Abstract
The microbiota is part of the gastrointestinal ecosystem. A more detailed understanding should provide insight into multiple human disease states. This study investigated inflammatory bowel disease (Crohn's disease and ulcerative colitis). Previous analyses have suggested a role of gram-negative bacteria including Escherichia coli. An integrated procedure is presented where gas chromatography-mass spectrometry is used to determine chemical markers of gram-negative bacterial lipopolysaccharide (3-hydroxy fatty acids with 10–18 carbon atoms) in faecal samples. Six patients with Crohn's disease (CD), five with ulcerative colitis (UC) and six healthy adults were chosen as groups of interest. Nine saturated straight-chain 3-OH fatty acids of 10–18 carbon chain lengths and six iso- and anteiso-branched-chain 3-OH fatty acids of 15–18 carbon chain lengths were detected. Significant differences were found in the 3-OH n-C17:0, 3-OH i-C18:0 and 3-OH n-C18:0 composition of faeces. The present study therefore confirms that alteration of the composition of the endogenous gram-negative microbiota may be of importance in inflammatory bowel disease and those alterations could be detected with a non-invasive chemical-analytical approach.
Introduction
The aetiologies of Crohn's disease (CD) and ulcerative colitis (UC) are still not clear Citation[1], although the microbial community of the bowel, the genetic susceptibility of the host and the mucosal immunity all seem to be involved in the development and recurrence of these diseases Citation[2], Citation[3]. The microbiota is part of the gastrointestinal ecosystem. It is known to play an active role in the pathogenesis of a range of inflammatory bowel diseases (IBD) Citation[4]. To date most of our knowledge on the microbial community in the gastrointestinal tract has been based on culturing from faecal material and/or the use of the 16S ribosomal ribonucleic acid (rRNA) genes Citation[5].
Culture-based studies have suggested an increase in gram-negative bacteria and a decrease in gram-positive bacteria in Crohn's disease Citation[6]. In the pathogenesis of Crohn's disease, failure of the innate immune system could be involved via reduced/impaired defence against gram-negative bacteria Citation[7]. The recently characterized Asp299Gly polymorphism in the lipopolysaccharide (LPS) receptor TLR4 is associated with impaired LPS signalling and increased susceptibility to gram-negative organisms in CD and UC Citation[8].
Both conventional culture methods and molecular techniques based on 16S rRNA genes (rDNA) have limitations when applied in studies on the microbiology of the intestine. To date > 50% of the total microbiota in faeces is assigned as ‘not culturable’. Furthermore, molecular techniques may have a detection limit as high as 106 bacteria per gram Citation[9]. In the present study we investigated a new chemical-analytical approach to provide information on both viable and non-viable gram-negative microorganisms.
All genuine gram-negative bacteria contain endotoxin (lipopolysaccharide, LPS) in the outer membrane. The lipid part of the LPS, lipid A, is responsible for the toxic effects of endotoxin and harbours a range of 3-hydroxy fatty acids (3-OH FAs), which can be used as chemical markers of LPS. Given that the LPS of different bacterial species may contain different 3-OH FAs in terms of chain length and branching, the 3-OH FA distribution, for example of a clinical sample, reflects the gram-negative bacterial population of the sample. Gas chromatography-mass spectrometry (GC-MS), and in particular GC-tandem MS (GC-MSMS), is the method of choice for this kind of analysis Citation[10] and represents a technique that detects (and quantifies) bacteria equally well regardless of whether they are culturable or non-culturable. To date this analytical approach has not been used to study the complex microbiota of the human intestinal tract.
The aim of the present investigation was to apply 3-OH FA analysis (using GC-MSMS) to faecal samples from hospitalized patients with CD, patients with UC, and healthy individuals (controls) in order (i) to characterize the gram-negative bacterial load of each individual and (ii) to investigate possible correlations between the gram-negative bacterial population (in terms of 3-OH FA composition) of faeces and clinical status.
Materials and methods
Patients
Six patients with CD, five patients with UC and six healthy individuals (controls) were enrolled (). All patients were hospitalized and had given their written consent. They came from the west part of France and had a normal western diet. Faecal samples were collected from all patients during active disease. In addition, a faecal sample was also collected from one of the CD patients during remission. None of the individuals had taken antibiotics and/or probiotics in the 4 weeks prior to providing the faeces. The samples (1000 mg) were stored at –80°C until analysed.
Table I. Characteristics of patients.
Sample preparation and analysis
The samples were transferred to sterile glass test tubes, lyophilized and subjected to methanolysis at 80°C overnight. The liberated fatty acid methyl esters were extracted with heptane and the hydroxylated fatty acid methyl esters were purified by column chromatography as described previously Citation[10]. Before analysis, the hydroxy fatty acid methyl esters were transformed to trimethylsilyl ethers. Samples thus obtained were kept at 4°C until analysis. A Varian model 1200 triple quadrupole GC-MS instrument (Varian, Walnut Creek, CA, USA) equipped with a fused-silica capillary column (CP-Sil 8 CB low bleed, 30 m by 0.25 mm ID, 0.25 µm film thickness) was used. The temperature of the column was programmed to rise from 90°C to 280°C, at a rate of 20°C min–1. The injector, transfer line and ion source were held at 250°C, 290°C and 200°C, respectively. Full mass spectra were recorded for identification of the 3-OH FAs, whereas GC-MSMS was used to calculate the relative percentage of each acid.
The statistical analyses were performed using either Mann-Whitney or t tests, depending on the number of variables.
Results
3-OH FA composition
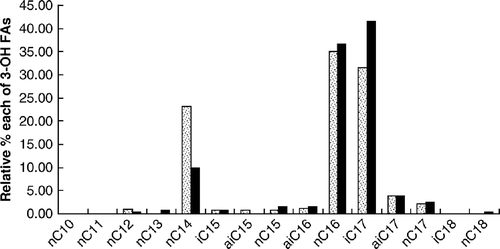
Nine saturated straight-chain 3-OH FAs of 10–18 carbon chain lengths (3-OH n-C10:0 to 3-OH n-C18:0) and six iso (i)- and anteiso (ai)-branched-chain 3-OH FAs of 15–18 carbon chain lengths were detected. The mass spectra of the derivatives of all 3-OH FAs (both straight- and branched-chain) showed prominent fragments at m/z 175 and m/z M+-15, as described previously Citation[11].
The relative percentages (mean, standard deviation) of the different 3-OH FAs in the faeces of the three groups of individuals are summarized in . The major 3-OH FAs in all samples were 3-OH n-C16:0 and 3-OH i-C17:0; together they made up > 50% of all acids found. The relative amounts of 3-OH ai-C15:0 were lower in faeces of controls than of patients with UC (p=0.009) and CD (non-significant, p>0.05) (). The relative amounts of 3-OH n-C17:0, 3-OH i-C18:0 and 3-OH n-C18:0 were higher in faeces of the controls than of the patients with CD (p=0.011, p=0.004 and p=0.017, respectively). Also, the relative amounts of 3-OH n-C17:0 and 3-OH i-C18:0 were higher in faeces of patients with UC than in patients with CD (p=0.026 in either case).
Figure 1. Relative amounts (%) of some selected 3-hydroxy fatty acids in faeces of healthy individuals (Controls), patients with Crohn's disease (CD) and patients with ulcerative colitis (UC). Upper left, 3-OH ai-C15:0 (*UC versus Controls, p=0.009); upper right, 3-OH n-C17:0 (*Controls versus CD, p=0.011, **UC versus CD, p=0.026); lower left, 3-OH i-C18:0 (*Controls versus CD, p=0.004, **UC versus CD, p=0.026); lower right, 3-OH n-C18:0 (*Controls versus CD, p=0.017).
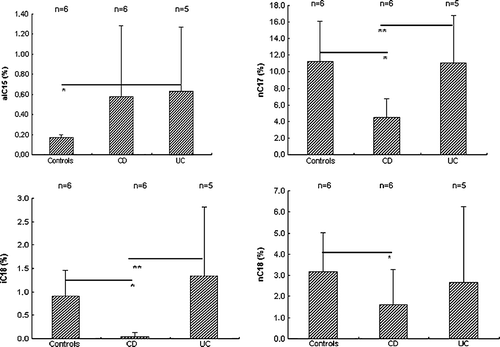
Table II. Relative percentages (mean) and standard deviations (SD) of saturated 3-hydroxy fatty acids, with indicated carbon chain lengths, in faeces of healthy individuals (C) and of patients with Crohn's disease (CD) and ulcerative colitis (UC).
Patient with active colonic CD and with colonic CD in remission
In faeces of this patient three 3-OH FAs (3-OH n-C13:0, 3-OH i-C18:0 and 3-OH n-C18:0) were found only during remission. Furthermore, the relative amount of 3-OH n-C14:0 in faeces was clearly higher during active CD than in remission, whereas the relative amounts of the other major 3-OH FAs, i.e. 3-OH n-C16:0 and 3-OH i-C17:0, were slightly lower during active CD than during remission ().
Discussion
This work was prompted by previous research where C10:0–C18:0 3-OH FAs have been used successfully as markers of LPS in clinical and environmental samples Citation[12], Citation[13]. The complexity of dysbiosis investigation in case of IBD Citation[4] warranted investigations for new approaches. Our study describes a non-invasive diagnostic approach. Patients were adults but for two control considered as elderly (aged 72 and 80 years, respectively). Earlier works indicated that human faeces vary with age as the number of different species (diversity) present increased Citation[14]. Nevertheless, in spite of the presence of the two elderly subjects in the control group, the here described chemical-analytical approach provided differentiating information on the gram-negative microbiota of faeces samples including cultivable and non-cultivable microorganisms as well as bacterial cell wall remnants.
Evidence is accumulating that the composition and activity of the intestinal bacterial community has a significant impact on the health of the host because of its influence on nutrition, bowel habit, the physiology of the mucin barrier and the ontogeny of the mucosal immune system Citation[15–17]. In addition, the composition and activity of the intestinal bacterial community is likely to influence the pathogenesis of colorectal cancer Citation[18] and IBD Citation[4], Citation[5].
Early gut microbial ecology work depended on cultivation methods, but to date a large majority of organisms remain unculturable, and therefore these methods do not reflect the true diversity of the gut bacteria Citation[16]. Our understanding of complex microbial communities has been greatly enhanced by the development of molecular techniques based on the 16S rRNA genes. A 16S rDNA clone library of a single faecal sample revealed extensive diversity Citation[19]. Thus, only 24% of sequences in this single sample were represented by cultured organisms, and 76% of analysed sequences belonged to unknown bacterial species. The numbers of microorganisms yet to be cultured may be much higher still Citation[20].
Using a molecular approach, Seksik et al. reported that the faecal microbiota in patients with both inactive and active colonic CD differed from the faecal microflora of healthy subjects, containing significantly more Enterobacteriaceae Citation[5]. Escherichia coli strains have been implicated in the pathogenesis of CD, especially some strains with particular adhesion properties which are increased in ileal lesions of CD Citation[21]. Bacteroides exhibited proinflammatory properties in several animals models of IBD Citation[22]. While some culture studies have suggested a possible increase in faecal Bacteroides in CD, Seksik et al. observed a decrease in the relative proportions of the Bacteroides phylogenetic group Citation[4], Citation[5]. Faecal microbiota composition in relation to frailty has been studied in elderly subjects (aged 70–100 years) with fluorescent in situ hybridization Citation[23]. It showed a significant reduction in percentage of the total number of Bacteroides for very frail volunteers Citation[23]. We therefore focused our analysis on markers specific for gram-negative bacteria.
The observed diversity in 3-OH FA composition of faeces between the three groups of individuals indicates differences in gram-negative bacterial community. Three of the four most abundant 3-OH FAs, i.e. 3-OH n-C16:0, 3-OH n-C17:0 and 3-OH i-C17:0, are constituents of the lipid A of Bacteroides spp. Citation[24], whereas the fourth most abundant 3-OH FA, 3-OH n-C14:0, is found in Enterobacteriaceae. The LPS 3-OH FAs are very consistent and do not change according to culture conditions Citation[24] and can therefore be useful in distinguishing gram-negative bacterial microflora, although the exact species identity remains unknown. Notably, and in accordance with the work of Seksik et al. Citation[5], 3-OH C14:0 (present, for example, in E. coli) was higher in CD than in UC and controls.
It is known that bacteria in the gut may influence the maturation of several components of the immune system. IBD is frequently characterized by a breakdown of immune tolerance towards resident gut bacteria. The mechanism remains unclear but has been linked by some researchers to the ‘hygiene hypothesis’ Citation[25]. According to this hypothesis the T-cell development is driven to the proinflammatory Th1 and Th2 phenotypes during childhood exposure to endotoxin, whereas early exposure to key antigens promotes the development of a regulatory phenotype (Tr) characterized by the production of interleukin IL-10 and transforming growth factor TGF-β. IBD presents a heavily skewed T-cell development towards the proinflammatory phenotypes. Therefore it is possible that exposure to at least some of the gut microbes and/or their endotoxins is crucial for the development of a normal adaptive immune system and the acquisition of tolerance to resident bacteria. Consequently it is imperative to characterize the microbiota of the intestine and the analytical methodology used herein represents a well adapted tool for this purpose.
Conclusion
The microbial community of the large intestine may vary considerably between healthy individuals Citation[9], Citation[17], Citation[20]. Clearly, it would be advantageous to study samples collected from the same individual patient(s) over time, both at different stages of active disease and during remission, to maximize the information. Although in the present study we only analysed one CD patient with both remission and active disease, a correlation between intestinal microbial community (as assessed by chemical markers) and IBD was found. In subsequent larger planned studies we will therefore focus on samples collected over time from given individuals versus controls from the same age group.
We express our gratitude to Prof. P. Marteau for his helpful advice, to Doct A. Bourreille for his patients and to T. Durand for his skilful technical help.
References
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004; 53: 685–93
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 2001; 411: 599–603
- Shanahan F. Inflammatory bowel disease: immunodiagnostics, immunotherapeutics, and ecotherapeutics. Gastroenterology 2001; 120: 622–35
- Marteau P, Lepage P, Mangin I, Suau A, Dore J, Pochart P, et al. Review article: gut flora and inflammatory bowel disease. Aliment Pharmacol Ther 2004; 20(Suppl 4)18–23
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 2003; 52: 237–42
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 1998; 115: 1405–13
- Klein W, Tromm A, Folwaczny C, Hagedorn M, Duerig N, Epplen J, et al. A polymorphism of the bactericidal/permeability increasing protein (BPI) gene is associated with Crohn's disease. J Clin Gastroenterol 2005; 39: 282–3
- Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, et al. Deficient host–bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut 2004; 53: 987–92
- Zoetendal EG, Ben-Amor K, Akkermans AD, Abee T, de Vos WM. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst Appl Microbiol 2001; 24: 405–10
- Sebastian A, Larsson L. Characterization of the microbial community in indoor environments: a chemical-analytical approach. Appl Environ Microbiol 2003; 69: 3103–9
- Hynes SO, Ferris JA, Szponar B, Wadstrom T, Fox JG, O'Rourke J, et al. Comparative chemical and biological characterization of the lipopolysaccharides of gastric and enterohepatic helicobacters. Helicobacter 2004; 9: 313–23
- Ferrando R, Szponar B, Sanchez A, Larsson L, Valero-Guillen PL. 3-Hydroxy fatty acids in saliva as diagnostic markers in chronic periodontitis. J Microbiol Methods 2005; 62: 285–91
- Saraf A, Park JH, Milton DK, Larsson L. Use of quadrupole GC-MS and ion trap GC-MS-MS for determining 3-hydroxy fatty acids in settled house dust: relation to endotoxin activity. J Environ Monit 1999; 1: 163–8
- Blaut M, Collins MD, Welling GW, Dore J, van Loo J, de Vos W. Molecular biological methods for studying the gut microbiota: the EU human gut flora project. Br J Nutr 2002; 87(Suppl 2)S203–S211
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 2002; 22: 283–307
- Wang X, Heazlewood SP, Krause DO, Florin TH. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol 2003; 95: 508–20
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science 2001; 291: 881–4
- Kado S, Uchida K, Funabashi H, Iwata S, Nagata Y, Ando M, et al. Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice. Cancer Res 2001; 61: 2395–8
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 1999; 65: 4799–807
- Zoetendal EG, Cheng B, Koike S, Mackie RI. Molecular microbial ecology of the gastrointestinal tract: from phylogeny to function. Curr Issues Intest Microbiol 2004; 5: 31–47
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004; 127: 412–21
- Rath HC, Ikeda JS, Linde HJ, Scholmerich J, Wilson KH, Sartor RB. Varying cecal bacterial loads influences colitis and gastritis in HLA-B27 transgenic rats. Gastroenterology 1999; 116: 310–19
- van Tongeren SP, Slaets JP, Harmsen HJ, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol 2005; 71: 6438–42
- Wilkinson MH. Model intestinal microflora in computer simulation: a simulation and modeling package for host-microflora interactions. IEEE Trans Biomed Eng 2002; 49: 1077–85
- Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol 2004; 12: 129–34