type 2 diabetic nephropathy in Chinese. Kidney Int 2003;63:1831–1835. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Racusen L C, Solez K, Colvin R B, , et al. The Banff 97 working classification of renal allograft pathology. Kidney Int 1999;55:713–723. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Levey A S, Bosch J P, Lewis J B, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of diet in renal disease study group. Ann Intern Med 1999;16:461–470. [CSA]
- Levey A S, Coresh J, Balk E, Kausz A T, Levin A, Steffes M W, Hogg R J, Perrone R D, Lau J, Eknoyan G. National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Ann Intern Med 2003;139:137–147. [PUBMED], [INFOTRIEVE], [CSA]
- Ruggeneti P, Schieppati A, Remuzzi G. Progression, remission, regression of chronic renal disease. Lancet 2001;357:1601–1608. [CSA], [CROSSREF]
- Mitch W E. Measuring the rate of progression of renal insufficiency. In: Mitch W E, ed. The Progressive Nature of Renal Disease. New York, NY: Churchill Livingstone, 1986:203–220.
- Eikmans M, Sijpkens Y W, Baelde H J, de Heer E, Paul L C, Bruijn J A. High transforming growth factor-beta and extracellular matrix mRNA response in renal allografts during early acute rejection is associated with absence of chronic rejection. Transplantation 2002;73:573–579. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Paul L C. Chronic allograft nephropathy—a model of impaired repair from injury?. Nephrol Dial Transplant 2000;15:141–151. [CSA]
- Lácha J, Hubáček J A, Potmìšil P, Viklicky O, Málek I, Vßtko S. TGF-β1 gene polymorphism in heart transplant recipients–effect on renal function. Ann Transplant 2001;6:39–43. [CSA]
- Yamada K, Hatakeyama E, Arita S, Sakamoto K, Kashiwabara H, Hamaguchi K. Prediction of chronic renal allograft dysfunction from evaluations of TGF β1 and the renin-angiotensin system. Clin Exp Nephrol 2003;7:238–242. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Melk A, Henne T, Kollmar T, Strehlau J, Latta K, Offner G, Jhangri G S, Ehrich J H, Von Schnakenburg C. Cytokine single nucleotide polymorphisms and intrarenal gene expression in chronic allograft nephropathy in children. Kidney Int 2003;64:314–320. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Hoffman S, Park J, Jacobson L M, Muehrer R J, Lorentzen D, Kleiner D, Becker Y T, Hullett D A, Mannon R, Kirk A D, Becker B N. Donor genomics influence graft events: the effect of donor polymorphisms on acute rejection and chronic allograft nephropathy. Kidney Int 2004;66:1686–1693. [CSA], [CROSSREF]