Abstract
Recently we demonstrated the effect of fenoldopam on ischemia/reperfusion (I/R) induced NFκB mediated pro-inflammatory signal transduction. However, the effect of fenoldopam on I/R-induced apoptosis is not known. We utilized a rat model of acute ischemic nephropathy to test the hypothesis that fenoldopam attenuates I/R-induced apoptosis. Sprague-Dawley rats were anesthetized by intraperitoneal administration of 50 mg/kg urethane and randomly allocated into 4 groups (n = 6 each): 1) sham-operated, 2) sham operation with infusion of 0.1 μg/kg/min fenoldopam, 3) unilateral renal ischemia followed by 4 h of reperfusion, and 4) I/R with fenoldopam infusion. Kidney samples were fixed and paraffin-embedded to measure apoptosis. Data were compared between groups using ANOVA with Bonferroni correction. RNA was extracted from each left kidney to probe cDNA microarray and measure gene expression as percent of positive control. Compared to the control group, I/R significantly (P < 0.001) induced apoptosis in both the cortex and medulla. Similarly, microarray analysis revealed that I/R induced 73 apoptosis-related genes. Treatment with fenoldopam significantly reduced (P < 0.001) I/R-induced apoptosis both in the cortex and medulla and attenuated all 73 I/R-induced apoptosis-related genes. Data from this rat model of ischemic nephropathy suggest that fenoldopam may attenuate I/R-induced apoptosis and apoptosis-related gene transcription.
INTRODUCTION
Ischemia/reperfusion (I/R) injury in kidney is one of the common causes of acute renal failure (ARF) that affects surgical intensive care unit patients.Citation[[1]] The depletion of high energy phosphate compounds, production of oxygen free radicals, intracellular calcium accumulation, neutrophil recruitment and activation, lipid peroxidation of cell membranes, and cytoskeletal dysfunction have been implicated in the pathogenesis of I/R injury.Citation[[2]], Citation[[3]] This complex and interrelated sequence of events results in injury and the eventual death of renal cells due to a combination of apoptosis and necrosis.Citation[[4]] Renal tubular apoptosis is an established feature of I/R injury in the kidney.Citation[[5]] The intracellular biochemical cascade of apoptosis involves proteolytic activation of caspases,Citation[[6]] which target various functional and structural intracellular proteins that ultimately result in cellular disassembly. Apoptosis is characterized by cell shrinkage, dense condensation of nuclear chromatin, nuclear fragmentation, dissolution of nuclear membrane, and plasma membrane blebbing that leads to membrane-bound cell remnants, apoptotic bodies.Citation[[7]] During renal I/R injury and other experimental conditions, these apoptotic bodies instigate inflammation.Citation[[8]]
Fenoldopam is a short-acting, parenteral vasodilator that lowers systemic vascular resistance by activating dopamine (DA1) receptors in mesenteric, coronary, cerebral, and renal arterioles.Citation[[9]] Stimulation of DA1 receptors initiates a G-protein coupled activation of adenylate cyclase, resulting in the release of cyclic adenosine monophosphate and arteriolar vasodilation and natriuresis.Citation[[10]], Citation[[11]] We recently demonstrated that fenoldopam significantly attenuated the I/R-induced NFκB DNA binding activity and modulated expression of I/R-induced NFκB signal transduction and pro-inflammatory genes.Citation[[12]] However, the effect of fenoldopam on I/R injury-induced apoptosis is unknown. Accordingly, we utilized a rat model of acute ischemic nephropathy to test the hypothesis that fenoldopam ameliorates I/R injury-induced apoptosis.
MATERIALS AND METHODS
Animals and Instrumentation
All experiments were carried out in accordance with guidelines laid down by the National Research Council,Citation[[13]] and were approved by the University of Texas M.D. Anderson Cancer Center institutional animal care and use committee. Twenty-four adult male Sprague-Dawley rats weighing 350–400 g each were acclimatized in standard animal quarters for at least 3 days prior to the study. Animals were randomly allocated (by sealed envelope) to one of four groups, and the technician performing the experiment was blinded to the nature of the infusate. Six rats were used for each group. We have generally found that six animals per group are sufficient to be able to differentiate true changes in experimental parameters between groups.Citation[[12]] In all animals, anesthesia was induced with an inhaled mixture of 3–5% isoflurane in oxygen. When the animal was sufficiently anesthetized (judged by loss of ear pinch reflex), a tracheotomy was performed, and the lungs were mechanically ventilated (Inspira ASV Ventilator, Harvard Apparatus Inc., Holliston, MA, USA). Anesthesia was maintained using intraperitoneal urethane (50 mg/kg). The inspired oxygen tension (FiO2) was maintained at 35% throughout the experiment. Muscle relaxation to permit mechanical ventilation (volume controlled, time cycled with tidal volume of 2 mL/100 g body weight and respiratory rate of 30 breaths per min and zero PEEP, Inspira, Harvard Apparatus) was achieved with intravenous boluses of pancuronium bromide (20 μg/100 g) as required. The carotid artery and jugular vein were cannulated (PE-20, Harvard Apparatus) to facilitate monitoring of arterial blood pressure and infusion of study fluids. An intravenous infusion of 0.9% saline was administered to prevent dehydration during the experiment (1 mL/100 g/hr). Core body temperature was maintained at 37°C by an automated rodent homeothermic blanket warming system (Harvard Apparatus).
Experimental Protocol
After instrumentation, and once the animal had reached a stable baseline, the randomization envelope was opened and the animal was assigned to one of the following four groups. Study infusions were begun immediately thereafter.
Saline Sham Group. After surgical preparation (including laparotomy), these rats (n = 6) underwent no further intervention. 0.9% saline was infused via the left jugular vein throughout.
Saline I/R Group. These animals (n = 6) underwent laparotomy, after which the left renal artery was dissected free and then occluded with a microvascular clamp for 60 min. The clamp was then removed to allow reperfusion of the kidney for 4 h. 0.9% saline was infused via the left jugular vein throughout.
Fenoldopam Sham Group. This group (n = 6) underwent the same surgical procedures (including laparotomy) as the saline sham group. In order to avoid unpredictable hypotension, we selected a clinically relevant fixed dose of fenoldopam 0.1 μg/kg/min and this was infused via the left jugular vein throughout.
Fenoldopam I/R Group. These animals (n = 6) were infused with fenoldopam (0.1 μg/kg/min) and underwent laparotomy followed by 60 min of left renal artery clamping and 4 h of reperfusion, as in the saline I/R group.
At the end of each experiment, the animals were killed by inhalation of 5% isoflurane and exsanguination via the arterial line. Kidney samples for histological analysis were immediately fixed in 10% buffered formaldehyde and for microarray and RT-PCR analysis the samples were snap frozen.Citation[[14]]
DNA Fragmentation Analysis
We used the Fluorescein-FragEL DNA fragmentation detection kit (Fluorescein-FragEL, Oncogene Research Products, Boston, MA, USA), an enzymatic detection method, to show nuclei of renal cells with DNA fragmentation. The principle of Fluorescein-FragEL is that terminal deoxynucleotidyl transferase (TdT) catalyzes the addition of fluorescein-labeled and unlabeled deoxynucleotide to the 3′-OH ends generated by endonucleases during apoptotic degeneration.
Paraffin-embedded sections of kidney tissue were deparaffinized and subjected to proteinase K digestion (20 μg/mL). After rinsing with 1X tris-buffered saline (TBS), the specimens were covered with 1 × TdT equilibration buffer for 20 min and then incubated in 60 μl of TdT labeling mixture containing 3 μl TdT enzyme for 1 h at 37°C. A kidney section incubated in 57 μl of TdT labeling mixture without TdT enzyme served as negative control. The specimens were rinsed with 1 × TBS and covered with mounting medium containing DAPI (4,6,-DiAmidino-2-PhenylIndole) as the fluorescent signal-sustaining component and covered with a cover slip. The slide was then observed with a Nikon D-FL EPI-Fluorescence microscope (Nikon Instruments Inc., Lewisville, TX, USA) and the photographs were taken with a Cool Snap-cf Monochrome camera (Roper Scientific GmbH Germany, Ottobrunn, Germany). Measurement by light microscopy was performed in a blinded fashion. Each tissue section was analyzed for DNA fragmentation in the nuclei of renal cells in at least 20 areas (10 in cortex and 10 in medulla) by two blinded investigators and the mean number of stained nuclei was calculated. The original magnification for nuclei counting was × 200.
Microarray Analysis
Gene array expression analysis was performed as described earlierCitation[[12]] using mouse apoptosis gene array (Super Array Bioscience Corporation, Bethesda, MD, USA) containing 96 genes representing 11 functional groups: 1) tumor necrosis factor (Tnf) ligand family, 2) Tnf receptor family, 3) B-cell leukemia/lymphoma 2 (Bcl2) family, 4) caspases, 5) inhibitor of apoptosis (Iap) proteins, 6) Tnf receptor associated (Traf) factors, 7) caspase recruitment domains (Card), 8) death domains, 9) death effector domains, 10) cell death-inducing DFFA-like effector (Cide) domains, and 11) P53 and ataxia telangiectasia mutated (Atm) pathway genes. We started with this highly selected microchip gene array instead of an all-encompassing gene array because the selected genes entail a well-characterized profile governing apoptosis; hence, facilitating interpretation of data, simplifying data acquisition and analysis, and avoiding genes not functionally characterized. Each array was also spotted with a negative control (pUC18 Plasmid DNA) and four housekeeping genes: β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), peptidylprolyl isomerase A (Ppia), and ribosomal protein L13a (Rpl13a). The relative abundance of a particular cDNA transcript was estimated by subtraction of the background spot intensity (average of three blanks) from the values recorded for each spot. Because the overall signal of different arrays may fluctuate substantially, the spot intensity was then normalized to the signal derived from GAPDH. The normalized data were then compared between the groups, and the relative expression level of each gene was expressed as a fold change. When comparing each gene's signal intensity between arrays, we used a two-fold or more increase in the experimental group compared with the control group to represent “stringent” criteria for up-regulation and an increase of less than two-fold to represent “less stringent” criteria. Classifying gene regulation criteria in this manner can provide an index of the reliability of the gene microarray data.
Reverse Transcriptase Polymerase Chain Reaction
For qualitative confirmation of our array results, we used reverse transcriptase polymerase chain reaction (RT-PCR) to confirm the expression of four genes, including Bcl2-associated X protein (Bax), Bcl-associated death promoter (Bad), Bcl2, and Bcl2-like 1 (Bclx), irrespective of their expression profile observed in the microarray analysis. GAPDH was used for the positive control, and a negative control without template RNA was also included. A quantity of 0.5 μg of RNA was added to a master mix consisting of RT-PCR buffer (Qiagen, Valencia, CA, USA) containing 2.5 mM MgCl2, dNTP mix (400 μM of each dNTP), RT-PCR enzyme mix, and the forward and reverse primers for either Bax, Bad, Bcl2, BclXCitation[[15]] or GAPDH.Citation[[16]] Then, 50 μL of this RT-PCR reaction mixture was subjected to RT for 30 min at 55°C, followed by the activation step for DNA polymerase (15 min at 95°C). The cDNA was amplified according to the following thermal cycling parameters: denaturation, annealing, and extension (conditions and primer sequences are shown in ) and a final 10 min extension period at 72°C. The PCR products were then subjected to agarose gel (2%) electrophoresis and visualized using ethidium bromide staining. One-dimensional gel analysis (Image Quant) was used to quantify each band. Pearson correlation was performed to compare the expression patterns of the genes on array analysis with those in the RT-PCR dataset (Prism Version 3.02, Graph Pad Software Inc., San Diego, CA, USA).
Table 1 Primer sequences and annealing conditions for polymerase chain reaction
RESULTS
Anesthesia and Surgical Data
There were no differences between study groups in mean volume of intravenous fluid infused, type of surgical instruments used (or procedural time), amount of anesthetic agent used (although the inhaled concentration of isoflurane was titrated against heart rate and systolic blood pressure), mean core temperature, or pulse oximetry oxygen saturation.
DNA Fragmentation Analysis
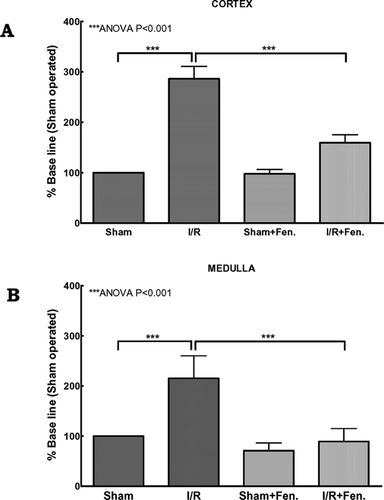
We used the Fluorescein FragEL DNA fragmentation detection kit, which is analogous to the TdT-mediated nick end-labeling (TUNEL) method to visualize DNA strand breakage in apoptotic renal cells after sham operation and I/R with or without fenoldopam infusion. In the kidney cortex, compared to the sham-operated group, 1 h of ischemia followed by 4 h of reperfusion significantly (P < 0.001) induced apoptosis (). Similarly, I/R significantly (P < 0.001) induced apoptosis in the medullary region (). Administration of low dose (0.1 μg/kg/min) fenoldopam has little effect on both the kidney cortex and medulla in the sham-operated individuals. However, under I/R conditions, the fenoldopam infusion significantly (P < 0.001) decreased the I/R-induced apoptosis in both the cortex and medulla ().
Figure 1 Effect of fenoldopam on ischemia/reperfusion induced apoptosis. Quantitative measurement of DNA fragmentation in nuclei of renal (A) cortex and (B) medulla. Each tissue section was analyzed in at least 20 areas (10 in cortex and 10 in medulla) by two blinded investigators, and the mean number of stained nuclei was calculated. The original magnification for nuclei counting was ×200. Group-wise differences were calculated using ANOVA.
Gene Array Analysis
Expression of Apoptosis-Related Genes during Kidney I/R
Of the 96 apoptosis-related genes analyzed, we observed increased expression of 89 genes, while 5 genes were down-regulated after I/R injury. Two genes showed no change. Seventy-three of the 89 up-regulated genes met stringent criteria for up-regulation (at least a two-fold increase over control). Similarly, 3 of 5 down-regulated genes showed complete suppression (stringent criteria).
Effect of Fenoldopam
Overall, fenoldopam suppressed 51 genes and up-regulated 42 genes. Forty-eight of 51 down-regulated genes were completely suppressed (stringent criteria). Similarly, expression of 33 of 42 up-regulated genes was distinct (stringent criteria). Three genes were not affected by the presence of fenoldopam.
Attenuating Effect of Fenoldopam on I/R- Modulated Genes
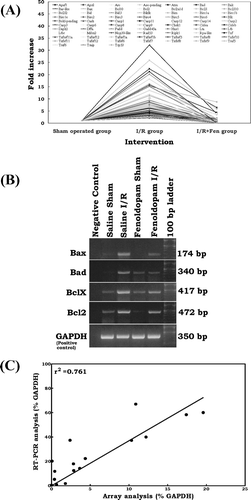
Microarray analysis of the fenoldopam I/R group compared with that of the saline I/R group showed that, using less stringent criteria, all 89 genes up-regulated by I/R injury were returned to the baseline level or less with fenoldopam. Using stringent criteria (≥ two-fold) we observed that all 73 up-regulated genes were attenuated by fenoldopam ().
Figure 2 (A) Effect of fenoldopam on ischemia/reperfusion-induced apoptosis signal transduction genes. All 73 genes up-regulated (stringent criteria ≥ two-fold) by I/R injury were returned to the baseline level or less with fenoldopam. In each group, total RNA from six individual animals were pooled and subjected to microarray analysis. Each spot in the figure represents the expression of the particular gene in each group. A connecting line is drawn for every single gene between groups to illustrate the level of induction and attenuation for each gene after I/R injury with saline and fenoldopam, respectively. (B) RT-PCR analysis of Bax, Bad, Bcl2, and Bclx mRNA expression in rat kidney after sham operation and ischemia/reperfusion with or without fenoldopam. GAPDH was used as positive control. Negative controls without template RNA were shown in lane 1. (C) Correlation analysis of the expression levels for Bax, Bad, Bcl2, and Bclx between the microarray analysis and RT-PCR analysis.
RT-PCR Validates the Microarray Analysis
The expression data from RT-PCR analysis were compared with the results of microarray analysis. All four genes analyzed by RT-PCR showed intervention-specific changes in expression (). Under saline sham and fenoldopam sham conditions, rat kidneys expressed normal levels of Bax mRNA, whereas the mRNA level was much higher following I/R. This I/R-induced Bax mRNA expression was markedly reduced with fenoldopam infusion. Similarly, normal baseline Bad mRNA expression was observed with sham controls with or without fenoldopam. We observed noticeably high Bad expression after I/R injury, which was reduced almost to baseline with fenoldopam. As with Bad, the mRNA levels of BclX were at baseline in the sham controls in the presence or absence of fenoldopam. Enhanced expression of BclX was evident after I/R injury and was reduced in the fenoldopam I/R group. Normal baseline levels of Bcl2 mRNA were observed after sham controls. However, Bcl2 mRNA expression was markedly enhanced after I/R injury. This enhanced Bcl2 expression after I/R injury was significantly reduced with fenoldopam. Good correlation (R2 = 0.76) between the microarray and RT-PCR analysis results was observed for all four genes studied ().
DISCUSSION
This study was designed to explore the potential of the selective dopamine (DA1) receptor agonist, fenoldopam, to protect against experimental surgical acute renal I/R injury. Apoptosis after I/R constitutes a potential trigger of inflammationCitation[[8]] and we studied the effect of fenoldopam on the apoptotic response in a rat model of surgical acute renal I/R injury. We found that I/R injury caused apoptosis both in the cortex and medullary regions of the kidney. Evidence for the early involvement of apoptosis is provided by the presence of DNA fragmentation. Induction of apoptosis has been reported to occur during the reperfusion phase.Citation[[7]] The mechanism of post-I/R apoptosis is attributed to the increased activity of endonuclease by increasing calcium entry into cells, inflammation, ATP depletion, or reactive oxygen species (ROS) release.Citation[[8]], Citation[[17]] ROS induced apoptosis by causing DNA damage, oxidation of lipid membranes, and activation of the proteins responsible for apoptosis.Citation[[18]] Daemen and colleagues demonstrated that inhibition of this I/R-induced apoptosis prevents subsequent inflammation and renal dysfunction.Citation[[8]] We recently established that fenoldopam significantly attenuated the I/R-induced NFκB DNA binding activity and differentially modulated expression of I/R-induced NFκB signal transduction and proinflammatory genes.Citation[[12]] In the present study, we demonstrated that treatment with fenoldopam resulted in a significant decrease of I/R injury-induced apoptosis.
To explore and further substantiate these findings, we studied the effect of fenoldopam on I/R injury-induced renal gene expression. We found that 1 h of ischemia followed by 4 h of reperfusion in the renal tissue activated the expression of Tnf ligand family genes and their receptors, Caspases, Trafs, death domain genes, death effector domain genes, Bcl-2 family genes, and others. A number of earlier studies established the induction of these apoptosis-related genes after I/R. Recently, Supaveikin and colleagues demonstrated that 45 min of ischemia followed by 0, 3, 12 and 24 h of reperfusion consistently up-regulated various members of both extrinsic and intrinsic apoptotic signaling pathway.Citation[[5]] Caspases are the molecular executioners of apoptosis and are activated by two converging signaling pathways, including the extrinsic Tnf ligand-receptor binding signaling and the intrinsic mitochondria mediated signaling.Citation[[19]] Binding of the Tnf ligands trimerizes the receptors and recruits death domain-containing molecules to their cytoplasmic regions of the receptors to form a death-inducing signaling complex. The interactions between the death effector domains lead to the recruitment of several pro-caspase-8 molecules, a key initiator caspase that instigates the downstream caspase cascade.Citation[[20]] On the other hand, the apoptotic signal transduction in response to an intrinsic signal involves Bcl2-mediated mitochondrial release of proapoptotic molecules.Citation[[21]] Based on their structure and functional similarities, Bcl2 family members are divided into the proapoptotic (Bax, Bcl2 homologous antagonist/killer (Bak), Bcl-2-related ovarian killer (Bok) protein), antiapoptotic (Bcl2, Bclx, Bcl2-like 2 (Bclw), Myeloid cell leukemia (Mcl) sequence 1), and death effector molecules (Bcl2 interacting mediator of cell death (Bim), BH3 interacting domain death agonist (Bid), Bcl2-interacting killer-like (Bik), BCL2/adenovirus E1B 19 kDa-interacting protein 1 (Bnip3), Bad), which can activate proapoptotic molecules or inactivate anti-apoptotic molecules. These pro-apoptotic and anti-apoptotic molecules homodimerize or heterodimerize and the heterodimerization of a pro-apoptotic and anti-apoptotic member can nullify the functions of each. The outcome of the cell that received an apoptotic stimulus depends on the ratio of the death promoter to the death suppressor.Citation[[19]] The induction of these molecules after I/R in the present study served as positive controls for our model.
In the present study, we demonstrated that infusion of low dose fenoldopam reduced the expression of all the 73 apoptosis-related genes induced by I/R. In the past, several strategies, including the use of antisense oligonucleotides, recombinant proteins, small molecules to disrupt protein-protein interactions, and caspase inhibitors were used to prevent the I/R-induced apoptosis. Caspase inhibition has shown remarkable efficacy in inhibiting apoptotic cell death in different models of I/R injury,Citation[[19]] including acute renal I/R.Citation[[8]] On the other hand, Varfolomeev and colleagues demonstrated in caspase-8 deficient mice that it is resistant to extrinsic apoptotic signaling, but susceptible to the mitochondrial death pathway.Citation[[22]] However, in the present study, we observed the reduced expression of both extrinsic and intrinsic apoptotic signaling pathway genes. The reduced expression of these I/R injury-induced Tnf ligand and receptor family genes, Bcl2 family genes, caspases, Iap family genes, Traf family genes, Card family genes, death domains, death effector domains, Cide family genes, and p53 and Atm pathway genes after fenoldopam administration demonstrates the inhibitory effect of fenoldopam on I/R-induced apoptosis and its subsequent inflammation.
The results presented here demonstrate that 1) renal I/R results in the induction of apoptosis and activation of apoptosis-related genes, and 2) for the first time in vivo, that fenoldopam attenuates the I/R-induced apoptosis and the expression of apoptosis-related genes.
The limitations of the present study include the following: the analysis of pooled RNA (we pooled our animals' RNA to increase the group-wise signal to noise ratio and contain costs), which could have masked a potentially heterogeneous (individual) response to injury. Despite this, there were no other suggestions that this was indeed the case. We sampled tissue at only one time point, which may have resulted in our missing changes in slow- and fast-response genes.
CONCLUSION
In conclusion, we investigated the effect of fenoldopam on I/R injury-induced apoptosis and on the expression of apoptosis signal transduction genes in a rat model of surgical ischemic ARF. We report that 1) I/R significantly induced apoptosis, 2) I/R stimulated the expression of 73 apoptosis signal transduction genes, 3) fenoldopam significantly reduced the associated apoptosis, and 4) fenoldopam attenuated all 73 I/R injury-induced genes. We believe our data suggest that fenoldopam has anti-apoptotic activity that is mediated via both extrinsic Tnf ligand–receptor and intrinsic Bcl2 mediated mitochondrial signaling pathways.
ACKNOWLEDGMENT
This work was supported by departmental funds and institutional research grant #1–8779701 to Dr. Andrew D. Shaw.
REFERENCES
- Thadhani R, Pascual M, Bonventre JV. Acute renal failure. N. Engl. J. Med. 1996; 334: 1448–1460, [PUBMED], [INFOTRIEVE], [CSA]
- Chatterjee PK, Zacharowski K, Cuzzocrea S, Otto M, Thiemermann C. Inhibitors of poly (ADP-ribose) synthetase reduce renal ischemia-reperfusion injury in the anesthetized rat in vivo. Faseb. J. 2000; 14: 641–651, [INFOTRIEVE], [CSA]
- Ozden A, Sarioglu A, Demirkan NC, Bilgihan A, Duzcan E. Antithrombin III reduces renal ischemia-reperfusion injury in rats. Res. Exp. Med. (Berl). 2001; 200: 195–203, [CSA]
- Lieberthal W, Levine JS. Mechanisms of apoptosis and its potential role in renal tubular epithelial cell injury. Am. J. Physiol. 1996; 271: F477–488, [INFOTRIEVE], [CSA]
- Supavekin S, Zhang W, Kucherlapati R, Kaskel FJ, Moore LC, Devarajan P. Differential gene expression following early renal ischemia/reperfusion. Kidney. Int. 2003; 63: 1714–1724, [INFOTRIEVE], [CSA], [CROSSREF]
- Thornberry NA, Lazebnik Y. Caspases: enemies within. Science 1998; 281: 1312–1316, [INFOTRIEVE], [CSA], [CROSSREF]
- Daemen MA, de Vries B, Buurman WA. Apoptosis and inflammation in renal reperfusion injury. Transplantation 2002; 73: 1693–1700, [INFOTRIEVE], [CSA], [CROSSREF]
- Daemen MA, van ‘t Veer C, Denecker G, Heemskerk VH, Wolfs TG, Clauss M, Vandenabeele P, Buurman WA. Inhibition of apoptosis induced by ischemia-reperfusion prevents inflammation. J. Clin. Invest. 1999; 104: 541–549, [INFOTRIEVE], [CSA]
- Tumlin JA, Dunbar LM, Oparil S, Buckalew V, Ram CV, Mathur V, Ellis D, McGuire D, Fellmann J, Luther RR. Fenoldopam, a dopamine agonist, for hypertensive emergency: a multicenter randomized trial. Fenoldopam Study Group. Acad. Emerg. Med. 2000; 7: 653–662, [INFOTRIEVE], [CSA]
- Horn PT, Murphy MB. Therapeutic applications of drugs acting on peripheral dopamine receptors. J. Clin. Pharmacol. 1990; 30: 674–679, [INFOTRIEVE], [CSA]
- O'Connell DP, Ragsdale NV, Boyd DG, Felder RA, Carey RM. Differential human renal tubular responses to dopamine type 1 receptor stimulation are determined by blood pressure status. Hypertension 1997; 29: 115–122, [INFOTRIEVE], [CSA]
- Aravindan N, Mohan NAndrew DS. Fenoldopam inhibits nuclear translocation of nuclear factor kappa B in a rat model of ischemic acute renal failure. J. Cardiothorac. Vasc. Anesth., 2005; in press, [CSA]
- National Research Council Guide for the Care and Use of Laboratory Animals. Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council. National Academy Press, WashingtonDC 1996; 1–118
- Maas W, de Graaf I, Schoen E, Koster HVan de Sandt J, Groten JP. Assessment of some critical factors in the freezing technique for the cryopreservation of precision-cut rat liver slices. Cryobiology 2000; 40: 250–263, [INFOTRIEVE], [CSA], [CROSSREF]
- Valks DM, Kemp TJ, Clerk A. Regulation of Bcl-xL expression by H2O2 in cardiac myocytes. J. Biol. Chem. 2003; 278: 25542–25547, [INFOTRIEVE], [CSA], [CROSSREF]
- Taniguchi A, Tajima T, Nonomura K, Shinohara N, Mikami A, Koyanagi T. Expression of vascular endothelial growth factor and its receptors Flk-1 and Flt-1 during the regeneration of autotransplanted adrenal cortex in the adrenalectomized rat. J. Urol. 2004; 171: 2445–2449, [INFOTRIEVE], [CSA], [CROSSREF]
- Lieberthal W, Menza SA, Levine JS. Graded ATP depletion can cause necrosis or apoptosis of cultured mouse proximal tubular cells. Am. J. Physiol. 1998; 274: F315–327, [INFOTRIEVE], [CSA]
- Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Today 1994; 15: 7–10, [CSA]
- Padanilam BJ. Cell death induced by acute renal injury: a perspective on the contributions of apoptosis and necrosis. Am. J. Physiol. Renal Physiol. 2003; 284: F608–627, [INFOTRIEVE], [CSA]
- Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science 1998; 281: 1305–1308, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Joza N, Kroemer G, Penninger JM. Genetic analysis of the mammalian cell death machinery. Trends Genet. 2002; 18: 142–149, [INFOTRIEVE], [CSA], [CROSSREF]
- Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P, Wallach D. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 1998; 9: 267–276, [INFOTRIEVE], [CSA], [CROSSREF]