Abstract
Objective: The aim of this study was to evaluate the presence of Enterobacteriaceae in different locations of the Evros-Ardas river ecosystem at the borderline of Greece-Turkey. Materials and methods: Usual filter methods were applied for the collection of water samples from the riverside subjected to waste and/or borderline polluted stream. Sampling was accomplished during 3 months, from January to March. A total of 150 strains were isolated and identified by API Mérieux. The antibiotic resistance profiles were performed according to CLSI; phylogeny analysis was performed by RAPD (random amplification polymorphic DNA analysis); integron class I, II and III were detected by PCR amplification of integrases I and II by using degenerated primers. The detection of the classes of integrons was performed after RsaI digestion of the PCR products and agarose gel electrophoresis. The antibiotic gene cassettes were also generated by PCR. Results: In all, 90% of the isolates were usual environmental strains without antibiotic resistance. Resistance to ampicilllin, amoxicillin, and/or ampicillin + sulbactam and several cephalosporins was detected in 12 strains. Resistance to tetracycline and chloramphenicol was observed in two isolates; resistance to nalidixic acid and to fluoroquinolones was detected in three strains. Finally, resistance to sulfamethoxazole and trimethoprim was observed in four isolates. Integron class I was detected in four isolates and class II in two of them. Several profiles were identified by RAPD and some unique types. Conclusion: It has been shown that plasmids containing β-lactamase, tetracycline and chloramphenicol genes as well as integron class I and II contribute to the pathogenic profiles of the isolates of the region. According to our results four locations represented environmental niches of pathogenic and antibiotic-resistant Enterobacteriaceae, namely the locations Ardas, Elaia bridge, Rizia and Kastanies and Evros Nea Vissa. Further investigation must be undertaken to control the microbial pollution of the riverside in Evros.
Introduction
Natural water bodies have been polluted as a result of receiving domestic sewage and effluents. As well as the obvious water pollution by discharges of chemicals and pesticides, a major part of pollution is associated with potentially health-threatening microorganisms. These microorganisms may be shed in faeces with subsequent contamination of aquatic environments Citation[1].
Maintenance of reduced microbial numbers in watery environments should play an important role in public health, as well as in the preservation of the ecosystem. Knowledge of both microbial diversity and chemical parameters in water bodies is important in explaining the potential roles of the microorganisms in the biosphere. On the other hand, it has been suggested that their large-scale dissemination is due to the extensive use of antibiotics in human and veterinary medicine as well as in agriculture worldwide Citation[2], Citation[3].
The persistence of antibiotic-resistant and multiresistant bacteria in the environment is a growing public health concern Citation[4]. The increased prevalence of resistant isolates raises many concerns, as the antibacterial value of many drugs is seriously threatened Citation[5]. Furthermore, resistance can easily be transferred among strains via R-factor plasmid vectors Citation[6].
Most studies on the occurrence of antimicrobial-resistant bacteria focus on bacteria of faecal origin because they are commonly used as indicators of faecal pollution in water and food and may be associated with infectious diseases Citation[7–9]. Specifically, Escherichia coli is a useful enteric bacterium as it is adapted to the gastrointestinal tracts of human and other warm-blooded animals, and is readily exposed to a variety of medical and veterinary antibiotic treatments Citation[10].
The aim of the present study was to describe and compare the fluctuation and antibiotic resistance profiles of Enterobacteriaceae, with a special note of E. coli, which is generally accepted as the classic bacterial indicator of faecal pollution in waters, at different locations of the Evros-Ardas river ecosystem at the borderline of Greece-Turkey.
Materials and methods
Geographical area
Nine stations of the Ardas-Evros ecosystem situated in the north-east part of Greece were entered in our study ().
Our ecosystem consists of two units. The first section of the ecosystem, the Ardas river, enters Greece from Bulgaria and after a short distance in this north part of Greece, it is oriented to Turkey (Edirne area) where it receives inputs of water from other ecosystems. Then, the second unit of our ecosystem, the Evros river, approaches Greece again and forms the physical border between the two countries.
Sampling stations
Nine different water locations were entered in the present study. One liter of water was collected in a sterilized bottle from each location. The water samples were placed in portable refrigerators and transported to the Microbiology Laboratory on ice within 2 h of collection. A total of 118 water samples in duplicate were taken from the nine designated locations: S1 (Komara), S2 (Elaia), S3 (Rizia), S4 (Kastanies), S5 (Dikaia), S6 (N. Vissa), S7 (Didimoticho), S8 (Lagina), S9 (Kipoi).
The samples were shaken vigorous by hand before analysis. At the laboratory, all samples were stored at 5±2°C until analyses were complete, which was always done within 24 h of sample collection.
Experimental protocol – classic bacteriological procedures
Membrane filtration equipment was used. All samples were alternatively passed through a membrane filter of 20 µm pore size, which was used for retention of soil impurities. Each water sample was analyzed for fecal coliforms, total coliforms, and total aerobic mesophilic microflora.
Samples (100 ml) were diluted (1:10) in accordance with the level of pollution and analysis was performed using the membrane filtration culture method in accordance with the standard procedures proposed by APHA in 1992 Citation[11].
The following growth media were used. Total coliforms (filtration method): m-ENDO agar (Difco) incubated at 36°C for 24 h. Confirmation was made by selection and culturing of 10 characteristic colonies in BGLB (brilliant green lactose broth) at 36°C for 24 h. Fecal coliforms: E. coli (filtration method) MFC agar (Difco) incubated at 44°C for 24 h. Confirmation was made by selection and culturing of 10 characteristic colonies in LTLSB (lactose-tryptone-lauryl-sulphate-broth) at 44°C for 24 h.
Standard classic procedures were performed for identification to species level of the aerobic microflora. Bacterial strains were isolated and identified by the API Mérieux. The antibiotic resistance profiles were performed according to the CLSI.
Total bacterial DNA was extracted with the Quiagen mini-prep kit. Random amplified polymorphic DNA (RAPD) was used as a rapid screening method to distinguish sample strains, and to exclude potential repeat isolates. Samples were distinguished by considering data from one PCR with the primer AP1290, 5′ CTGGATGCGA 3′ as described previously Citation[12].
Integrons of class I, II and III were detected by PCR amplification of the integrases I and II by using degenerated primers. The detection of the classes of integrons was performed after RsaI digestion of the PCR products and agarose gel electrophoresis Citation[13]. Since no class integron class III was detected after Rsa digestion of the PCR products, the strains were not examined further for this antibiotic gene cassette. The antibiotic gene cassettes were also generated by PCR for both class I Citation[14] and class II as described previously Citation[15], by using 60–100 ng of DNA.
Results
Microbiological and chemical profile
The Ardas river ecosystem coming from Bulgaria has low levels of all the above bacteria. After its passage through Turkey the newly originated river ecosystem called Evros seems to collect an important bacteriological load, as higher numbers of E. coli (5×103 cfu/100 ml) and total coliforms (5×103 cfu/100 ml) are present at the first Greek station (). These numbers showed an increasing profile following the flow of the Evros river to the sea ().
Figure 2. Mean concentration of indicator bacteria following the physical flow of the Ardas and Evros rivers to the sea (from north to south).
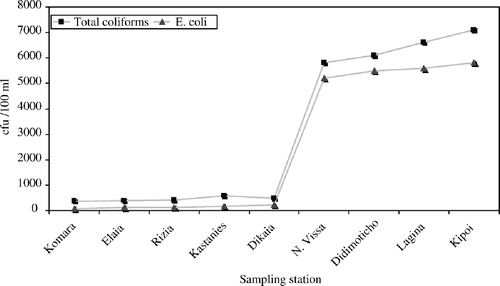
Table I. Mean concentration of indicator bacteria at all sampling stations (n = 118).
Considering the chemical parameters, Zn levels were always negative and Cu levels were found to be positive only twice and at very low concentrations. Iron levels varied from 6 to 14.5 µg/l following the studied stations. The Ardas ecosystem seems to sustain higher levels of Fe (12–14.5 µg/l) than the Evros ecosystem (6–9.5 µg/l) and this is in accordance with the conductivity levels, which were shown to berelatively higher in the Ardas ecosystem ().
Table II. Mean concentration of chemical parameters at all sampling stations (n=118).
Analysis of antibiotic resistance profiles, phylogeny and integrons
In all, 90% of the isolates were usual environmental strains without antibiotic resistance. As can be observed in , resistance to ampicilllin, amoxicillin, and/or ampicillin + sulbactam or ampicillin + clavulanic acid, and several cephalosporins was detected in 12 strains. Moreover, resistance to tetracycline and chloramphenicol was shown in three isolates, and resistance to nalidixic acid and to fluoroquinolones was detected in three strains. Finally, resistance to sulfamethoxazole and trimethoprim was observed in four isolates. Integron class I was detected in four isolates and class II in two of them. All the strains containing integrons were multiresistant. Several profiles were identified by RAPD and some unique types, thus confirming that the strains were unrelated and that the sampling was accurate. Strains 1–10 () were isolated in Ardas, whereas strains 11–14 were isolated from Evros.
Table III. Antibiotic resistance profiles of integron class I and II and gene cassettes of 14 strains isolated in ecosystems of Ardas and Evros.
Discussion
Understanding the acquatic bacterial ecology and estimation of bacterial antibiotic resistance profiles can be an important tool for those who are responsible for public health and environmental protection and are charged with reducing pollution, protecting public health, and improving water quality. In the present study, Enterobacteriaceae and especially E. coli were chosen routinely as representative of the fecal contamination indicator Citation[16].
Although no consistent relationship between any precise microbiological indicator of water quality associated directly with disease has been confirmed, many undertaken on the field indicate the possible link between microbiological water pollution and measurable health effects. Total coliform bacteria concentrations are generally assumed to be replaced by fecal coliform and other fecal bacteria concentrations as indicators of water quality Citation[17], Citation[18]. Furthermore, fecal coliforms have been reported to be the most sensitive indicators of the seasonal fluctuation of water pollution Citation[19], Citation[20]. Total coliform and E. coli were present in all nine stations of the Ardas-Evros river.The same bacterial pattern was observed for all the above.
The first part of our ecosystem (Ardas river) showed lower bacterial populations (101–102 usually) compared with the second part of the ecosystem (Evros river) (>103). Moreover, all bacterial populations showed a similar peak in their counts, at the same station of the Evros river, called Didimoticho (S7).
It has been reported that peak concentrations of fecal coliforms are coincident with the increased phosphorus and nitrogen concentrations in the water body for the same periods Citation[21], Citation[22].
Evidently, numbers and survival of fecal coliforms were closely associated and depended on the trophic conditions of the aquatic environment, as well as on other physicochemical parameters Citation[17], Citation[23], Citation[24]. Additionally, some important industries and municipal wastewater treatment plants are located in the region.
In epidemiological investigations of the source of water contamination, Rhodococcus coprophilus was used Citation[25] as a specific indicator of pollution with animal feces. More specifically, C. perfringens serves as an indicator of fecal pollution of remote origin in many aquatic environments Citation[1], Citation[7], Citation[8], Citation[22], Citation[26].
The framework of the present study focused on screening the resistance of Enterobacteriaceae isolates from the Ardas-Evros ecosystem to a variety of commonly used antimicrobial compounds.
RAPD with appropriate statistical analysis proved to be suitable for the discrimination of the E. coli isolates. Several profiles were identified by RAPD and some unique types, confirming that the sampling was accurate and that the strains were distinct.
About 90% of the isolates were usual environmental strains without antibiotic resistance. Resistance to ampicilllin, amoxicillin, and/or ampicillin + sulbactam and several cephalosporins was detected in 12 strains. Moreover, resistance to tetracycline and chloramphenicol was shown in three isolates; resistance to nalidixic acid and to fluoroquinolones was detected in three strains Finally, resistance to sulfamethoxazole and trimethoprim was observed in four isolates. All of them contained integrons class I and II.
Constructively, taking into account our parameters, we can accept that plasmids containing β-lactamase, tetracycline and chloramphenicol genes contribute to the pathogenic profiles of the isolates of the region. Integrons class I and II consistently were associated with antibiotic resistances. Although the 1600 bp cassette from class I integron was not sequenced, and because of its length and the concomitant resistances, the cassettes seem to belong to class aad and dfr genes Citation[27–31].
According to our results, four locations represented environmental niches of pathogenic and antibiotic-resistant Enterobacteriaceae, namely the locations Ardas, Elaia bridge, Rizia and Kastanies and Evros Nea Vissa.
Further investigation has to be undertaken to control the microbial pollution of the riverside in Evros. Clearly, there are certain factors that affect the fecal indicator bacteria more in very eutrophic areas than in other sampling areas Citation[1] and so it is necessary to use very sensitive indicators to evaluate the quality of the water being used.
Moreover, development of antibiotic resistance and dissemination of the resistant or multiresistant bacteria via integron or plasmid in the watery environment affect the natural ecosystem as well as the effectiveness of the antibiotic therapies in humans and animals.
Although the dissemination of antibiotic-resistant bacteria seems to take place in the aquatic environment of the river Ardas and Evros, the phenomenon, first, was not identified to be as expanded as expected. This may be due to the previous period of inundation, meaning the consequent clearance. Second, the antibiotic resistance profiles and the presence of integrons and gene cassettes seem consistent with the spread of both plasmid and integron.
It is clear, however, that we need a better understanding of the mechanisms of action of preservative systems, particularly factors commonly used to control the growth of microorganisms. These studies will include identification of sites of action, lag times and rates of bacterial growth, and biovariability, together with exhaustive environmental knowledge and intervening factors.
Attention must be paid to human activities situated by the riverside to preserve the water ecosystem and so controlling on one hand the microbial pollution and its effect, on the other hand, when resistance is exhibited, upon humans. To protect public health it is imperative to determine the precise source of the fecal material and toxic pollutants, especially if monitoring and management plans are to be implemented.
The project is co-funded by the European Social Fund & National Resources – EPEAEK II – PYTHAGORAS II.
References
- Bezirtzoglou E, Dimitriou D, Panagiou A. Occurrence of Clostridium perfringens in river water by using a new procedure. Anaerobe 1996; 2: 169–73
- Voidarou, C, Tzora, A, Fotou, K, Giza, E, Alexopoulos, A, Bezirtzoglou, E. Food shelf-life as indicator of the hygienic processing in vacuum-packed cold butchery. Food Technology and Biotechnology. 2006, (in press).
- Levidiotou S, Bezirtzoglou E, Gesouli E, Papatheodorou M, Antoniadis G. Occurrence of Salmonella serotypes in Greece. Med J Inf Parasit Dis 1993; 18: 203–6
- Armstrong JL, Shigeno DS, Calomiris JJ, Seidler RJ. Antibiotic-resistant bacteria in drinking water. Appl Environ Microbiol 1981; 42: 277–83
- Grabow WOK, Prozesky OW. Drug resistance of coliform bacteria in hospital and city sewage. Antimicrob Agents Chemother 1973; 3: 175–80
- Venieri, D, Komninou, G, Bezirtzoglou, E, Papapetropoulou, M. Profiles of antibiotic resistance in Escherichia coli strains isolated from municipal tap water and raw sewage samples in Greece. Proceedings of International Conference on Sustainable Management and Development of Mountainous and Island Areas. ISBN: 960-89345-2-4. 2006; 2:353–361.
- Bezirtzoglou E, Panagiou A, Savvaidis I, Maipa V. Distribution of C. perfringens in polluted lake environments. Anaerobe 1997; 3: 169–72
- Bezirtzoglou E, Dimitriou D, Panagiou A, Karalou I, Demoliates Y. Distribution of C. perfringens in different aquatic environments in Greece. Microbiol Res 1994; 149: 129–34
- Edge TA, Hill S. Occurrence of antibiotic resistance in Escherichia coli from surface waters and fecal pollution sources near Hamilton, Ontario. Can J Microbiol 2005; 51: 1–5
- Kay D, Fleisher JM, Salmon RL, Wyer MD, Godfree AF, Zelenauch-Jacquotte Z, et al. Predicting likelihood of gastroenteritis from sea bathing: results from randomized exposure. Lancet 1994; 344: 905–9
- APHA. Standard methods for the examination of water and waste water, 17th edn. Washington, 1990.
- Maurer JJ, Lee MD, Lobsinger C, Brown T, Maier M, Thayer SG. Molecular typing of avian Escherichia coli isolates by random amplification of polymorphic DNA. Avian Dis 1998; 42: 431–51
- White PA, McIver CJ, Deng Y, Rawlinson WD. Characterisation of two new gene cassettes, aadA5 and drfA17. FEMS Microbiol Lett 2000; 82: 265–9
- Lévesque C, Roy PH. PCR analysis of integrons. Diagnostic molecular microbiology: principles and applications, DH Persing, TF Smith, FC Tenover, TJ White. American Society for Microbiology, Washington, DC 1993; 590–4
- White PA, McIver C J, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 2001; 45: 2658–61
- Bezirtzoglou E, Maipa V, Voidarou C, Tsiotsias A, Papapetropoulou M. Foodborne intestinal bacterial pathogens. Microb Ecol Health Dis 2000; S2: 96–105
- Hirn J, Viljamaa H, Raevuori M. The effect of physicochemical, phytoplankton and seasonal factors on faecal indication bacteria in northern brackish water. Water Res 1996; 285: 25–9
- Hiraishi A, Saheki K, Horie S. Relationships of total coliforms, fecal coliform and organic pollution levels in the Tamagawa river. Bull J Soc Sci Fisheries 1984; 50: 991–7
- Morozzi G, Cenci G, Scarabattoli P. Bacteriological and chemical variations and their inter-relationships in a slightly polluted water-body. Intern J Environ Studies 1984; 23: 121–9
- Matsumoto, J, Omura, I. Some factors affecting the survival of fecal indicator bacteria in sea water. Technology Reports. Tohokw University, 1980; vol. 45, no. 2.
- Romero, JR, Imberger, J. Lake Pamvotis project, final report. Western Australia: CWR, 1999.
- Savvaidis I, Kegos Th, Papagianis C, Voidarou C, Evagelou A, Bezirtzoglou E. Bacterial indicators and metal ions in high mountains lake environments. Microb Ecol Health Dis 2001; 13: 147–52
- Sorensen DL, Eberl SG, Dicksa RA. C. perfringens as a point source polluted streams. Water Res 1989; 23: 191–7
- Pinfold JV. Faecal contamination of water and fingertip-rinses as a method for evaluating the effect of low cost water supply and sanitation activities on faecal-oral disease transmission I-II. A hygiene intervention study in rural north-east Thailand. Epidemiol Infect 1990; 105: 363–9
- Oragui JL, Mara DD. Investigation of the survival characteristics of Rhodococcus coprophilus and certain faecal indicator bacteria. Appl Environ Microbiol 1983; 46: 356–60
- Kagalou I, Tsimarakis, Bezirtzoglou E. Inter-relationships between bacteriological and chemical variations in Lake Pamvotis-Greece. Microb Ecol Health Dis 2002; 14: 37–41
- Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 1999; 63: 507–22
- Bass L, Liebert CA, Lee MD, Summers AO, White BDG, Thayer SG, et al. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother 1999; 43: 2925–9
- Yu HS, Lee JC, Kang HY, Ro DW, Chung JY, Jeong YS, et al. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J Clin Microbiol 2003; 41: 5429–33
- Burchall JJ, Hitchings GH. Inhibitor binding analysis of dihydrofolate reductases from various species. Mol Pharmacol 1965; 1: 126–36
- Amyes SG, Smith JT. R-factor trimethoprim resistance mechanism: an insusceptible target site. Biochem Biophys Res Commun 1974; 58: 412–18