Abstract
Background
Emerging evidence suggests that gut microbiota dysbiosis may play a critical role in the development of lupus nephritis (LN). However, the specific characteristics of the gut microbiota in individuals with LN have not been fully clarified.
Methods
The PubMed, Web of Science, and Embase databases were systematically searched for clinical and animal studies related to the relationship between LN and gut microbiota from inception until October 1, 2023. A semiquantitative analysis was used to assess the changes in gut microbial profiles.
Results
A total of 15 clinical studies were selected for analysis, which included 138 LN patients, 441 systemic lupus erythematosus patients, and 1526 healthy controls (HCs). Five different types of LN mouse models were included in 5 animal studies. The alpha diversity was decreased in LN patients compared to HCs. A significant decrease in the Firmicutes/Bacteroidetes (F/B) ratio is considered a hallmark of pathological conditions. Specifically, alterations in the abundance of the phylum Proteobacteria, genera Streptococcus and Lactobacillus, and species Ruminococcus gnavus and Lactobacillus reuteri may play a critical role in the pathogenesis of LN. Remarkably, the gut taxonomic chain Bacteroidetes-Bacteroides-Bacteroides thetaiotaomicron was enriched in LN patients, which could be a crucial characteristic of LN patients. The increased level of interleukin-6, imbalance of regulatory T cells and T helper 17 cells, and decreased level of the intestinal tight junction proteins zonula occludens-1 and claudin-1 also might be related to the pathogenesis of LN.
Conclusions
Specific changes in the abundance of gut microbiota such as decreased F/B ratio, and the level of inflammatory indicators, and markers of intestinal barrier dysfunction may play a crucial role in the pathogenesis of LN. These factors could be effective diagnostic and potential therapeutic targets for LN.
Introduction
Systemic lupus erythematosus (SLE) is a well-known systemic autoimmune disorder characterized by the production of pathogenic autoantibodies and immune complexes, which contribute to the damage of various organs and tissues [Citation1,Citation2]. The incidence and prevalence of SLE are higher in Asian, Hispanic, and Black populations, and SLE predominantly impacts young women [Citation3]. Approximately 50% of patients with SLE experience kidney damage, which is characterized by symptoms such as hematuria, proteinuria, edema, or decreased renal function, known as lupus nephritis (LN) [Citation4,Citation5]. The prevalence of LN is reported to be 33-82% in Asian individuals with SLE and 34-51% in African American individuals [Citation6]. Currently, LN is considered the most significant risk factor for the incidence and mortality of SLE [Citation5]. Meanwhile, the management of LN relies on steroids and nonselective immunosuppressive drugs, but only 50-70% of patients with LN achieve clinical remission, and 10-20% of patients progress to end-stage kidney disease within 5 years of the initial diagnosis [Citation4,Citation7]. The prognosis suggests that the pathogenesis and treatment of LN remain an unresolved challenge.
A growing body of evidence suggests that gut dysbiosis may be associated with LN by contributing to dysregulated immunity in its latent pathogenesis [Citation6]. The human gut microbiota includes a complex assemblage of 100 trillion microorganisms, which have the ability to influence the metabolism of host enterocytes and impact both local and systemic immunity [Citation8–10]. An increasing number of studies have reported a correlation between gut dysbiosis and immune-related diseases, including allergies, multiple sclerosis, and rheumatoid arthritis [Citation11–13]. In particular, the connection between SLE and the gut microbiota has been extensively demonstrated. In SLE, there is often an imbalance in the gut microbiota, known as gut dysbiosis, which is characterized by an increased presence of Enterococcus gallinarum and Enterobacteriaceae, and a decreased presence of Ruminococcaceae [Citation14,Citation15]. The translocation of gut bacteria, the promotion of systemic inflammation, epitope spreading, and molecular simulation are the main mechanisms underlying the pathogenesis of gut microbiota dysbiosis in the development of LN [Citation16]. In another aspect, the gut microbiota plays a critical role in the pathogenesis of chronic kidney disease (CKD) [Citation17,Citation18]. The gut microbiota is a vital component of the gut-kidney axis, which is a complicated interaction between the gut and kidney [Citation19]. Dysregulation of the gut microbiota can disrupt the integrity of tight junctions, resulting in increased epithelium permeability, commonly referred to as ‘leaky gut’, which can lead to disorders in the intestinal mucosal immune system and inflammation and contribute to the development and progression of immune-related nephropathies [Citation9,Citation20]. Moreover, gut dysbiosis may lead to an imbalance between immune tolerance and immune responses, causing the abnormal proliferation and differentiation of B and T cells with the production of autoantibodies and inflammatory factors that could contribute to CKD onset and progression [Citation21,Citation22]. Therefore, we hypothesize that the gut microbiota may play a role in the progression of SLE to LN. Recent cohort studies and animal experiments have provided initial insights into the significant impact of gut dysbiosis in LN. However, due to variations in participant selection and animal models, the findings have not been uniformly consistent. As a result, the specific characteristics of the gut microbiota in LN remain to be fully determined.
Herein, we systematically summarized the alterations in the gut microbiota in LN patients, identified the key gut microbiota and metabolites involved in the occurrence and progression of LN, and searched for the possible mechanisms by which the gut microbiota participates in the onset and progression of LN with the goal of identifying bacterial targets for the prevention and diagnosis of LN. Meanwhile, we aimed to investigate whether modulation of the gut microbiota could serve as an effective therapeutic strategy for LN.
Materials and methods
This systemic review was registered in the PROSPERO database (CRD42022379603) in advance and adhered to the preferred reporting project methods in the Systematic Review and Meta-Analysis (PRISMA) guidelines.
Study selection and quality assessment
We conducted a systematic search of PubMed, Web of Science, and Embase databases from their inception date to October 1, 2023, without any language or publication date limitations, to search for studies on the gut microbiome characteristics of patients with LN. The study search strategy involved the crossing of LN studies (Title, Abstract, Keywords, [Mesh] “Lupus Glomerulonephritis” or “Lupus nephritis”) and microbiome studies (Title, Abstract, Keywords, [Mesh] “Microbiota”) and the search strings utilized were capable of being discovered in Table S1. The title, abstract, and full-text views of articles were screened independently by two reviewers to ensure consistency. Disagreements were settled by consultation with the third investigator. The inclusion criteria for studies were as follows: (1) studies that investigated the correlation between LN and gut microbiota composition, (2) studies on the intervention of gut microbiota, and (3) clinical studies related to SLE and gut microbiota that enrolled a subset of SLE patients with renal involvement. The exclusion criteria were as follows: (1) studies on SLE and microbiota that did not mention renal involvement in the SLE patients included, (2) study protocols, and (3) review articles. Moreover, the Newcastle–Ottawa scale was used to assess the quality of the shortlisted studies [Citation23].
Data extraction and statistical description
The key components extracted for our study included the features of the study, analysis method, alpha diversity, beta diversity, differential abundance of gut microbiota, and functional characterization of gut microbiota. Indefinite information was acquired by contacting the relevant authors. The alpha diversity represented the community diversity (Simpson and Shannon) and richness (Chao1 and ACE), and the beta diversity represented the similarity of the species diversity within the community. For the relative abundance of gut microbiota, we conducted a semiquantitative analysis in our systemic review. A changed parameter [Citation24] was defined, whereby a significant change was deemed noteworthy if it consistently occurred in the same direction in more than two studies without contradictory results. Additionally, if three or more studies reported a consistent result, it was considered likely specific to LN. Proportions (n/N) were used to represent the alterations in gut microbiota profiles.
Results
Study characteristics
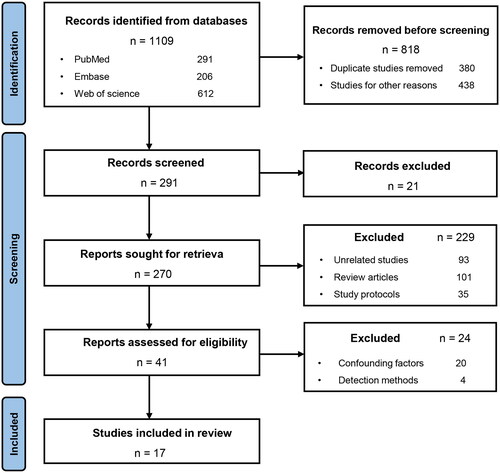
A total of 291 potentially eligible studies were acquired from the literature search. After screening the titles and abstracts, and excluding duplicate search results, 15 relevant clinical studies [Citation25–39] and 7 animal studies [Citation28,Citation38,Citation40–44] were included in this systematic review (). The studies included 138 LN patients, 441 SLE patients, and 1526 healthy controls (HCs) () as well as five different types of LN mouse models (). Four (4/15) clinical studies [Citation25,Citation31,Citation34,Citation38] only enrolled female LN patients, while the remaining 11 (11/15) [26–30,32,33,35–37,39] had no gender restriction. Twelve (12/15) clinical studies [Citation25–27,Citation30–34,Citation36–39]and six (6/7) animal studies [Citation28,Citation38,Citation41–44] analyzed the composition of the gut microbiota using 16S rRNA sequencing techniques, while three (3/15) clinical studies [Citation28,Citation33,Citation35] utilized metagenomic sequencing. Polymerase chain reaction (PCR) was used in one (1/15) clinical study [Citation29] and one (1/7) animal study [Citation40].
Table 1. Characteristics of clinical studies included in this systemic review.
Table 2. Characteristics of animal studies included in this systemic review.
Quality of included studies
After evaluating the 15 clinical studies using the Newcastle–Ottawa scale [Citation23], five (5/14) clinical studies [Citation26,Citation27,Citation32,Citation34,Citation39] were rated with a score of 7, eight (8/15) [Citation25,Citation28–31,Citation33,Citation35,Citation38]with a score of 6, and two (2/15) [Citation36,Citation37] with a score of 5, indicating that the overall quality of the included studies was universally high (Table S2).
Differences in gut microbiota diversity in LN
In eight (8/15) of the clinical studies, alpha diversity was found to be lower in LN patients than in HCs, mainly by the Chao1 [Citation25,Citation36,Citation37,Citation39], Shannon indexes [Citation26,Citation28,Citation33,Citation35,Citation37,Citation39] and Simpson indexes [Citation26] and in three (3/15) clinical studies [Citation31,Citation32,Citation45], no significant differences were found. In terms of beta diversity, seven (7/15) clinical studies [Citation25,Citation26,Citation28,Citation30,Citation31,Citation33,Citation39] found that there was a difference in the diversity between the two groups, while four (4/15) clinical studies [27,Citation32,Citation36,Citation37] reported no significant difference (Table S3).
Differences in the gut microbiota composition in LN
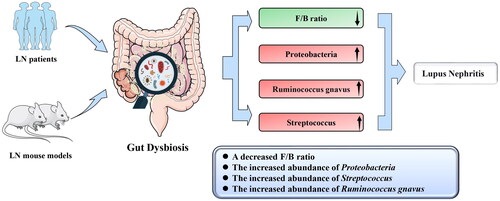
At the phylum level, six (6/15) clinical studies [Citation29–32,Citation34,Citation36] found a lower Firmicutes/Bacteroidetes (F/B) ratio in LN patients than in the HCs (). Four (4/14) clinical studies [Citation32,Citation33,Citation36,Citation37] and one (1/7) animal study [Citation43] found a higher abundance of Proteobacteria in LN patients than in HCs. At the genus level, three (3/14) clinical studies [27,Citation35,Citation39] found a higher abundance of Streptococcus in LN patients than in the HCs. At the species level, two (2/14) clinical studies [Citation25,Citation28] found a higher abundance of Ruminococcus gnavus (R. gnavus, RG) in LN patients than in HCs. Additionally, the abundance of Ruminococcus torques spp. (R. torques spp.) was found to be higher in the LN group than in the control group in one (1/14) clinical study [Citation28] and two (2/5) animal studies [Citation28,Citation41], but one (1/7) animal study [Citation44] found a lower abundance. The genus Lactobacillus and species Lactobacillus reuteri (L. reuteri) were mentioned as having changes in abundance in two (2/14) clinical studies [Citation32,Citation38] and three (3/7) animal studies [Citation38,Citation42,Citation43]. The results with consistent variation in bacterial abundance were extracted to draw a phylogenetic diagram (Figure S1). From the phylogenetic diagram, it was observed that the gut taxonomic chain Bacteroidetes–Bacteroides–Bacteroides thetaiotaomicron (B. theta) was significantly enriched in LN. And changes in the abundance of gut microbiota are independent of geography ().
Figure 2. Changes in the abundance of the gut microbiota in the LN in the clinical and animal studies. The abundance of the phylum Proteobacteria, genus Streptococcus, and species Ruminococcus gnavus were enriched in LN. The F/B ratio was decreased in the LN. F/B, Firmicutes/Bacteroidetes. LN, lupus nephritis.
Changes in inflammatory indicators and intestinal barrier dysfunction markers in LN
Two (2/14) clinical studies [Citation25,Citation34] and five (5/7) animal studies [Citation38,Citation40,Citation41,Citation43,Citation44] reported changes in inflammatory markers and intestinal barrier dysfunction markers. In both clinical and animal studies [Citation25,Citation36,Citation44], the inflammatory indicator interleukin (IL)-6 was found to be increased in patients with LN but decreased after Lactobacillus treatment [Citation40]. The level of calcium-protective protein, an intestinal barrier deficiency biomarker was higher in patients with LN, and the expression of the gut tight junction protein claudin-1 (Cldn1) and Zonula Occludens-1 (ZO-1) was downregulated in LN model mice compared with controls [Citation41]. However, the expression of Cldn1 and ZO-1 was upregulated after Lactobacillus treatment [Citation40]. The macrophage chemoattractant MCP-1 was increased in both clinical and animal studies [Citation34,Citation41]. Increases in immune cells, such as T helper 17 (Th17) cells [Citation41], group 3 innate lymphoid cells (ILC3s) [Citation41], and plasmacytoid dendritic cells (pDCs) [Citation38], were found in two (2/5) animal studies. IgG anti-RG2 antibody was found to be increased in LN patients and mouse models in a clinical study [Citation25] and an animal study [Citation42].
Functional characteristics of the gut microbiota in LN
Two (2/15) clinical studies reported that the sulfur metabolism pathway [Citation28], some glycan degradation pathways [Citation31], and oxidative phosphorylation processes [Citation31] were enriched in the gut microbiota of LN patients, as determined by KEGG analyses. One (1/7) animal study [Citation38] found that the type I interferon (IFN) pathway was enriched in animal models of LN.
Discussion
We conducted a comprehensive analysis of the available evidence on the characteristics of microbial taxa in LN patients to identify specific gut microbiota that may contribute to the development and progression of the disease. To our knowledge, this is the first effort to systemically explore the alteration and effect of the gut microbiota on the pathogenesis of LN. The present study revealed a decrease in the Chao1 and Shannon indices in LN patients, indicating a compromised richness and diversity of gut microbiota. Furthermore, we observed a decreased F/B ratio and an increased abundance of the phylum Proteobacteria, genus Streptococcus, and species R. gnavus, which may be closely associated with LN (). Of particular significance, the gut taxonomic chain Bacteroidetes-Bacteroides-B. theta was significantly enriched in LN, as indicated by the analysis of the phylogenetic diagram, suggesting its pivotal role in the pathogenesis of LN. This review provides novel insights not only into understanding the pathogenesis of LN but also into the development of targeted therapeutic methods.
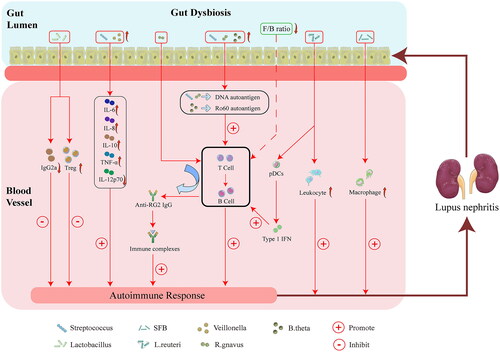
Figure 3. Possible mechanism linking gut microbiota dysbiosis to LN. Alteration of specific microbial taxa may contribute to the pathogenesis and progression of LN through the following four factors. First, the alteration of specific microbial taxa can induce LN by promoting kidney M2-like macrophage infiltration and leukocyte recruitment. Second, the gut microbiota may contribute to LN by enhancing the autoimmune response. Third, Streptococcus combined with Veillonella can enhance the autoimmune response, including by increasing IL-6, IL-8, IL-10, and TNF-α levels, whereas decreased IL-12p70 may induce LN. Fourth, the alteration of specific microbial taxa can increase the abundance of Tregs, while the decrease in the deposition of IgG2a may alleviate LN. SFB, Segmented Filamentous Bacteria. L. reuteri, Lactobacillus reuteri. R. gnavus, Ruminococcus gnavus. B. theta, Bacteroides thetaiotaomicron. F/B, Firmicutes/Bacteroidetes. pDCs, plasmacytoid dendritic cells. IL-12p70, interleukin-12p70. IL-10, interleukin-10. IL-8, interleukin-8. IL-6, interleukin-6. TNF-α, tumor necrosis factor α. Tregs, regulatory T cells. LN, lupus nephritis.
A decreased F/B ratio plays a crucial role in the pathogenesis of LN
A growing body of evidence indicates that a change in the F/B ratio may be a hallmark of pathological conditions [Citation45,Citation46]. A decreased F/B ratio can lead to an imbalance in regulatory T cells (Tregs) and Th17 cells, which seems likely to aggravate preexisting gut inflammation [Citation8,Citation47]. A reduced F/B ratio in patients with SLE is associated with a decreased level of IFN-γ, which is a primary characteristic of SLE dysbiosis [Citation31]. The dysbiosis caused by a reduced F/B ratio in lupus patients contributes to the dysfunction of ILC3s, a key contributor to barrier immunity [Citation16]. Moreover, the imbalance of the F/B ratio is in accord with elevated levels of plasma lipopolysaccharide (LPS) in SLE patients; thereby enhancing the activation of B cells and promoting progression of SLE in mouse models [Citation33,Citation40]. The above evidence suggests that a decreased F/B ratio may be an indispensable component of the pathogenesis of LN, primarily due to abnormal immune responses, and play a crucial role in LN by affecting the levels of IFN-γ, causing imbalances in Treg and Th17 cells, and activating B cells.
However, the exact gut microbiota responsible for the lower F/B ratio remains unclear. In our results, the abundance of the gut taxa chain Bacteroidetes-Bacteroides-B. theta was higher in individuals with LN. This increase in the genus Bacteroides, which belongs to the phylum Bacteroidetes, may directly contribute to the decrease in the F/B ratio. Additionally, B. theta contains an epitope protein homolog that is similar to the Ro60 autoantigen, a common factor in SLE, which can bind to B and T cells and potentially induce inappropriate autoimmunity [Citation30,Citation48]. Hence, we propose that B. theta may be the key gut microbiota responsible for the lower F/B ratio in LN.
An increased abundance of Proteobacteria may be a specific hallmark of LN patients
The increased abundance of Proteobacteria in the gut microbiota may be a significant component in the pathogenesis of LN, perhaps based on the autoimmune and inflammatory responses. Elevated levels of Proteobacteria have also been observed in inflammatory bowel diseases (IBD), diabetic kidney disease, and idiopathic membranous nephropathy [Citation49–51]. Thus, we hold the view that the increase in Proteobacteria in LN patients is indicative of potential alterations in the gut environment and disruption of intestinal barrier integrity, which leads to systemic inflammation, hastens the development of kidney disease, and results in more damage [Citation18]. Proteobacteria may have a significant impact on the pathogenesis of LN in two following aspects. First, LPS obtained from Escherichia coli, a member of the phylum Proteobacteria, can stimulate the production of IL-6 through the TLR4–NF-κB and TLR4–p38MAPK pathways, leading to the initiation of an inflammatory response [Citation52]. Coincidentally, animal studies have demonstrated that IL-6 plays a significant role in promoting LN [Citation53]. Second, the abundance of Enterobacteriaceae, a member of the phylum Proteobacteria, is associated with T cells, which are crucial in the immune response [Citation14]. This suggests that changes in the abundance of Enterobacteriaceae may contribute to the pathogenesis and progression of LN. Consequently, the increase in Proteobacteria may serve as a specific marker for alterations in the gut microbiota and may contribute to LN development by modulating the levels of IL-6 and T cells.
Enhanced autoimmune responses induced by Streptococcus may be associated with LN
An increased abundance of Streptococcus can potentially alter the levels of inflammatory cytokines and trigger inappropriate immune responses that influence the development of immune-related disease. Existing studies have found that Streptococcus has a crucial effect on immune-related nephropathies, such as Henoch-Schönlein purpura nephritis, the severity of which is positively associated with Streptococcus abundance [Citation54]. Similarly, the genus Streptococcus was positively correlated with lupus activity in patients with SLE [Citation55]. An enhanced autoimmune response due to streptococcal infection may be an important causative factor for LN. First, the existing evidence has indicated that the combination of Streptococcus and Veillonella increased the production of IL-6, IL-10, IL-8, and tumor necrosis factor α (TNF-α) and inhibits the production of interleukin-12p70 [Citation56]. Interestingly, Azzouz et al. [Citation25] found an expansion of the species Veillonella in patients with SLE, further confirming that the synergy of Streptococcus along with Veillonella may play a significant role in the pathogenesis of SLE. Second, the polysaccharide of Streptococcus pneumonia possesses the same epitope as the pentapeptide found in the anti-dsDNA antibody [Citation57]. In fact, antibodies produced against bacterial antigens can cross-react with host tissue through a process known as ‘molecular mimicry’, and certain species of Streptococcus may utilize antigen presentation to trigger the particular CD4+ T cells and initial stimulation of B cells [Citation58,Citation59]. Molecular mimicry of bacterial orthologs against human autoantigens has been reported to trigger cross-reactive responses of T and B lymphocytes and activate autoimmunity, which is one of the possible pathogeneses of SLE [Citation27]. Thus, the increase in Streptococcus may be involved in the pathogenesis of SLE based on the enhancement of autoimmunity, which could also be a potential mechanism in LN. However, the definite pathogenesis of LN has not been fully elucidated, and further studies are required to identify the role of Streptococcus in LN patients.
R. gnavus exerts important effects that impact the occurrence of LN
R. gnavus is an anaerobic gram-positive taxon belonging to the phylum Firmicutes and family Lachnospiraceae [Citation60]. R. gnavus may exert a pathogenic role by eliciting an inappropriate immune response. R. gnavus is associated with ankylosing spondylitis and IBD [Citation61,Citation62]. Previous investigators pointed out that the abundance of R. gnavus was not only associated with the activity of lupus disease and LN but also inversely correlated with C3 and C4 complement levels [Citation25]. The expansion of R. gnavus is linked to the production of serum antibodies toward strain RG2, and active LN can manifest as a high concentration of IgG anti-RG2 antibodies in patients [Citation25]. In addition, the cell wall lipoglycans of the RG2 strain have antigenic properties that can induce the production of anti-dsDNA antibodies, which can cross-react with lupus anti-dsDNA antibodies and lead to inappropriate immune reactions [Citation25]. Another study showed that the abundance of R. torques spp. was higher in SLE patients and that this species shared a common ancestor with R. gnavus, suggesting a close relationship between these two bacteria [Citation28,Citation41]. Therefore, R. torques spp. may also be important in the pathogenesis of lupus apart from R. gnavus. Although only one animal study [Citation44] appears to have contradictory results for R. torques spp., we believe that the variation in R. torques spp. abundance in LN patients may also be closely related to LN occurrence. However, the specific mechanism remains to be further explored. In summary, molecular mimicry between lipoglycan expressed on the cell wall of intestinal RG2 and native DNA molecules may be the cause of SLE and the progression of SLE to LN.
The dual role of the genus Lactobacillus and species L. reuteri in the pathogenesis of LN
The role of the genus Lactobacillus and species L. reuteri in the pathogenesis of LN is currently a subject of controversy. Our observations indicate that both the genus Lactobacillus and species L. reuteri play a significant role in LN in partially shortlisted studies. On the positive side, Mu et al. [Citation40] discovered that MRL/lpr mice with a ‘leaky gut’ which were treated by increased Lactobacillus colonization. Lactobacillus treatment contributed to fostering an intestinal environment without inflammation, increasing the serum level of IL-10 and restoring the balance of Treg and Th17 cells inside the kidney [Citation40]. On the negative side, Lactobacillus may become harmful under certain genetic or environmental conditions [Citation38]. L. reuteri gavage increased splenomegaly and pDCs accumulation in the spleen and Peyer’s patches (PP) and exacerbated leukocyte recruitment to the kidney [Citation38]. Furthermore, L. reuteri-monocolonized mice showed increased accumulation of pDCs in the spleen and mesenteric lymph nodes, as well as worsening signs of LN. Moreover, short-chain fatty acids (SCFAs) derived from starchy diets inhibited L. reuteri in vitro and in vivo [Citation38]. Based on the above discussion, the genus Lactobacillus and species L. reuteri have an impact on the progression of LN, but further in-depth studies are needed to determine whether this effect is beneficial or harmful.
The role of inflammatory and intestinal barrier dysfunction markers in the pathogenesis of LN
The alteration of gut permeability may be an integral part of the process by which the gut microbiota leads to the occurrence of LN. Increased levels of intestinal tight junction proteins [Citation41] and immune cells [Citation38,Citation41] were observed in animal models. Segmented Filamentous Bacteria (SFB) may decrease the expression of the intestinal tight junction proteins Cldn1 and ZO-1 while Lactobacillus treatment can upregulate their expression, suggesting that changes in the gut microbiota can affect intestinal permeability [Citation40,Citation41]. ‘Leaky gut’ was found to increase LPS levels, thereby enhancing B-cell activation and promoting lupus progression in an animal study [Citation40]. Additionally, increased levels of IL-6 along with an imbalance of Tregs and Th17 cells were observed in SFB-colonized LN mouse models, which may enhance the inflammatory response in mouse models and was reversed by Lactobacillus treatment [Citation40,Citation41]. Furthermore, Toll-like receptor 7 (TLR7)-dependent translocation of L. reuteri increases the activation of pDCs and the production of type I IFN, thereby deteriorating autoimmune manifestations of SLE [Citation38]. Hence, gut disorders may lead to the downregulation of intestinal tight junction protein expression, resulting in increased intestinal permeability and subsequently promoting the occurrence of inflammatory responses and immune disorders in the host. This may be a critical factor in the pathogenesis of LN.
The underlying effect of bacterial metabolites in the pathogenesis of LN
SCFAs are the most frequently mentioned bacterial metabolites that have beneficial effects on many nephropathies [Citation63–65]. The phylum Bacteroidetes was capable of producing high amounts of acetate and propionate, while the phylum Firmicutes generated a significant amount of butyrate [Citation16]. A previous study found that SCFAs are capable of regulating the gut microbiota and host metabolism [Citation66]. Researchers have also discovered that changes in SCFAs production are associated with gut dysbiosis in SLE and that an increase in fecal SCFAs levels is linked to a reduced F/B ratio [Citation34]. Animal models of SLE have shown that SCFAs supplementation can alleviate the lupus phenotype [Citation67]. Moreover, SCFAs may restrain B-cell activation-induced expression of cytidine deaminase and Blimp1, restrict the differentiation of plasma cells and systemic class-switched autoantibodies, and prevent the deposition of IgG1/IgG2a in the kidney [Citation67]. In summary, SCFAs may play a significant role in the progression of SLE to LN and could be an effective strategy to hinder LN progression.
The microbiota-based therapeutic prospects of LN
Successful treatment of LN through diet, probiotics, and fecal microbiota transplantation (FMT) may provide new evidence for understanding the relationship between LN and the gut microbiota. First, nutritional intervention has been demonstrated to ameliorate LN in mouse models [Citation68]. Second, probiotic therapy is another treatment option for LN. Oral administration of Bacteroidetes fragilis can effectively relieve LN [Citation69]. The mechanism involves in decreasing the level of autoantibodies in LN patients, renovating the immune response of B lymphocytes, relieving intestinal inflammation, and restoring the balance of Tregs and Th17 cells in LN mouse models [Citation69]. Third, fecal microbial content from healthy donors was transferred to patients in a procedure known as FMT [Citation70]. Currently, the safety and efficacy of FMT in the treatment of SLE and CKD have been confirmed [Citation71,Citation72]. In the case of SLE treatment, reductions in the level of serum anti-dsDNA antibody, inflammation-related gut microbiota, and the level of IL-6 in the peripheral blood, were observed after 12 weeks of FMT intervention, as well as increases in bacterial taxa producing SCFAs and the production of SCFAs. These compelling results indicated that FMT is a viable treatment regimen for SLE [Citation72]. Therefore, considering the effect of FMT in lupus, FMT could be an available therapeutic strategy for LN. However, this application will require further investigation for validation in the future.
This study aims to provide evidence regarding the underlying mechanisms linking gut dysbiosis and LN, as well as insights into potential strategies for preventing and treating LN. However, it is important to acknowledge the limitations of this systematic review. First, further meta-analysis was not available due to insufficient data and heterogeneity between the enrolled studies and qualitative reporting of the consequences. Second, the critical gut microbiota tightly linked to the progression or onset of LN were not completely determined on account of the nature of the observational studies included in this review. Therefore, it is necessary to conduct more high-quality, multicenter, multinational clinical studies to validate the findings of this study.
Conclusions
The present systemic review supports the notion that a decreased F/B ratio may be a particular hallmark of the gut microbiota in LN patients, which is associated with decreased levels of IFN-γ, imbalances in Treg and Th17 cells, and the activation of B cells. In addition, an increased abundance of the phylum Proteobacteria was also a specific hallmark of the gut microbiota in LN patients, which may be linked to the levels of IL-6 and T cells. Notably, the gut taxonomic chain Bacteroidetes-Bacteroides-B. theta was elevated in LN, further supporting the aforementioned speculation. In addition, the enrichment of the genus Streptococcus and species R. gnavus may also play a role in the pathogenesis of LN by enhancing autoimmunity and molecular mimicry. These findings contribute to a better understanding of how the gut microbiota influences LN and its unrecognized role in the prevention and treatment of LN.
Author contributions
Study design: S. Sun. Data acquisition: A. Wang, Y. Qin. Data analysis: A. Wang, X. Ning, J. Zhao. Quality control: S. Sun, J. Zhao, X. Ning. Created figure: A. Wang, Y. Xing. Modify the manuscript: J. Zhao, Y. Qin, Y. Zhang, Y. Wang, Z. Yu, J. Yan, M. Han, G. Yuan, Y. Hui, S. Guo. Each author contributed significant intellectual content during the drafting or revision of the manuscript and assumed responsibility for the overall work.
Funding sources
This work was supported by National Natural Science Foundation of China grants (reference number: 82170722, 82270715), Clinical research project of Fourth Military Medical University (reference number: 2021LC2205), The Xijing Hospital discipline promoting plan (reference number: XJZT21L15), Key Research and Development Plan of Shaanxi Province grant(reference number: No.2023-ZDLSF-15), and Postdoctoral Lan Jian Sustentation Fund of the Fourth Military Medical University (reference number: LJ20220102).
Statement of ethics
The study type does not require an ethics statement and does not use human or animal subjects or materials.
Supplemental Material
Download PDF (332.1 KB)Acknowledgment
The authors would like to acknowledge all statisticians for participating in this study. Figures were partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
Disclosure statement
The authors have no relevant financial or non-financial interests to disclose.
Data availability statement
All data generated or analyzed in the course of this study are included in the document and its supplementary materials. Further inquiry can be available from the corresponding author.
References
- Kim JW, Kwok SK, Choe JY, et al. Recent advances in our understanding of the link between the intestinal microbiota and systemic lupus erythematosus. Int J Mol Sci. 2019;20(19):1. doi:10.3390/ijms20194871.
- Kiriakidou M, Ching CL. Systemic lupus erythematosus. Ann Intern Med. 2020;172(11):ITC81–13. doi:10.7326/AITC202006020.
- Barber MRW, Drenkard C, Falasinnu T, et al. Global epidemiology of systemic lupus erythematosus. Nat Rev Rheumatol. 2021;17(9):515–532. doi:10.1038/s41584-021-00668-1.
- Yu F, Haas M, Glassock R, et al. Redefining lupus nephritis: clinical implications of pathophysiologic subtypes. Nat Rev Nephrol. 2017;13(8):483–495. doi:10.1038/nrneph.2017.85.
- Almaani S, Meara A, Rovin BH. Update on lupus nephritis. Clin J Am Soc Nephrol. 2017;12(5):825–835. doi:10.2215/CJN.05780616.
- Anders HJ, Saxena R, Zhao MH, et al. Lupus nephritis. Nat Rev Dis Primers. 2020;6(1):7. doi:10.1038/s41572-019-0141-9.
- Rovin BH, Adler SG, Barratt J, et al. Executive summary of the KDIGO 2021 guideline for the management of glomerular diseases. Kidney Int. 2021;100(4):753–779. doi:10.1016/j.kint.2021.05.015.
- Rahbar Saadat Y, Hejazian M, Bastami M, et al. The role of microbiota in the pathogenesis of lupus: dose it impact lupus nephritis? Pharmacol Res. 2019;139:191–198. doi:10.1016/j.phrs.2018.11.023.
- Lau WL, Chang Y, Vaziri ND. The consequences of altered microbiota in immune-related chronic kidney disease. Nephrol Dial Transplant. 2021;36(10):1791–1798. doi:10.1093/ndt/gfaa087.
- Shi N, Li N, Duan X, et al. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. 2017;4:14. doi:10.1186/s40779-017-0122-9.
- Wopereis H, Sim K, Shaw A, et al. Intestinal microbiota in infants at high risk for allergy: effects of prebiotics and role in eczema development. J Allergy Clin Immunol. 2018;141(4):1334–1342 e5. doi:10.1016/j.jaci.2017.05.054.
- Cignarella F, Cantoni C, Ghezzi L, et al. Intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cell Metab. 2018;27(6):1222–1235 e6. doi:10.1016/j.cmet.2018.05.006.
- Mei L, Yang Z, Zhang X, et al. Sustained drug treatment alters the gut microbiota in rheumatoid arthritis. Front Immunol. 2021;12:704089. doi:10.3389/fimmu.2021.704089.
- Xiang S, Qu Y, Qian S, et al. Association between systemic lupus erythematosus and disruption of gut microbiota: a meta-analysis. Lupus Sci Med. 2022;9(1):e000599. doi:10.1136/lupus-2021-000599.
- Manfredo Vieira S, Hiltensperger M, Kumar V, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359(6380):1156–1161. doi:10.1126/science.aar7201.
- Chen Y, Lin J, Xiao L, et al. Gut microbiota in systemic lupus erythematosus: a fuse and a solution. J Autoimmun. 2022;132:102867. doi:10.1016/j.jaut.2022.102867.
- Hu X, Ouyang S, Xie Y, et al. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad Med. 2020;132(6):495–505. doi:10.1080/00325481.2020.1744335.
- Zhao J, Ning X, Liu B, et al. Specific alterations in gut microbiota in patients with chronic kidney disease: an updated systematic review. Ren Fail. 2021;43(1):102–112. doi:10.1080/0886022X.2020.1864404.
- Evenepoel P, Poesen R, Meijers B. The gut-kidney axis. Pediatr Nephrol. 2017;32(11):2005–2014. doi:10.1007/s00467-016-3527-x.
- Vaziri ND, Zhao YY, Pahl MV. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol Dial Transplant. 2016;31(5):737–746. doi:10.1093/ndt/gfv095.
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi:10.1038/nature18848.
- Thaiss CA, Zmora N, Levy M, et al. The microbiome and innate immunity. Nature. 2016;535(7610):65–74. doi:10.1038/nature18847.
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi:10.1007/s10654-010-9491-z.
- Nikolova VL, Smith MRB, Hall LJ, et al. Perturbations in gut microbiota composition in psychiatric disorders: a review and meta-analysis. JAMA Psychiatry. 2021;78(12):1343–1354. doi:10.1001/jamapsychiatry.2021.2573.
- Azzouz D, Omarbekova A, Heguy A, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis. 2019;78(7):947–956. doi:10.1136/annrheumdis-2018-214856.
- Azzouz DF, Chen Z, Izmirly PM, et al. Longitudinal gut microbiome analyses and blooms of pathogenic strains during lupus disease flares. Ann Rheum Dis. 2023;82(10):1315–1327. doi:10.1136/ard-2023-223929.
- Bellocchi C, Fernandez-Ochoa A, Montanelli G, et al. Identification of a shared microbiomic and metabolomic profile in systemic autoimmune diseases. J Clin Med. 2019;8(9):1291. doi:10.3390/jcm8091291.
- Chen BD, Jia XM, Xu JY, et al. An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol. 2021;73(2):232–243. doi:10.1002/art.41511.
- Gerges MA, Esmaeel NE, Makram WK, et al. Altered profile of fecal microbiota in newly diagnosed systemic lupus erythematosus Egyptian patients. Int J Microbiol. 2021;2021:9934533–9934537. doi:10.1155/2021/9934533.
- Greiling TM, Dehner C, Chen XG, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med. 2018;10(434):eaan2306. doi:10.1126/scitranslmed.aan2306.
- Hevia A, Milani C, Lopez P, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5(5):e01548-14. doi:10.1128/mBio.01548-14.
- Li BZ, Wang H, Li XB, et al. Altered gut fungi in systemic lupus erythematosus – a pilot study. Front Microbiol. 2022;13:1031079. doi:10.3389/fmicb.2022.1031079.
- Luo XM, Edwards MR, Mu Q, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol. 2018;84(4):e02288-17. doi:10.1128/AEM.02288-17.
- Rodriguez-Carrio J, Lopez P, Sanchez B, et al. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Front Immunol. 2017;8:23. doi:10.3389/fimmu.2017.00023.
- Tomofuji Y, Maeda Y, Oguro-Igashira E, et al. Metagenome-wide association study revealed disease-specific landscape of the gut microbiome of systemic lupus erythematosus in Japanese. Ann Rheum Dis. 2021;80(12):1575–1583. doi:10.1136/annrheumdis-2021-220687.
- van der Meulen TA, Harmsen HJM, Vila AV, et al. Shared gut, but distinct oral microbiota composition in primary Sjogren’s syndrome and systemic lupus erythematosus. J Autoimmun. 2019;97:77–87. doi:10.1016/j.jaut.2018.10.009.
- Yu M, Li L, Ren Q, et al. Understanding the gut-kidney axis in antineutrophil cytoplasmic antibody-associated vasculitis: an analysis of gut microbiota composition. Front Pharmacol. 2022;13:783679. doi:10.3389/fphar.2022.783679.
- Zegarra-Ruiz DF, El Beidaq A, Iniguez AJ, et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25(1):113–127 e6. doi:10.1016/j.chom.2018.11.009.
- Wang X, Shu Q, Song L, et al. Gut microbiota in systemic lupus erythematosus and correlation with diet and clinical manifestations. Front Med (Lausanne). 2022;9:915179. doi:10.3389/fmed.2022.915179.
- Mu Q, Zhang H, Liao X, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5(1):73. doi:10.1186/s40168-017-0300-8.
- Valiente GR, Munir A, Hart ML, et al. Gut dysbiosis is associated with acceleration of lupus nephritis. Sci Rep. 2022;12(1):152. doi:10.1038/s41598-021-03886-5.
- Johnson BM, Gaudreau MC, Al-Gadban MM, et al. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin Exp Immunol. 2015;181(2):323–337. doi:10.1111/cei.12609.
- Chancharoenthana W, Kamolratanakul S, Yiengwattananon P, et al. Enhanced lupus progression in alcohol-administered Fc gamma receptor-IIb-deficiency lupus mice, partly through leaky gut-induced inflammation. Immunol Cell Biol. 2023;101(8):746–765. doi:10.1111/imcb.12675.
- Wang C, Lin Y, Chen L, et al. Gut microbiota mediated the effects of high relative humidity on lupus in female MRL/lpr mice. Adv Rheumatol. 2023;63(1):24. doi:10.1186/s42358-023-00306-2.
- Sanz Y, Moya-Perez A. Microbiota, inflammation and obesity. Adv Exp Med Biol. 2014;817:291–317. doi:10.1007/978-1-4939-0897-4_14.
- Santisteban MM, Qi Y, Zubcevic J, et al. Hypertension-linked pathophysiological alterations in the gut. Circ Res. 2017;120(2):312–323. doi:10.1161/CIRCRESAHA.116.309006.
- Lopez P, de Paz B, Rodriguez-Carrio J, et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep. 2016;6(1):24072. doi:10.1038/srep24072.
- McClain MT, Heinlen LD, Dennis GJ, et al. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med. 2005;11(1):85–89. doi:10.1038/nm1167.
- Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi:10.1016/j.tibtech.2015.06.011.
- He X, Sun J, Liu C, et al. Compositional alterations of gut microbiota in patients with diabetic kidney disease and type 2 diabetes mellitus. Diabetes Metab Syndr Obes. 2022;15:755–765. doi:10.2147/DMSO.S347805.
- Li M, Wei L, Sun J, et al. Association of gut microbiota with idiopathic membranous nephropathy. BMC Nephrol. 2022;23(1):164. doi:10.1186/s12882-022-02797-5.
- Qiu C, Yuan Z, He Z, et al. Lipopolysaccharide preparation derived from Porphyromonas gingivalis induces a weaker Immuno-Inflammatory response in BV-2 microglial cells than Escherichia coli by differentially activating TLR2/4-Mediated NF-kappaB/STAT3 signaling pathways. Front Cell Infect Microbiol. 2021;11:606986. doi:10.3389/fcimb.2021.606986.
- Yao X, Huang J, Zhong H, et al. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther. 2014;141(2):125–139. doi:10.1016/j.pharmthera.2013.09.004.
- Tan J, Zhong Z, Tang Y, et al. Intestinal dysbiosis featuring abundance of Streptococcus associates with Henoch-Schonlein purpura nephritis (IgA vasculitis with nephritis) in adult. BMC Nephrol. 2022;23(1):10. doi:10.1186/s12882-021-02638-x.
- Li Y, Wang HF, Li X, et al. Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin Sci (Lond). 2019;133(7):821–838. doi:10.1042/CS20180841.
- van den Bogert B, Meijerink M, Zoetendal EG, et al. Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota. PLoS One. 2014;9(12):e114277. doi:10.1371/journal.pone.0114277.
- Kotzin BL, Kozora E. Anti-DNA meets NMDA in neuropsychiatric lupus. Nat Med. 2001;7(11):1175–1176. doi:10.1038/nm1101-1175.
- Blank M, Barzilai O, Shoenfeld Y. Molecular mimicry and auto-immunity. Clin Rev Allergy Immunol. 2007;32(1):111–118. doi:10.1007/BF02686087.
- Pisetsky DS. The role of bacterial DNA in autoantibody induction. Curr Top Microbiol Immunol. 2000;247:143–155. doi:10.1007/978-3-642-59672-8_10.
- Peene I, Elewaut D. Changing the wolf from outside: how microbiota trigger systemic lupus erythematosus. Ann Rheum Dis. 2019;78(7):867–869. doi:10.1136/annrheumdis-2019-215221.
- Breban M, Tap J, Leboime A, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76(9):1614–1622. doi:10.1136/annrheumdis-2016-211064.
- Hall AB, Yassour M, Sauk J, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9(1):103. doi:10.1186/s13073-017-0490-5.
- Everard A, Cani PD. Gut microbiota and GLP-1. Rev Endocr Metab Disord. 2014;15(3):189–196. doi:10.1007/s11154-014-9288-6.
- Andrade-Oliveira V, Amano MT, Correa-Costa M, et al. Gut bacteria products prevent AKI induced by Ischemia-Reperfusion. J Am Soc Nephrol. 2015;26(8):1877–1888. doi:10.1681/ASN.2014030288.
- Jiang S, Xie S, Lv D, et al. A reduction in the butyrate producing species roseburia spp. and Faecalibacterium prausnitzii is associated with chronic kidney disease progression. Antonie Van Leeuwenhoek. 2016;109(10):1389–1396. doi:10.1007/s10482-016-0737-y.
- Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11(10):577–591. doi:10.1038/nrendo.2015.128.
- Sanchez HN, Moroney JB, Gan H, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. 2020;11(1):60. doi:10.1038/s41467-019-13603-6.
- Hall AV, Parbtani A, Clark WF, et al. Abrogation of MRL/lpr lupus nephritis by dietary flaxseed. Am J Kidney Dis. 1993;22(2):326–332. doi:10.1016/s0272-6386(12)70326-8.
- Li D, Pan Y, Xia X, et al. Bacteroides fragilis alleviates the symptoms of lupus nephritis via regulating CD1d and CD86 expressions in B cells. Eur J Pharmacol. 2020;884:173421. doi:10.1016/j.ejphar.2020.173421.
- Chen D, Wu J, Jin D, et al. Fecal microbiota transplantation in cancer management: current status and perspectives. Int J Cancer. 2019;145(8):2021–2031. doi:10.1002/ijc.32003.
- Zhao J, Bai M, Yang X, et al. Alleviation of refractory IgA nephropathy by intensive fecal microbiota transplantation: the first case reports. Ren Fail. 2021;43(1):928–933. doi:10.1080/0886022X.2021.1936038.
- Huang C, Yi P, Zhu M, et al. Safety and efficacy of fecal microbiota transplantation for treatment of systemic lupus erythematosus: an EXPLORER trial. J Autoimmun. 2022;130:102844. doi:10.1016/j.jaut.2022.102844.