Abstract
Objective: This study was undertaken to determine whether Butyrivibrio fibrisolvens, a butyrate-producing bacterium present in the rumen of cattle, could be detected in the feces of healthy southern Indians. Subjects and methods: Feces were collected from 25 children and 25 adults from a village and from 25 children living in a town in southern India. Four cow dung samples were also collected from the village. DNA was extracted and real-time PCR was carried out using primers targeted at 16S rDNA sequences peculiar to B. fibrisolvens. Amplicons were subjected to temporal temperature gradient electrophoresis (TTGE) and DNA sequencing. Phylogenetic analysis was done. Results:B. fibrisolvens rDNA was detected in feces of 13 adults (52%) and 15 children (60%) from the village, but not in urban children. In TTGE, amplicons from children and from cattle migrated as a single band, with only one sample showing a second band. Sequencing revealed 12 novel sequences (accessed in GenBank, 8 from children, 4 from dung) closely similar to the ATCC strain of B. fibrisolvens. Conclusions:B. fibrisolvens, a microorganism found in cattle, colonizes the human gastrointestinal tract in rural southern India.
Introduction
Many species of bacteria have evolved and learnt to adapt themselves to live and grow in the human intestine, which contains approximately 1014 bacteria belonging to more than 400 species. In humans the intestinal flora is formed after birth and is considered to remain generally stable after the first years of life. Differences in the flora between individuals are likely to be determined by dietary preferences and environment. Reducing or increasing specific dietary carbohydrates significantly alters the composition of the bowel flora in humans and experimental animals Citation[1], Citation[2]. The role of environmental factors in altering the bowel flora is not quite as clear. Pathogenic bacteria from cattle cause asymptomatic infection or disease in human beings Citation[3], Citation[4]. Evidence to suggest that the normal commensal flora of the human intestine is changed by close or prolonged contact with animals is tenuous Citation[5].
In India, more than 70% of the population lives in rural areas in conditions intimately connected to the village natural resource base. Residents of villages in southern India live in close proximity to cattle. They use cattle dung to plaster the walls and floors of their dwellings, and dry the dung on the house or compound walls to make patties used as fuel. This close contact between humans and animals prompted us to examine whether ruminal commensals could be found in the intestinal flora of humans. Butyrivibrio fibrisolvens, a hemicellulolytic butyrate-producing bacterium, is commonly isolated from the rumen of domestic and wild animals Citation[6]. Butyrate, a short chain fatty acid, plays an important role in the physiology and metabolism of the intestine and intestinal mucosa. In addition to serving as a preferred energy source for colonocytes, butyrate has been implicated in protection against colon cancer and ulcerative colitis Citation[7]. Other butyrate-producing bacteria, Clostridium butyricum and Eubacterium limosum, have been used in humans to ameliorate bowel inflammation Citation[8]. Administration of C. butyricum suppressed chemically induced aberrant crypt foci (ACF), putative pre-neoplastic lesions, in the rat colon and experimental colitis in rats, an effect ascribed to increased production of butyrate Citation[9]. B. fibrisolvens is expected to have similar attributes and hence has the potential to be used as a probiotic. We hypothesized that a close association between humans and cattle in rural southern India might lead to natural colonization of the human intestine with B. fibrisolvens, a butyrate-producing ruminal bacterium, and the present study was designed to examine this hypothesis.
Subjects and methods
The participants were recruited from three overlapping villages approximately 20 km from Vellore. Twenty-five families in these villages were chosen for their characteristic of proximity of human dwelling to animal shelter and presence of a child under the age of 3 years in the house. Each of these families lived in mud houses with cattle sheds adjoining the house. In all, 25 children under the age of 5 years and 25 adults (aged 20–40 years) were chosen from these families. In addition, 25 consecutive healthy children attending an urban well baby clinic in a tertiary care hospital were selected as controls. All these children were resident in an urban district headquarter town and did not have domestic exposure to cattle or cow dung. Any individual who had received antibiotic therapy within the past month was excluded from the study. Informed consent was obtained from the participants or their parents. The study was approved by the institutional research committee.
Fresh feces was collected from each participant and transported on ice within 30 min to the laboratory, where the samples were labeled and stored in aliquots at −80°C for molecular analyses. Cultures were done immediately on receipt of samples in the laboratory in a medium containing glucose (0.05%), maltose (0.05%), cellobiose (0.05%), peptone (0.05%), yeast extract (0.05%), resazurine solution (0.4%), salt solution (50 ml), L-cystine (0.05%), rumen fluid (30%), sodium carbonate (0.4%), and agar (2%). The inoculated plates were incubated in an anaerobic chamber (atmosphere N2 80%, CO2 10%, H2 10%) at 37°C for 72 h. Cow dung and fecal samples from rural children were plated on the above-mentioned culture plates. The standard strain of B. fibrisolvens (ATCC 19171) was obtained from LGC Promochem (Bangalore, India). Intestinal isolates and a range of other standard intestinal bacterial strains were used as controls in testing the specificity of the PCR primers.
Fecal samples were processed in batches. DNA was extracted from approximately 250 mg (wet weight) of fecal sample using the QIAamp DNA stool mini kit (QIAgen, Germany). The DNA was eluted in a final volume of 200 µl and was stored at −20°C. Oligonucleotide primers were targeted at 16S ribosomal DNA sequences and were designed to amplify a sequence specific for B. fibrisolvens. The forward primer was 5′ccttatgatttgggccacac3′ and reverse primer sequence was 5′tccttacggttaggccactg3′. To ensure specificity, the selected primers were compared to all available 16S rDNA sequences using the BLAST database search program (www.ncbi.nlm.nih.gov/blast), and PCR was performed using target and a variety of non-target test bacteria. Primers were purchased from Sigma Genosys (Bangalore, India). Primers were also used to amplify a conserved 16S rDNA sequence present in all bacteria, the universal primer set, the amplification of which served as the denominator against which amplification of other bacterial nucleic acid was compared Citation[10]. The PCR amplification was initially optimized with gradient PCR in a Chromo 4 system (Biorad, USA) using SYBR Green master mix (Eurogentec, Belgium). All PCRs were performed in duplicate in a volume of 20 µl, using high profile tubes and ultraclear sealing caps (Biorad, USA). Fecal B. fibrisolvens was quantified relative to universal bacterial amplification using the Opticon 3.1 software provided with the Chromo 4 real-time PCR. Colony PCR was performed on all the isolated single colonies grown on the selective media, to confirm the presence of B. fibrisolvens.
TTGE
The PCR products using forward primer 5′ccttatgatttgggccacac3′ and reverse primer 5′tccttacggttaggccactg3′ were separated by temporal temperature gradient electrophoresis (TTGE), using a CBS electrophoresis system (CBS Scientific, USA). TTGE was performed in a 9% polyacrylamide gel (160×160×0.75 mm) (37.5:1 acrylamide:bisacrylamide) and 8.5 M urea (Medox, India). Premigration was realized at 20 V and 66°C over a 30 min period. Gels were run overnight at 80 V with the temperature increasing at 1°C/h from 66 to 72°C. The DNA fragments were stained with ethidium bromide and the gel was scanned using a gel documentation apparatus (ChemiSmart, Vilber-Lourmat, France).
Sequencing
All the TTGE bands were excised from the gel and stored overnight at 4°C in 50 µl of sterile water. Amplifications were carried out using the above-mentioned primers and the products were sequenced without cloning using the above-mentioned primers. Sequencing was done by the dye terminator cycle method on a Beckman CEQ8000 at Bioserve (Hyderabad, India). These sequences were manually corrected using Chromas version 1.45. A search of the GenBank nucleotide database was conducted using the BLAST algorithm to determine the closest relative of the partial 16S rRNA gene sequences. Then 16S rRNA sequences were aligned using the Clustal X 2.0.5 program. Estimates of phylogenetic relatedness among different sequences were determined using neighbor-joining (N-J) analyses of 16S rRNA. Multiple alignment parameters were set as: gap opening penalty = 10.0, gap extension penalty = 0.20, delay divergent sequences (%) = 30. N-J trees were constructed in Clustal X using the IUB matrix and viewed using an NJplot program.
Statistics
Values were expressed as median (interquartile range) and significance of differences was assessed using the Kruskal-Wallis test with post hoc comparisons using the Dunn test. Two-sided p values < 0.05 were considered significant.
Results
Bacterial colonies similar in colony morphology to that produced by the standard strain of B. fibrisolvens were cultured from cow dung samples, whereas plating of human fecal samples on the enrichment medium led to overgrowth of the plates with lactobacilli and bifidobacteria. All the colonies were sampled for PCR and compared to the standard strain. The PCR product was a single band of 240 bp producing a single peak on melt curve analysis.
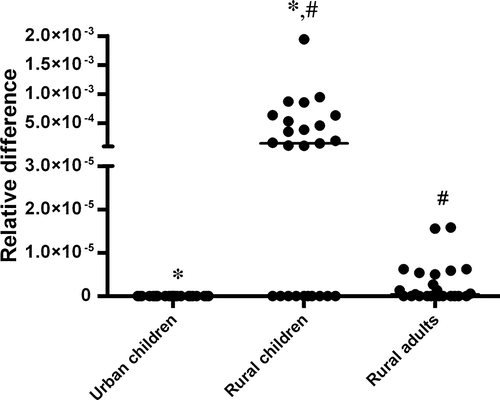
B. fibrisolvens rDNA was detected by real-time PCR in both cattle dung and human feces, being present in the feces of 15 of the 25 children (60%) from the village and 13 of the 25 adults (52%) from the village, but in none of the 25 children from an urban area. The levels in feces of rural-dwelling children were significantly higher than in rural adults, and they were not detectable by PCR in the feces of urban-residing children ().
Figure 1. Quantitation of fecal B. fibrisolvens levels in the three study groups. Amplification of specific rDNA was expressed relative to the amplification of universal rDNA sequence and is shown as relative difference. The individual values are represented for each participant and the median value is shown by the bar. *p<0.001, #p<0.05.
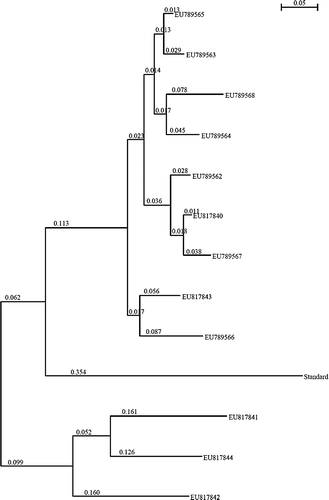
To determine the relationship between the human and cattle isolates and standard strain, all PCR products from feces of children (n=15) and from dung (n = 4) were amplified by a second round of PCR and the products of second-round PCR were subjected to TTGE. The TTGE profile of all human isolates, except for one, showed patterns comparable with that of the standard (). Twenty bands were found in total, which were isolated and amplified again with PCR, and of these 16 bands were available in sufficient quantity and quality for sequencing. Of the 16, 2 were the standard strain of B. fibrisolvens, which was identical to strain 19171 in the NCBI database. Sequence data of the remaining 14 bands were compared against existing databases using BLAST search. Twelve novel sequences (eight from feces of children and four from cow dung) were detected and were submitted to GenBank (accession nos EU789562–EU789568, and EU817840–817844). shows the phylogenetic tree constructed using the neighbor-joining method showing isolates from children, dung, and the standard ATCC strain (standard).
Figure 2. TTGE profile of B. fibrisolvens from human feces, cow dung, and standard strain. A single band was noted.
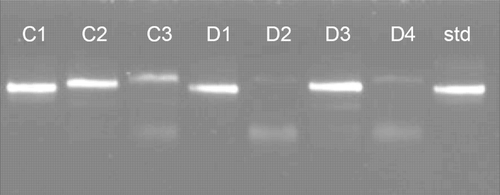
Figure 3. Phylogenetic mapping of the Butyrivibrio amplicons from this study. GenBank accession numbers EU789562, EU789563, EU789564, EU789565, EU789566, EU789567, EU789568, and EU817844 represent amplicons obtained from feces of rural children. EU8171840, EU817481, EU817842, and EU817843 represent amplicons from cow dung.
Discussion
This study demonstrated the nucleic acids of a ruminal bacterium of cattle in the feces of healthy children living in southern Indian villages, but not in urban children. In humans the composition of the intestinal flora appears to be determined shortly after birth, matures over the first few years of life, and remains fairly stable thereafter. The present study suggests that a commensal bacterium found in the environment can colonize the human intestine. In this particular case, it is likely to be a consequence of the close interaction between humans and cattle in the rural setting. Particularly in rural southern India, in the lower socio-economic group, the houses are made of mud walls and have mud flooring and thatched roofing. As their livelihood depends on cattle rearing, the cattle sheds are more often than not built very close to the already cramped living quarters of the people. The floors and walls of the living quarters are plastered with dilute cow dung, which serves to bind the mud. Sun-dried cow dung patties are used as a source of fuel. This proximity is likely to lead to bowel colonization with cattle bacteria, particularly in young children, as shown in this study. The lack of this bacterium in the feces of children living in an urban area, where cattle are generally not found in proximity to human dwellings, suggests that this is indeed the case.
B. fibrisolvens and methanogens are among the predominant ruminant flora Citation[11]. Methanogens are normally known to be part of the human gut flora. Bacteria belonging to the genus Butyrivibrio are occasionally reported from human feces Citation[12], but not B. fibrisolvens, which resides in the gastrointestinal tract of animals Citation[13]. Quantitative PCR showed that the levels of this organism in rural children were higher than in adults from the same background and they were not present in the feces of healthy urban children. This, together with the sequence data showing relatedness of both human and dung isolates to the standard strain of B. fibrisolvens, suggests that the organisms were derived from domestic cattle and that they had colonized the human intestine in this geographic region. Culture of human feces for this organism did not yield the organisms, but we ascribe this to the fastidious requirements, low numbers, and the predominance of other commensals in the fecal samples.
Several probiotic bacteria are used in the expectation that they will alter metabolic parameters and immune status in the individual who ingests these probiotics. The ruminal bacterium B. fibrisolvens is one such potential probiotic microorganism. It is a carbohydrate-fermenting bacterium that produces extracellular xylanases that cleave xylans Citation[14] and cell-associated alpha-amylases that digest starch, resulting in the production of short chain fatty acids, particularly butyrate Citation[15]. Butyrate is a molecule of particular relevance to colonic health and disease, in particular having a protective effect against colon cancer. Its presence in normal humans indicates that it can adapt to the milieu of the human intestine and survive without causing any apparent illness. It therefore fulfills one criterion for a candidate probiotic.
B. fibrisolvens produces conjugated linoleic acid from linoleic acid in animals, which is postulated to benefit health in many ways including prevention of carcinogenesis, atherosclerosis, and tumorigenesis, and improvement of hyperinsulinemia and immune functions Citation[16]. Probiotics may exert their beneficial effects partially through their influence on carbohydrate fermentative processes Citation[17], Citation[18]. B. fibrisolvens is a potential probiotic candidate, influencing the severity of colitis or development of colon cancer in animals Citation[19], Citation[20]. The detection of this ruminant bacterium in the feces of healthy children and adults in rural southern Indians suggests that the organism can coexist peacefully in the human intestine and raises the possibility that this bacterium may be considered a candidate probiotic microorganism for human use.
Acknowledgements
Dr Balamurugan was supported by a Senior Research Fellowship from the Indian Council of Medical Research. The Laboratory was supported by a FIST grant from the Department of Science and Technology, Government of India. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 2007; 73: 1073–8
- Ramakrishna BS. The normal bacterial flora of the human intestine and its regulation. J Clin Gastroenterol 2007; 41((Suppl 1))S2–S6
- Serna, A, 4th, Boedeker, EC. Pathogenesis and treatment of Shiga toxin-producing Escherichia coli infections. Curr Opin Gastroenterol 2008;24:38–47.
- Waddell LA, Rajić A, Sargeant J, Harris J, Amezcua R, Downey L, et al. The zoonotic potential of Mycobacterium avium spp. paratuberculosis: a systematic review. Can J Public Health 2008; 99: 145–55
- Bujanover Y, Peled Y, Gilat T. Is human methane production affected by long-term occupational exposure to a methanogenic flora?. Isr J Med Sci 1986; 22: 16–18
- Hespell RB, Kato K, Costerton JW. Characterization of the cell wall of Butyrivibrio species. Can J Microbiol 1993; 39: 912–21
- Ramakrishna BS, Roediger WEW. Bacterial short chain fatty acids: their role in gastrointestinal disease. Dig Dis 1990; 8: 337–45
- Kanauchi O, Matsumoto Y, Matsumura M, Fukuoka M, Bamba T. The beneficial effects of microflora, especially obligate anaerobes, and their products on the colonic environment in inflammatory bowel disease. Curr Pharm Des 2005; 11: 1047–53
- Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T. Oral administration of Butyrivibrio fibrisolvens, a butyrate-producing bacterium, decreases the formation of aberrant crypt foci in the colon and rectum of mice. J Nutr 2005; 135: 2878–83
- Balamurugan R, Janardhan HP, George S, Raghava MV, Muliyil J, Ramakrishna BS. Molecular studies of fecal anaerobic commensal bacteria in acute diarrhea in children. J Pediatr Gastroenterol Nutr 2008; 46: 514–19
- Koike S, Yoshitani S, Kobayashi Y, Tanaka K. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol Lett 2003; 229: 23–30
- Rumney CJ, Duncan SH, Henderson C, Stewart CS. Isolation and characteristics of a wheatbran-degrading Butyrivibrio from human faeces. Lett Appl Microbiol 1995; 20: 232–6
- Rafii F, Franklin W, Cerniglia CE. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol 1990; 56: 2146–51
- Hespell RB, Whitehead TR. Physiology and genetics of xylan degradation by gastrointestinal tract bacteria. J Dairy Sci 1990; 73: 3013–22
- Ramsay AG, Scott KP, Martin JC, Rincon MT, Flint HJ. Cell-associated alpha-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology 2006; 152: 3281–90
- Fukuda S, Suzuki Y, Murai M, Asanuma N, Hino T. Isolation of a novel strain of Butyrivibrio fibrisolvens that isomerizes linoleic acid to conjugated linoleic acid without hydrogenation, and its utilization as a probiotic for animals. J Appl Microbiol 2006; 100: 787–94
- Araki Y, Andoh A, Takizawa J, Takizawa W, Fujiyama Y. Clostridium butyricum, a probiotic derivative, suppresses dextran sulfate sodium-induced experimental colitis in rats. Int J Mol Med 2004; 13: 577–80
- Heselmans M, Reid G, Akkermans LM, Savelkoul H, Timmerman H, Rombouts FM. Gut flora in health and disease: potential role of probiotics. Curr Issues Intest Microbiol 2005; 6: 1–7
- Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T. Effect of oral administration of Butyrivibrio fibrisolvens MDT-1 on experimental enterocolitis in mice. Clin Vaccine Immunol 2006; 13: 1231–6
- Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T. Effect of oral administration of Butyrivibrio fibrisolvens MDT-1, a gastrointestinal bacterium, on 3-methylcholanthrene-induced tumor in mice. Nutr Cancer 2007; 59: 92–8
