Abstract
Background
Dysregulated circular RNAs (circRNAs) are involved in osteoarthritis (OA) progression.
Objective
We aimed to explore the effect of hsa_circ_0044719 (circTRIM25) on the ferroptosis of chondrocytes.
Methods
Chondrocytes were treated with interleukin (IL)-1β to generate cell model. Cellular behaviours were measured using cell counting kit-8, enzyme-linked immunosorbent assay, relevant kits, propidium iodide staining, and immunofluorescence assay. Quantitative real-time polymerase chain reaction was performed to examine the expression of circTRIM25, miR-138-5p, and cAMP responsive element binding protein 1 (CREB1), and their interactions were assessed using luciferase reporter analysis and RNA pull-down assay.
Results
CircTRIM25 was upregulated in OA tissues and IL-1β-stimulated chondrocytes. Knockdown of circTRIM25 facilitated the viability and suppressed ferroptosis and inflammation of IL-1β-induced cells. CircTRIM25 served as a sponge of miR-138-5p, which directly targets CREB1. Downregulation of miR-138-5p abrogated the effect induced by knockdown of circTRIM25. Furthermore, enforced CREB1 reversed the miR-138-5p induced effect. Moreover, knockdown of circTRIM25 attenuated cartilage injury in vivo.
Conclusion
Silencing of circTRIM25 inhibited ferroptosis of chondrocytes via the miR-138-5p/CREB axis and thus attenuated OA progression.
Introduction
Osteoarthritis (OA) is a common chronic musculoskeletal disorder with an increased prevalence to 527.81 million cases in 2019 worldwide [Citation1]. The incidence rate of OA is increasing in conjunction with the ageing population, leading to a decline in quality of life and an increase in social burden [Citation2]. OA is now known to be a type of inflammation caused by severe degeneration of articular cartilage [Citation3]. OA commonly occurs in weight-bearing joints, including the knee, hip, cervical spine, and lumbar spine, and manifests as pain, stiffness, instability, and deformities [Citation4]. There is no cure for OA and current treatments only help patients relieve pain and maintain physical function [Citation3,Citation5]. Intra-articular injections of drugs, such as non-steroidal anti-inflammatory drugs (NSAIDs), analgesics, and corticosteroids are used for the treatment of OA; however, these drugs have little long-term value, and opioids may have ominous consequences [Citation6]. Hence, it is necessary to understand the pathogenesis of OA to investigate novel therapeutic strategies.
Abnormal structure and function of articular cartilage are the primary causes of OA [Citation7]. Cartilage damage results in an imbalance between bone resorption and remodelling, which in turn induces OA [Citation8]. However, because the mechanism of cartilage matrix degradation remains unclear, there is no effective strategy to delay cartilage degradation or slow OA progression [Citation9]. Therefore, studying the mechanism that promotes chondrogenesis may help decelerate the progression of OA.
Circular RNAs (circRNAs) are single-stranded non-coding RNAs that are widely found in all species. CircRNA is a covalent cyclic structure formed by splicing at specific 3′ and 5′ sites of mRNA [Citation10]. In human cells, circRNAs modulate biological functions, including proliferation, differentiation, cell death, and metastasis [Citation11]. Accumulating evidence shows that circRNAs perform biological functions by functioning as transcriptional regulators, microRNAs (miRs), or protein sponges [Citation12]. Numerous circRNAs are dysregulated in OA joint tissues, acting as diagnostic biomarkers, and are associated with the pathogenesis of OA [Citation13,Citation14]. However, the functions of circRNAs in OA remain largely unknown.
Ferroptosis is a newly discovered type of iron-dependent programmed cell death that plays a crucial role in numerous human diseases, promoting or inhibiting disease development. In ferroptosis, unsaturated fatty acids are catalysed to produce lipid peroxides by Fe2+ or ester oxygenase stimulation, thereby inducing cell death [Citation15]. It has broad application prospects in the treatment of cancer, neurological diseases, and inflammation [Citation16–18]. Inflammation is inhibited by ferroptosis owing to its immunogenicity. Ferroptosis is a potential pathogenesis of OA. High concentration of iron promotes joint degeneration, thus facilitating OA development [Citation19]. Previous research has revealed that inhibition of ferroptosis in chondrocytes contributes to preventing or attenuating OA [Citation20,Citation21]. Moreover, ferroptosis-related genes, such as GPX4, system xc-, SLC7A11, are involved in the progression of OA [Citation22,Citation23]. However, the involvement of circRNAs in the regulation of ferroptosis in OA remains poorly understood.
In this study, we identified an abnormally expressed circRNA (has_circ_0044719, termed circTRIM25) in patients with OA and aimed to clarify its functional role in OA progression and the underlying mechanisms. Depletion of the circTRIM25/miR-138-5p/CREB1 axis suppresses ferroptosis in chondrocytes. These data provide a theoretical basis for the involvement of circTRIM25 in OA pathogenesis.
Materials and methods
Microarray assay
To investigate which circRNA participates in OA progress, differentially expressed circRNAs in patients with knee OA and healthy controls using microarray analysis. A microarray (GSE175959) was searched in the Gene Expression Omnibus (GEO) database. The data were analyzed using the R language. All circRNAs were normalized to log2 values. Differentially expressed circRNAs were defined as |Log2 FC|≥1 and p < 0.05.
Cartilage tissue collection
Human cartilage tissue collection was approved by The Ethics Committee of First Affiliated Hospital of Gannan Medical University (Approval No. LLSC-2022122602). Informed consent was obtained from all subjects. Normal cartilage tissues (n = 40) were obtained during total knee arthroplasty (TKA) from patients without OA (range of age: 50–74 years old, mean age: 66.525 years old, and male: female = 3:7). Pathological cartilage tissues (n = 40) were obtained from patients with end-stage symptomatic knee OA during TKA (range of age: 52–79 years old, mean age: 66.45 years old, and male: female = 2:3). The inclusion criteria were patients diagnosed with OA according to the American College of Rheumatology (ACR) classification criteria [Citation24]. Patients with rheumatoid arthritis or other conditions affecting the joints were excluded from the study. The cartilage tissues of articular surface from the lateral femur knee joint were selected and washed with PBS to remove the blood and tissue fluid. The cartilages were wrapped in saline gauze and cut into a cylinder with a diameter of about 1 cm. Then, the tissues were placed in Dulbecco’s modification of Eagle’s medium (DMEM) and subpackaged in frozen storage tube, which were frozen in liquid nitrogen immediately and then stored at −80 °C until subsequent analyses. The whole sampling process were carried out under strict aseptic conditions.
Chondrocyte collection
Primary chondrocytes were isolated according to the previous description with minor changes [Citation25]. Cartilage samples were cut into pieces and digested with 0.2% collagenase II (Thermo Fisher Scientific). The resulting precipitated cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco) at 37 °C and 5% CO2. The medium was replaced daily, and the cells were washed with PBS. The cells were subcultured using trypsin (Thermo Fisher Scientific). Second-passage cells maintained in chondrocyte growth medium (PromoCell) were used for transfection. Chondrocytes were treated with 5 ng/mL IL-1β (Sigma-Aldrich) for 24 h to establish the OA cell injury model.
Cell transfection
Short hairpin RNA (sh)-circTRIM25 1# (CCGGAACACAATTATGAAGTTGGGACTCGAG), sh-circTRIM25 2# (CCGGACACAATTATGAAGTTGGGATCTCGAG), sh-circTRIM25 3# (CCGGCACAATTATGAAGTTGGGATACTCGAG), and sh-NC (CCGGGGUACACCCUCCAUGGUAAUUCTCGAG) were cloned into the pLKO1 puro vectors. The recombinant plasmids were transfected into HEK293T cells with psPAX2 and pMD2.G vectors. The lentiviruses were collected. Vector (pcDNA3.1) and overexpressing CREB1 vector (pcDNA3.1-CREB1) were acquired by Genepharma (Shanghai, China). MiR-138-5p mimic, inhibitor, NC mimic and NC inhibitor were purchased from RiboBio (Guangzhou, China). Lipofectamine 3000 (Invitrogen, ThermoFisher Scientific) was utilized for cell transfection of lentiviruses or plasmids [Citation26]. After 48 h, the cells were harvested.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from chondrocytes and cartilage tissues using TRIzol reagent (Invitrogen). The miRNA 1st strand cDNA was synthesized using a one-step PrimeScript miRNA cDNA synthesis kit (Takara, Dalian, China), whereas the cDNA synthesis kit (Takara) was evaluated using the 1st strand cDNA of mRNA, using 1 μg total RNA. qPCR (50 ng/μl cDNA) was conducted using SYBR Premix Ex TaqII (Perfect Real Time) (Takara) on a LightCycler real-time PCR system (Roche, Basel, Switzerland). The reaction conditions were 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The relative expression of miR-138-5p was normalized to that of U6, and mRNA was normalized to that of GAPDH. The 2-ΔΔCt formula was used to calculate the expression. The primers used are shown in .
Table 1. Primer sequences used for quantitative PCR.
RNase R treatment
To verify the stability of circTRIM25 molecular structure, RNase R digestion experiment was performed [Citation27]. RNase R can slice RNA to dinucleotides and trinucleotides. So, it can digest almost all linear RNA, but it is difficult to digest circRNA. Total RNA (2.5 μg) was incubated with or without 10 U RNase R (Geneseed, Guangzhou, China) at 37 °C for 30 min. Then the expression of circTRIM25 and TRIM25 was tested using qRT-PCR.
Actinomycin D treatment
To assess the stability of circTRIM25, actinomycin D (2 mg/ml; Sigma-Aldrich) was used to block transcription for 0, 4, 8, and 12 h as previously described [Citation27], and the circTRIM25 or TRIM25 levels were also tested using qRT-PCR.
Fluorescence in situ hybridization (FISH)
To evaluate the location of circTRIM25 and miR-138-5p in cells, FISH was performed as previously described [Citation28]. Cells were immobilized with 4% paraformaldehyde (1 mL; Sigma-Aldrich) and treated with 0.25 mg/ml protease K (Sigma-Aldrich) at 37 °C for 20 min. After dehydration with a gradient concentration of alcohol, a fluorescent-labelled probe for circTRIM25 was degenerated at 73 °C for 3 min and added onto a slide for hybridization at 42 °C overnight. The probe was then degenerated at 73 °C for 3 min. The slides were then washed with formamide and 2 × SSC buffer. 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) was used to stain the nuclei. Fluorescence signals were photographed using a laser confocal microscope (Leica, Mannheim, Germany).
Cell viability
The cells (2000 cells/well) plated onto 96-well plates were incubated at 37 °C for 24 h. Next, the cells were incubated with 10 μl CCK-8 solution (Sangon, Shanghai, China) for 4 h [Citation27]. The absorbance at 450 nm was examined with a microplate reader (ThermoFisher Scientific).
Enzyme-linked immunosorbent assay (ELISA)
The levels of inflammation related factor were assessed using tumor necrosis factor (TNF)-α, interleukin (IL)-6 or IL-10 ELISA kits (BBI, Shanghai, China) based on the manufacturer’s protocol. Briefly, cell culture medium was centrifuged at 1000 g for 20 min to collect the supernatant. The samples were incubated with 100 μl corresponding biotin-labelled antibody solution at 37 °C for 60 min, washed with 350 μl washing buffer, incubated with 100 μl HRP-labelled streptavidin working solution at 37 °C for 30 min, washed with 350 μl washing buffer, and incubated with 90 μl color developing reagent at 37 °C for 15 min in the dark. After adding 50 μl reaction stop buffer, the absorbance at 450 nm was examined.
Western blot
Radio-immunoprecipitation assay (RIPA) buffer was used to split the cells on the ice for 15 min, and protein was harvested after determining the concentration using the bicinchoninic acid (BCA) method. Equal amounts of protein (40 μg) were added to each lane and separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After protein transfer, the membranes were blocked with 5% skimmed milk for 1 h. Membranes were incubated at 4 °C overnight with primary antibodies (Abcam, Cambridge, MA, USA) including anti-MMP13 (ab39012, 1:6000), anti-ADAMTS-5 (ab182795, 1:1000), anti-collagen II (ab188570, 1:5000), and anti-GAPDH (ab9485, 1:2500), followed by incubation at room temperature for 1 h with a secondary antibody (ab6721, 1:3000, Abcam). Signals were visualized using enhanced chemiluminescence (ECL) reagent (Sangon). GAPDH was used as an internal control. Protein levels were quantified using the ImageJ software.
Reactive oxygen species (ROS) assay
As previously described [Citation29], ROS levels were determined using a DCFHDA ROS assay kit (BestBio, Shanghai, China). Briefly, cells were plated into 6-well plates (2 × 106 cells) and incubated with serum-free diluted DCFHDA at 37 °C for 20 min. The cells were then washed with serum-free medium, and fluorescence was determined using a laser confocal microscope.
Lipid ROS assay
To evaluate the lipid peroxidation and antioxidant properties in living cells, lipid ROS levels were examined using a C11 BODIPY 581/591 kit (ABclonal, Wuhan, China) [Citation30]. The cells were then incubated with 50 μM C11 BODIPY 581/591 for 1 h. After washing with PBS, fluorescence was determined using a laser confocal microscope.
Malondialdehyde (MDA) activity
The determination of MDA is widely used as an indicator of lipid oxidation, a lipid peroxidation MDA assay kit (Beyotime, Shanghai, China) was used to detect MDA levels [Citation30]. Briefly, cells (1 × 106 cells) were lysed on ice. Subsequently, 0.2 ml MDA detection solution was mixed with the cell homogenate and allowed to react at 100 °C for 15 min. After cooling, the specimens were centrifuged at 1000 × g for 10 min. Optical density was measured using a microplate reader at 532 nm.
Prodium iodide (PI) staining
To view cell death, PI staining was performed. The PI staining kit (Sangon) was used to assess cell death. The cells (1 × 106 cells) were suspended in assay buffer and incubated with 5 μl PI solution at 4 °C for 0.5 h. DAPI was used to stain nuclei. Finally, fluorescence signals were captured using a fluorescence microscope (Leica).
Iron assay
Fe2+ content was evaluated using an iron assay kit (Abcam, Cambridge, USA) [Citation29]. Cell samples (1 × 106 cells) were washed with PBS and diluted with 100 μL iron assay buffer. Each well sample was incubated with 5 μl iron reducer at 37 °C for 0.5 h, followed by incubation with 100 μL iron probe at 37 °C for 1 h in the dark. Optical density was measured at 593 nm using a microplate reader.
Immunofluorescence assay
The experiment was performed as previously described [Citation28]. To evaluate the GPX4 levels, cells (1 × 106 cells) were immobilized with 4% paraformaldehyde for 15 min, followed by permeabilization with 0.2% Triton X100 for 15 min. After blocking with normal goat serum for 1 h, the cells were incubated with the primary antibody (anti-GPX4: ab125066, 1:100, Abcam) at 4 °C overnight. The cells were then incubated with Alexa Fluor-conjugated secondary antibody (ab150077, 1:1000, Abcam) at 37 °C for 1 h. Images were captured using a fluorescent microscope.
Bioinformatics
The targeted miRNAs and the binding sites between circTRIM25 and miR-138-5p were predicted using the Starbase database. The downstream target mRNAs of miR-138-5p were predicted using the three datasets including Starbase, TargetScan, and miRDB. The signal pathway of miR-138-5p-target genes enrichment was analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment on the DAVID database with language R.
Luciferase reporter analysis
Luciferase reporter analysis was carried out to assess the targeting relation between miR-138-5p and circTRIM25 or CREB1 [Citation31]. Wild-type (WT) and mutated (MUT) 3′-UTR fragments of circTRIM25 or CREB1 were inserted into reporter vectors. The cells were co-transfected with mimic or NC mimic together with WT or MUT vectors for 48 h. A dual-luciferase assay kit (Solarbio, Beijing, China) was used to determine luciferase activity. Renilla luciferase activity was used as an internal standard.
RNA pull-down assay
To assess the relation between miR-138-5p and circTRIM25 or CREB1, RNA pull-down was conducted [Citation31]. cells were lysed using lysis buffer on ice for 10 min, and the lysates were collected by centrifuging at 10,000 g for 15 min. A part of lysates was incubated with biotin-labelled miR-138-5p probes and biotin-labelled NC. Lysates were used as the input group. Streptavidin Dynabeads (Invitrogen) were incubated with the lysates at 4 °C overnight. After washing, the results were analyzed using qRT-PCR.
Animal study
The animal study was approved by The Ethics Committee of First Affiliated Hospital of Gannan Medical University (Approval No.22SC-2022122602). The OA model was established using the destabilization of the medial meniscus (DMM) surgery on C57BL/6 mice (male, 8 weeks old, weight 22–25 g). Mice were housed under specific pathogen-free conditions with 12 h/h light/dark cycles, 24 °C temperature, 50–60% humidity, and enough food and water. After a week of acclimatization, all mice were divided into 3 groups: sham, DMM, and DMM + sh-circTRIM25, with 5 mice per group. To perform DMM surgery [Citation32], after anesthetizing using 1% pentobarbital sodium, the medial meniscotibial ligament (MMTL) was transected on right knee joints of mice. The wound was sutured using 5-0 silk thread. The mice in the sham group were only exposed MMTL at the same location. The mice in the DMM + sh-circTRIM25 were intra-articular injected with 5 μL sh-circTRIM25 (109 PFU) one week after model establishment, once per week [Citation33]. Mice were sacrificed after 4 weeks of surgery by injecting (i.p.) overdose pentobarbital sodium. The right knee joint was collected and fixed in 4% paraformaldehyde for 36 h. After decalcification, the joint tissues were embedded in paraffin and sectioned along the sagittal plane (4 μm of thickness).
Histological evaluation
Safranin O-fast green staining was used for general histological evaluation [Citation34]. The paraffin sections were dewaxed and rehydrated. Iron hematoxylin was added to incubate with nucleus for 20 s and differentiated with ethanol hydrochloride solution. Then, the sections were stained with 0.02% fast green for 6 min, followed by stained with 1% safranin O for 10 min. After sealing, the sections were observed under a light microscope. The severity of OA was estimated using the Osteoarthritis Research Society International (OARSI) scoring system via estimating the highest score from the medial femoral condyle and the medial tibial plateau following a previous study [Citation35].
Immunohistochemistry staining
As previously described [Citation30], To evaluate the levels of collagen II and GPX4 in the joint tissues of mice, the sections were quenched endogenous peroxidase activity and blocked with normal goat serum. Then, the sections were incubated with primary antibody (Abcam) including anti-collagen II (ab34712, 1:100) and anti-GPX4 (ab125066, 1:100) at 4 °C overnight, followed by incubated with the secondary antibody (ab6721, 1:1000, Abcam) at 37 °C for 1 h. The results were observed under a light microscope.
Statistical analysis
All experiments were performed in triplicate. The results were analyzed using GraphPad Prism 7 software and are presented as mean ± SD. Comparisons were analyzed using Student’s t-test and one-way analysis of variance followed by Tukey’s post hoc test, which was used to identify significant differences. Statistical significance was set at p < 0.05.
Results
circTRIM25 expression was increased in OA
Numerous circRNAs were significantly dysregulated between healthy subjects and patients with OA (). Hsa_circ_0044719 was significantly upregulated in OA. The gene symbol of hsa_circ_0044719 is TRIM25, and the information of hsa_circ_0044719 and TRIM25 is shown in . Therefore, we named hsa_circ_0044719 as circTRIM25 below. Subsequently, circTRIM25 and hsa_circ_0002643 were tested in cartilage tissues and chondrocytes. circTRIM levels were significantly higher in OA cartilage tissues and chondrocytes than in healthy control tissues, whereas hsa_circ_0002643 showed no significant difference between the normal and OA groups (). OA chondrocytes stimulated with IL-1β showed a marked increase in circTRIM expression (). RNase R treatment induced degradation of linear TRIM25 transcripts but not circTRIM25 (). Actinomycin D treatment data revealed that circTRIM25 had a half-life of > 12 h, whereas linear TRIM25 had a half-life of less than 8 h (). We found that circTRIM25 was located on chr17, encoded by 5744 bp and cyclized exons 1-9 ( and ). Moreover, the FISH results indicated that circTRIM25 mainly existed in the cytoplasm (), consistent with circRNA localization in cells, suggesting that we studied circRNA TRIM25 rather than linear TRIM25.
Figure 1. circTRIM25 is upregulated in osteoarthritis (OA). Differentially expressed circRNAs in healthy controls and patients with OA from the microarray GSE175959 are shown using (A) a heatmap and (B) a volcano plot. (C) Information on the TRIM25 gene. The expression of (D) circTRIM25 and (E) circ_0002643 was measured in cartilage tissues of normal participants and patients with OA using quantitative real-time PCR (qRT-PCR). The expression of (F) circTRIM25 and (G) circ_0002643 was examined using qRT-PCR in chondrocytes isolated from normal or OA participants. (H) The circTRIM25 levels were tested using qRT-PCR following chondrocytes treated with or without IL-1β. (I) qRT-PCR was used to examined linear TRIM25 and circTRIM25 levels after RNase R treatment. (J) qRT-PCR was used to examined linear TRIM25 and circTRIM25 levels after actinomycin D treatment for 0, 4, 8, and 12 h. (K) The cyclic structure of circTRIM25. (L) The location and the formation of circTRIM25. (M) Cellular localisation of circTIRM25 was assessed using fluorescence in situ hybridisation (FISH) analysis. **p < 0.01. ***p < 0.001. ns indicates no significant difference.
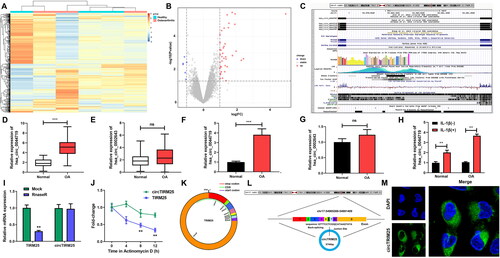
Knockdown of circTRIM25 suppresses ferroptosis of IL-1β-induced chondrocytes
To investigate the effect of circTRIM25 on chondrocyte cellular behaviors, we designed three shRNA sequences targeting circTRIM25 and constructed lentivirus containing these shRNAs, named sh-circTRIM25 1#, 2#, and 3#. The circTRIM25 expression was highly reduced following transfection with sh-circTRIM25 1#, 2#, and 3# (). Cell viability was inhibited by IL-1β and reversed by knockdown of circTRIM25 (). Additionally, IL-1β significantly elevated TNF-α and IL-6 levels, but reduced IL-10 levels. However, loss of circTRIM25 reversed the effects induced by IL-1β (). IL-1β significantly increased the levels of MMP13 and ADAMTS-5 but decreased the levels of collagen II. Depletion of circTRIM25 abrogated IL-1β effects (). ROS, lipid ROS, and MDA levels were significantly decreased by knockdown of circTRIM25 in IL-1β-induced cells (). The content of Fe2+ was significantly increased by IL-1β but was rescued by sh-circTRIM25 (). The PI staining assay demonstrated that circTRIM25 loss downregulated cell death in IL-1β-stimulated cells (). GPX4 expression was significantly reduced by IL-1β and stopped by circTRIM25 depletion ().
Figure 2. Knockdown of circTRIM25 suppresses ferroptosis of interleukin (IL)-1β-induced chondrocytes. (A) circTRIM25 expression was tested using quantitative real-time PCR (qRT-PCR) after chondrocytes transfected with sh-circTRIM25 1#, 2#, and 3#. (B) Cell viability was assessed by cell counting kit-8 (CCK-8) assay. (C) The tumor necrosis factor (TNF)-α, (D) IL-6 and (E) IL-10 levels were tested by enzyme-linked immunosorbent assay (ELISA). The MMP13, ADAMTS-5 and collagen II levels were examined using (F) qRT-PCR and (G and H) western blot. (I) Reactive oxygen species (ROS) and (J) lipid ROS levels were tested using a ROS assay kit. (K) The malonaldehyde (MDA) content was tested using a lipid peroxidation MDA assay kit. (L) Fe2+ content was analysed iron assay kit. (M) Cell death was analysed using propidium iodide (PI) staining assay, and (N) percentage of cell death was quantified. (O) GPX4 expression was evaluated using immunofluorescence (IF) assay, and (P) relative fluorescence intensity was quantified. *p < 0.05. **p < 0.01. ***p < 0.001.
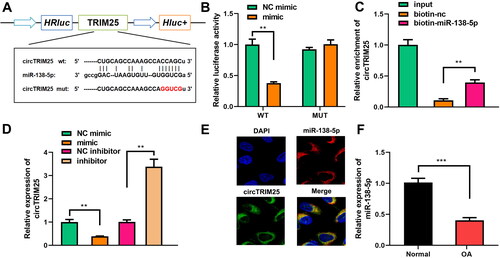
circTRIM25 directly targets miR-138-5p
Bioinformatic results indicated that circTRIM25 could directly bind to miR-138-5p (). miR-138-5p mimics significantly reduced luciferase activity in the WT group compared to that in the MUT group (). circTRIM25 was precipitated by biotin-labelled miR-138-5p () and the expression of circTRIM25 was negatively correlated with that of miR-138-5p (). As shown in , circTRIM25 and miR-138-5p were localized in the cytoplasm, further suggesting that circTRIM25 acted as a sponge of miR-138-5p in the cytoplasm. Compared to healthy subjects, downregulation of miR-138-5p was observed in OA chondrocytes ().
Figure 3. circTRIM25 is a miR-138-5p sponge. (A) Sequences of circTRIM25 and miR-138-5p binding sites were predicted using Starbase database. (B) Luciferase reporter analysis and (C) RNA pull down assay were used to confirm the interaction between circTRIM25 and miR-138-5p. (D) Quantitative real-time PCR (qRT-PCR) was performed to determine circTRIM25 levels when miR-138-5p was overexpressed or inhibited. (E) Fluorescence in situ hybridisation (FISH) was conducted to identify the intracellular location of circTRIM25 and miR-138-5p. (F) miR-138-5p was examined in cartilage tissues of healthy controls and patients with osteoarthritis (OA). **p < 0.01. ***p < 0.001.
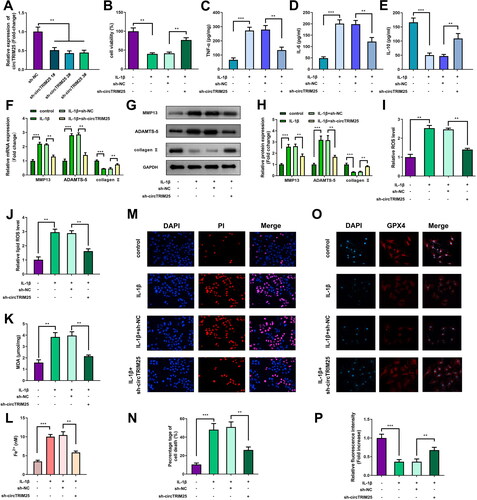
circTRIM25 directly targets miR-138-5p to regulate ferroptosis induced by IL-1β
miR-138-5p levels were markedly decreased when cells were transfected with miR-138-5p inhibitor (). Inhibition of miR-138-5p reversed the enhanced cell viability induced by sh-circTRIM25 (). The effect of circTRIM25 knockdown on TNF-α, IL-6, and IL-10 levels was significantly reversed by miR-138-5p inhibition (). Depletion of circTRIM25 significantly decreased MMP13 and ADAMTS-5 levels and increased collagen II levels, while inhibition of miR-138-5p abrogated this effect (). The decreased levels of ROS, lipid ROS, MDA, and Fe2+ and cell death were elevated by inhibiting miR-138-5p (). The upregulation of GPX4 expression induced by knockdown of circTRIM25 was largely rescued by inhibiting miR-138-5p ().
Figure 4. Interfering of circTRIM25 suppressed ferroptosis of interleukin (IL)-1β-stimulated cells via targeting miR-138-5p. (A) miR-138-5p was examined by quantitative real-time PCR (qRT-PCR) post-transfection. (B) Cell counting kit-8 (CCK-8) assay was used to determine cell viability. (C) The tumor necrosis factor (TNF)-α, (D) IL-6 and (E) IL-10 levels were examined by enzyme-linked immunosorbent assay (ELISA). (F) qRT-PCR and (G and H) western blot were used to evaluate the levels of MMP13, ADAMTS-5, and collagen II. (I) Reactive oxygen species (ROS) and (J) lipid ROS levels were tested using a ROS assay kit. (K) The malonaldehyde (MDA) content was tested using a lipid peroxidation MDA assay kit. (L) Fe2+ content was analysed using an iron assay kit. (M) Cell death was analysed using propidium iodide (PI) staining assay, and (N) percentage of cell death was quantified. (O) GPX4 expression was evaluated using immunofluorescence (IF) assay, and (P) relative fluorescence intensity was quantified. *p < 0.05. **p < 0.01. ***p < 0.001.
miR-138-5p directly targets CREB1
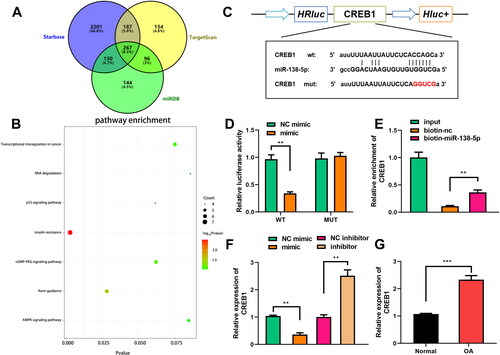
miRNA serves the functions by binding to downstream target mRNAs. Therefore, the targets of miR-138-5p were necessary to be investigated. We predicted the potential targets of miR-138-5p using Starbase, TargetScan, and miRDB databases. A total of 267 potential targets could be predicted using all these databases (). Pathway enrichment analyses demonstrated that these targets were associated with several pathways, including insulin resistance, axon guidance, transcriptional misregulation, RNA degradation, p53, cGMP-PKG, and AMPK pathways (). CREB1 is a factor insulin resistance, cGMP-PGK, and AMPK pathways, therefore, it was selected for further study. CREB1 was directly bound to miR-138-5p (). miR-138-5p markedly reduced luciferase activity when co-transfected with the WT reporter vector, while NC and miR-138-5p did not influence luciferase activity when co-transfected with the MUT vector (). CREB1 was markedly downregulated by biotin-labelled miR-138-5p (). Overexpression of miR-138-5p decreased CREB1, whereas inhibition of miR-138-5p increased CREB1 (). CREB1 levels were significantly higher in OA tissues than in normal tissues ().
Figure 5. miR-138-5p directly targets CREB1. (A) The miR-138-5p targets were predicted by Starbase, TargetScan, and miRDB datasets, and these targets are shown in a Venn diagram. (B) The pathways related to the 267 targets of miR-138-5p were predicted using Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis using DAVID software. (C) Sequences of CREB1 and miR-138-5p binding sites are shown. The interaction between miR-138-5p and CREB1 was affirmed by (D) luciferase reporter analysis and (E) an RNA pull down assay. (F) The levels of CREB1 were examined by quantitative real-time PCR (qRT-PCR) when miR-138-5p was overexpressed or inhibited. (G) CREB1 was examined in normal or osteoarthritic cartilage tissues. **p < 0.01. ***p < 0.001.
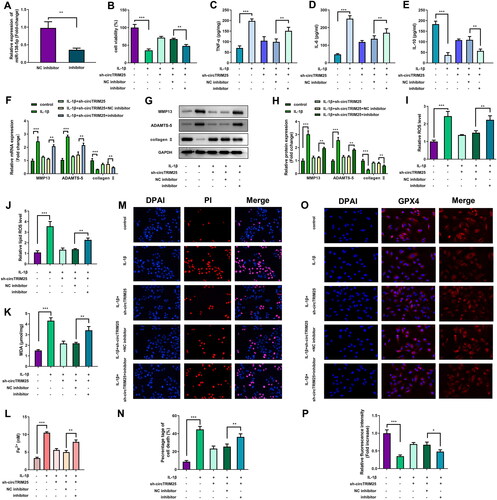
miR-138-5p targets CREB1 to suppress ferroptosis induced by IL-1β
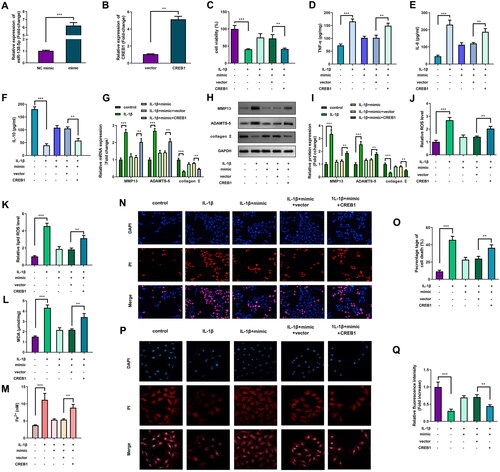
miR-138-5p and CREB1 were markedly elevated after transfecting mimic and CREB1 overexpressing vector (). The promotion of cell viability by miR-138-5p of IL-1β-stimulated cells was markedly reversed by CREB1 (). Overexpression of miR-138-5p significantly decreased TNF-α, IL-6 levels and significantly increased IL-10 levels, while CREB1 significantly abrogated miR-138-5p effect (). Additionally, increased CREB1 expression markedly abrogated the effect of miR-138-5p on MMP13, ADAMTS-5, and collagen II levels (). miR-138-5p markedly rescued the IL-1β-induced upregulation of ROS levels, lipid ROS levels, MDA content, cell death, and Fe2+ content, whereas CREB1 markedly the effect of rescued miR-138-5p (). The expression of GPX4, which was markedly elevated by miR-138-5p, was abrogated by CREB1 ().
Figure 6. Overexpression of miR-138-5p inhibits ferroptosis of interleukin (IL)-1β-induced cells by targeting CREB1. (A) The miR-138-5p and (B) CREB1 levels were examined using quantitative real-time PCR (qRT-PCR) post-transfection. (C) Cell viability was assessed by cell counting kit-8 (CCK-8) assay. Enzyme-linked immunosorbent assay (ELISA) was conducted to measure the (D) tumor necrosis factor (TNF)-α, (E) IL-6, and (F) IL-10 levels. The MMP13, ADAMTS-5, and collagen II levels were evaluated by (G) qRT-PCR and (H and I) western blot. (J) Reactive oxygen species (ROS) and (K) lipid ROS levels were examined using a ROS assay kit. (L) The malonaldehyde (MDA) content was tested using a lipid peroxidation MDA assay kit. (M) Fe2+ content was analysed iron assay kit. (N) Cell death was analysed using propidium iodide (PI) staining assay, and (O) percentage of cell death was quantified. (P) The GPX4 level was evaluated via immunofluorescence assay, and (P) relative fluorescence intensity was quantified. **p < 0.01. ***p < 0.001.
Knockdown of circTRIM25 alleviated DMM-induced OA
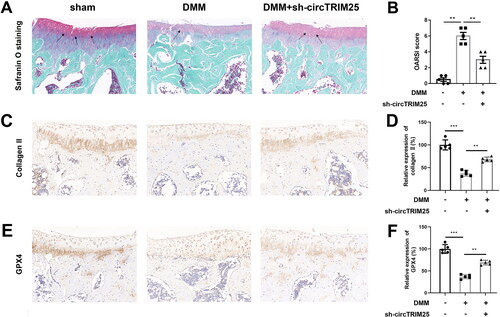
Finally, we evaluated the role of circTRIM25 in vivo. As shown in , the mice in the DMM group represented OA-like features, including proteoglycan content loss, cartilage fibrillation, and erosion, whereas knockdown of circTRIM25 attenuated OA-like manifestations (). The OARSI score was increased by DMM surgery and was decreased by circTRIM25 knockdown (). In addition, the levels of collagen II and GPX4 were decreased in the DMM group, which were reversed after silencing circTRIM25 ().
Figure 7. Knockdown of circTRIM25 alleviated destabilisation of the medial meniscus (DMM)-induced osteoarthritis (OA). (A) The cartilage destruction was visualised using safranin O-fast green staining and was shown using solid arrow. (B) The severity of OA in articular cartilage was evaluated using the Osteoarthritis Research Society International (OARSI) scoring system. The levels of (C and D) collagen II and (E and F) GPX4 were measured in the cartilage tissues using immunohistochemistry (IHC) assay. **p < 0.01. ***p < 0.001.
Discussion
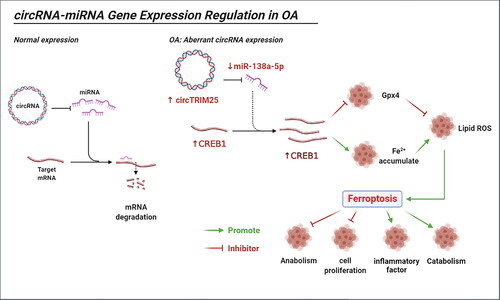
In the present study, we explored the role of circTRIM25 in OA. First, we found that depletion of the circTRIM25/miR-138-5p/CREB1 axis suppressed ferroptosis and therefore decelerated the progression of OA ().
Figure 8. Illustration of the mechanism by which the circTRIM25/miR-138-5p/CREB1 axis regulates ferroptosis in osteoarthritis (OA). The combination of miRNA and mRNA leads to mRNA degradation, and normal expressed circRNA does not affect this process. circTRIM25, an aberrant expressed circRNA in OA, acts as a sponge of miR-138-5p, which targets CREB1 to inhibit GPX4 and promotes Fe2+ accumulation, leading to lipid peroxidation and induced ferroptosis. Ferroptosis suppresses anabolism and cell proliferation but promotes inflammation and catabolism.
OA is a global public health burden that significantly affects life quality of patients and may even cause disability. circRNAs are dysregulated in OA joint tissues and are associated with the pathogenesis of OA. In the present study, microarray was performed to screen dysregulated circRNAs in OA. We demonstrated, for the first time, that circTRIM25 was upregulated in OA cartilage tissues and chondrocytes, suggesting that circTRIM25 may be involved in the progression of OA. However, how circTRIM25 affects the progression of OA remains unknown, Hence, we focused on the role of circTRIM25 in OA.
The levels of inflammatory cytokines control OA. Pro-inflammatory cytokines, such as TNF-α, IL-6, IL-15, promote inflammation response, whereas anti-inflammatory cytokines, such as IL-4, IL-10, and IL-13, attenuate OA [Citation36]. A previous study showed that apigenin reduces IL-6 and TNF-α levels, and increases IL-10 levels to inhibit the progression of OA [Citation37], suggesting that the change trend of IL-6 and TNF-α is opposite to that of IL-10. We found that knockdown of circTRIM25 decreased TNF-α and IL-6 levels but increased IL-10 levels, suggesting that inflammation was alleviated.
Cartilage is a tissue composed of chondrocytes. OA alters the proliferation, activity, and secretion of chondrocytes [Citation38]. MMP13, ADAMTS-5, and collagen II play pivotal roles in chondrogenesis. Increased MMP-13 levels lead to increased degradation of collagen II, resulting in chondrocyte damage [Citation39]. ADAMTS-5 acts as a target in OA and is responsible for the degradation of aggregates, which are the main proteoglycans in articular cartilage [Citation40]. Previous studies have reported that circRNAs involved in OA are regulated by a balance between cartilage formation and degradation. For instance, the loss of circPDE4D upregulates MMP13 and ADAMTS-5, thereby promoting the progression of OA [Citation41]. circ_0001236 suppresses OA progression by promoting cartilage formation through upregulating collagen II and SOX9 and downregulating MMP13 levels [Citation42]. In the present study, we demonstrated, for the first time, that circTRIM25 was upregulated in OA cartilage tissues and chondrocytes. Knockdown of circTRIM25 decreased TNF-α and IL-6 levels but increased IL-10 levels, suggesting that inflammation was alleviated. Moreover, the knockdown of circTRIM25 reduced MMP13 and ADAMTS-5 levels but increased collagen II levels, suggesting that circTRIM25 silencing protected against OA by promoting chondrogenesis.
Programmed cell death, such as apoptosis, pyroptosis, necroptosis, ferroptosis, autophagy, and cuproptosis, are associated with the occurrence and development of OA [Citation43]. Chondrocyte death occurs in the processes of OA, thereby, targeting cell death may be a potential target for OA therapy [Citation44]. Ferroptosis, a form of cell death, is involved in numerous diseases, including OA. Unlike other programmed cell death pathways, ferroptosis is characterized by the accumulation of lipid peroxides and iron dependence [Citation45,Citation46]. Lipoxygenases are important enzymes that catalyze the production of lipid peroxides, which can be transferred by labile iron [Citation47]. ROS are involved in the reaction between H2O2 and Fe2+ during lipid peroxidation without enzymes [Citation48]. Additionally, GPX4 plays a pivotal role in blocking ferroptosis by converting lipid hydroperoxides into nontoxic lipid alcohols [Citation49]. The expression of GPX4 is decreased in OA cartilage than that in undamaged cartilage. Ferroptosis and inhibition of GPX4 contribute to OA progression [Citation50]. Inactivation of GPX4, caused by the ferroptosis-initiated factor RSL3, leads to the accumulation of lipid peroxides, which is associated with cartilage damage in patients with OA [Citation51,Citation52]. MDA is a biomarker of lipid peroxidation, and intensified ferroptosis refers to the accumulation of MDA [Citation53]. circRNAs regulate ferroptosis in human diseases. For example, circ-clARS silencing inhibits ferroptosis by suppressing autophagy and ferritinophagy in hepatocellular carcinoma [Citation54]. circTTBK2 dysregulation is related to glioma cell proliferation, metastasis and ferroptosis [Citation55]. However, little is known regarding the involvement of circRNAs in ferroptosis in OA. In this study, circTRIM25 knockdown inhibited ROS levels, lipid ROS levels, MDA content, and Fe2+ content, but elevated GPX4 levels, suggesting that depletion of circTRIM25 suppressed ferroptosis in IL-1β-treated chondrocytes. Moreover, we explored the role of circTRIM25 in vivo. OA animal model can be established using several methods, such as spontaneous model [Citation56], DMM surgery [Citation32], and anterior cruciate ligament transection [Citation21]. Here, mice were performed DMM surgery to establish the OA model as it is a common method for model generation, and the results showed that knockdown of circTRIM25 inhibited OA-like lesions and increased collagen II and GPX4 expression in DMM mice, suggesting that silencing of circTRIM25 attenuates OA by promoting chondrogenesis and suppressing ferroptosis. Taken together, silencing circTRIM25 promotes chondrogenesis by promoting viability and suppressing ferroptosis, thereby inhibiting the development of OA.
The underlying mechanisms were also explored. We found that circTRIM25 is highly expressed in the cytoplasm. This demonstrates that circTRIM25 plays a functional role in OA through the ceRNA mechanism. miR-138-5p was affirmed be a circTRIM25 target. miR-138-5p is involved in numerous diseases, including malignancies, cardiovascular diseases, and metabolic disorders [Citation57–59]. Recent research has shown that downregulation of miR-138-5p facilitates cartilage degradation [Citation60]. Moreover, miR-138-5p has been reported to regulate ferroptosis of retinal pigment epithelial cells [Citation61]. Nevertheless, whether miR-138-5p is involved in chondrocyte ferroptosis remains unknown. In the present study, we found that miR-138-5p was expressed at lower levels in OA. Inhibition of miR-138-5p abrogated the effects on viability, ferroptosis, and inflammation induced by circTRIM25 deficiency, suggesting that circTRIM25 silencing attenuated OA progression by sponging miR-138-5p by suppressing ferroptosis.
miRNAs regulate the incidence and progression of diseases by binding to target mRNAs and modulating post-transcriptional gene expression. Multiple databases have been developed to predict targets of miRNAs, including Starbase, TargetScan, and miRDB databases. we predicted 267 targets of miR-138-5p. These targets were enriched in several pathways, particularly insulin resistance. Emerging data showed that OA is associated with type 2 diabetes, which involving insulin resistance. Thus, we chose CREB1, a member in the insulin pathway, for study. CREB1 is a transcription factor that is a member of the basic leucine zipper family. AMPK is located downstream of CREB1, which can be phosphorylated by AMPK [Citation62]. Numerous miRNAs can interact with CREB1 [Citation63], and abnormal CREB1 expression is associated with OA. Wang et al. reported that decreased expression of CREB1 suppresses cell growth and accentuates inflammation [Citation64]. Xue et al. revealed that lactoferrin promotes the phosphorylation of CREB1, which further facilitates the repair of articular cartilage damage [Citation65]. Moreover, CREB1 is associated with ferroptosis in lung cancer; however, whether it can regulate ferropotosis in OA is not understood [Citation66]. In this study, CREB1 levels are elevated in OA, which reverses the effects of miR-138-5p. These data suggest that silencing of circTRIM25 attenuates OA progression by sponging miR-138-5p/CREB1.
In conclusion, silencing the circTRIM25/miR-138-5p/CREB1 axis suppressed ferroptosis of chondrocytes and attenuated DMM-induced OA in vivo. circTRIM25 has potential for OA therapy by regulating ferroptosis.
Ethical approval
The human and animal studies were all approved by The Ethics Committee of the First Affiliated Hospital of Gannan Medical University.
Informed consent from participants
Informed consent was obtained from all subjects. The animal study was performed according to the guide for the care and use of the laboratory animals.
Authors’ contributions
CH and YY conceived the study; ZZ, SY and QW conducted the experiments; XL and CL analyzed the data; CH wrote the manuscript; ZZ, SL and WZ revised the data; all the authors read and approved the final version of the manuscript.
Acknowledgments
Not applicable.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Long H, Liu Q, Yin H, et al. Prevalence trends of Site-Specific osteoarthritis from 1990 to 2019: findings from the Global Burden of Disease Study 2019. Arthritis Rheumatol. 2022;74(7):1–15.
- Pereira D, Ramos E, Branco J. Osteoarthritis. Acta Med Port. 2015;28(1):99–106.
- Abramoff B, Caldera FE. Osteoarthritis: pathology, diagnosis, and treatment options. Med Clin North Am. 2020;104(2):293–311.
- Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376–387.
- Taruc-Uy RL, Lynch SA. Diagnosis and treatment of osteoarthritis. Prim Care. 2013;40(4):821–836, vii. vii.
- Siddiq MAB, Clegg D, Jansen TL, et al. Emerging and new treatment options for knee osteoarthritis. Curr Rheumatol Rev. 2022;18(1):20–32.
- Roseti L, Desando G, Cavallo C, et al. Articular cartilage regeneration in osteoarthritis. Cells. 2019;8(11):1305.
- Marchev AS, Dimitrova PA, Burns AJ, et al. Oxidative stress and chronic inflammation in osteoarthritis: can NRF2 counteract these partners in crime? Ann N Y Acad Sci. 2017;1401(1):114–135.
- Xia B, Di Chen, Zhang J, Hu S, Jin H, Tong P. Osteoarthritis pathogenesis: a review of molecular mechanisms. Calcif Tissue Int, 2014;95(6):495–505.
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172.
- Liu D, Liang YH, Yang YT, et al. Circular RNA in osteoarthritis: an updated insight into the pathophysiology and therapeutics. Am J Transl Res. 2021;13:11–23.
- Ehrlich GD. Circular RNAs as diagnostic biomarkers for osteoarthritis. Genet Test Mol Biomarkers. 2019;23(10):701–702.
- Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–143.
- Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12(1):34.
- Weiland A, Wang Y, Wu W, et al. Ferroptosis and its role in diverse brain diseases. Mol Neurobiol. 2019;56(7):4880–4893.
- Sun Y, Chen P, Zhai B, et al. The emerging role of ferroptosis in inflammation. Biomed Pharmacother. 2020;127:110108.
- Yang J, Hu S, Bian Y, et al. Targeting cell death: pyroptosis, ferroptosis, apoptosis and necroptosis in osteoarthritis. Front Cell Dev Biol. 2021;9:789948.
- Yao X, Sun K, Yu S, et al. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J Orthop Translat. 2020;27:33–43.
- Zhou X, Zheng Y, Sun W, et al. D-mannose alleviates osteoarthritis progression by inhibiting chondrocyte ferroptosis in a HIF-2alpha-dependent manner. Cell Prolif. 2021;54(11):e13134.
- He Q, Yang J, Pan Z, et al. Biochanin a protects against iron overload associated knee osteoarthritis via regulating iron levels and NRF2/system xc-/GPX4 axis. Biomed Pharmacother. 2023;157:113915.
- Ruan Q, Wang C, Zhang Y, et al. Ruscogenin attenuates cartilage destruction in osteoarthritis through suppressing chondrocyte ferroptosis via Nrf2/SLC7A11/GPX4 signaling pathway. Chem Biol Interact. 2024;388:110835.
- Altman R, Asch E, Bloch D, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and therapeutic criteria committee of the American rheumatism association. Arthritis Rheum. 1986;29(8):1039–1049.
- Wang S, Geng L, Zhao G, et al. Effectiveness of selenium on chondrocyte glycoprotein glycosylation which play important roles in the pathogenesis of an endemic osteoarthritis, kashin-beck disease. Biol Trace Elem Res. 2022;200(4):1531–1537.
- Zhao C, Gu Y, Wang Y, et al. miR-129-5p promotes osteogenic differentiation of BMSCs and bone regeneration via repressing Dkk3. Stem Cells Int. 2021;2021:7435605–7435618.
- Guo X, Wang Z, Deng X, et al. Circular RNA CircITCH (has-circ-0001141) suppresses hepatocellular carcinoma (HCC) progression by sponging miR-184. Cell Cycle. 2022;21(15):1557–1577.
- Peng QS, Cheng YN, Zhang WB, et al. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit hippo signaling pathway. Cell Death Dis. 2020;11(2):112.
- Yang W, Wang Y, Zhang C, et al. Maresin1 protect against ferroptosis-induced liver injury through ROS inhibition and Nrf2/HO-1/GPX4 activation. Front Pharmacol. 2022;13:865689.
- Jiang Y, Zhao J, Li R, et al. CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis. J Exp Clin Cancer Res. 2022;41(1):307.
- Zhang J, Zhang Z. Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression. Open Life Sci. 2021;16(1):495–510.
- Wang BW, Jiang Y, Yao ZL, et al. Aucubin protects chondrocytes against IL-1β-induced apoptosis in vitro and inhibits osteoarthritis in mice model. Drug Des Devel Ther. 2019;13:3529–3538.
- Kim D, Song J, Jin EJ. BNIP3-Dependent mitophagy via PGC1α promotes cartilage degradation. Cells. 2021;10(7):1839.
- Yan J, Feng G, Ma L, et al. Metformin alleviates osteoarthritis in mice by inhibiting chondrocyte ferroptosis and improving subchondral osteosclerosis and angiogenesis. J Orthop Surg Res. 2022;17(1):333.
- Glasson SS, Chambers MG, Van Den Berg WB, et al. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr Cartil. 2010;18 Suppl 3: S 17–23.
- Wojdasiewicz P, Poniatowski ŁA, Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm. 2014;2014:561459–561419.
- Ji X, Du W, Che W, et al. Apigenin inhibits the progression of osteoarthritis by mediating macrophage polarization. Molecules. 2023;28(7):2915.
- Charlier E, Deroyer C, Ciregia F, et al. Chondrocyte dedifferentiation and osteoarthritis (OA). Biochem Pharmacol. 2019;165:49–65.
- Xie XW, Wan RZ, Liu ZP. Recent research advances in selective matrix metalloproteinase-13 inhibitors as anti-osteoarthritis agents. ChemMedChem. 2017;12(15):1157–1168.
- Santamaria S. ADAMTS-5: a difficult teenager turning 20. Int J Exp Pathol. 2020;101(1-2):4–20.
- Wu Y, Hong Z, Xu W, et al. Circular RNA circPDE4D protects against osteoarthritis by binding to miR-103a-3p and regulating FGF18. Mol Ther. 2021;29(1):308–323.
- Mao G, Xu Y, Long D, et al. Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res Ther. 2021;12(1):389.
- Liu S, Pan Y, Li T, et al. The role of regulated programmed cell death in osteoarthritis: from pathogenesis to therapy. Int J Mol Sci. 2023;24(6):5364.
- Del Carlo M, Jr, Loeser RF. Cell death in osteoarthritis. Curr Rheumatol Rep. 2008;10(1):37–42.
- Liu LL, Cao ZH, He CL, et al. Ferric ion induction of triggering receptor expressed in myeloid cells-2 expression andPI3K/akt signaling pathway in preosteoclast cells to promote osteoclast differentiation. Orthop Surg. 2020;12(4):1304–1312.
- Xu T, Ding W, Ji X, et al. Molecular mechanisms of ferroptosis and its role in cancer therapy. J Cell Mol Med. 2019;23(8):4900–4912.
- Shah R, Shchepinov MS, Pratt DA. Resolving the role of lipoxygenases in the initiation and execution of ferroptosis. ACS Cent Sci. 2018;4(3):387–396.
- Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35(6):830–849.
- Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692.
- Miao Y, Chen Y, Xue F, et al. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine. 2022;76:103847.
- Maiorino M, Conrad M, Ursini F. GPx4, lipid peroxidation, and cell death: discoveries, rediscoveries, and open issues. Antioxid Redox Signal. 2018;29(1):61–74.
- Mitrovic DR, Uzan M, Quintero M, et al. Lipid peroxides in human articular cartilage. Rheumatol Int. 1984;5(1):33–37.
- Tsikas D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal Biochem. 2017;524:13–30.
- Liu Z, Wang Q, Wang X, et al. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6(1):72.
- Zhang HY, Zhang BW, Zhang ZB, et al. Circular RNA TTBK2 regulates cell proliferation, invasion and ferroptosis via miR-761/ITGB8 axis in glioma. Eur Rev Med Pharmacol Sci. 2020;24(5):2585–2600.
- Kyostio-Moore S, Nambiar B, Hutto E, et al. STR/ort mice, a model for spontaneous osteoarthritis, exhibit elevated levels of both local and systemic inflammatory markers. Comp Med. 2011;61(4):346–355.
- He Z, Ruan X, Liu X, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in glioma. J Exp Clin Cancer Res. 2019;38(1):65.
- Mao Q, Liang XL, Zhang CL, et al. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138-5p/Sirt1 axis. Stem Cell Res Ther. 2019;10(1):393.
- Chen Z, Zhao F, Liang C, et al. Silencing of miR-138-5p sensitizes bone anabolic action to mechanical stimuli. Theranostics. 2020;10(26):12263–12278.
- Yuan Y, Zhang GQ, Chai W, et al. Silencing of microRNA-138-5p promotes IL-1beta-induced cartilage degradation in human chondrocytes by targeting FOXC1: miR-138 promotes cartilage degradation. Bone Joint Res. 2016;5(10):523–530.
- Tang X, Li X, Zhang D, et al. Astragaloside-IV alleviates high glucose-induced ferroptosis in retinal pigment epithelial cells by disrupting the expression of miR-138-5p/Sirt1/Nrf2. Bioengineered. 2022;13(4):8240–8254.
- Dai W, Xu Y, Mo S, et al. GLUT3 induced by AMPK/CREB1 axis is key for withstanding energy stress and augments the efficacy of current colorectal cancer therapies. Signal Transduct Target Ther. 2020;5(1):177.
- Wang YW, Chen X, Ma R, et al. Understanding the CREB1-miRNA feedback loop in human malignancies. Tumour Biol. 2016;37(7):8487–8502.
- Wang Z, Ni S, Zhang H, et al. Silencing SGK1 alleviates osteoarthritis through epigenetic regulation of CREB1 and ABCA1 expression. Life Sci. 2021;268:118733.
- Xue H, Tu Y, Ma T, et al. Lactoferrin inhibits IL-1beta-induced chondrocyte apoptosis through AKT1-induced CREB1 activation. Cell Physiol Biochem. 2015;36(6):2456–2465.
- Wang Z, Zhang X, Tian X, et al. CREB stimulates GPX4 transcription to inhibit ferroptosis in lung adenocarcinoma. Oncol Rep. 2021;45(6):88.
